Abstract
Arabidopsis ROS1 belongs to a family of plant 5-methycytosine DNA glycosylases that initiate DNA demethylation through base excision. ROS1 displays the remarkable capacity to excise 5-meC, and to a lesser extent T, while retaining the ability to discriminate effectively against C and U. We found that replacement of the C5-methyl group by halogen substituents greatly decreased excision of the target base. Furthermore, 5-meC was excised more efficiently from mismatches, whereas excision of T only occurred when mispaired with G. These results suggest that ROS1 specificity arises by a combination of selective recognition at the active site and thermodynamic stability of the target base. We also found that ROS1 is a low-turnover catalyst because it binds tightly to the abasic site left after 5-meC removal. This binding leads to a highly distributive behaviour of the enzyme on DNA substrates containing multiple 5-meC residues, and may help to avoid generation of double-strand breaks during processing of bimethylated CG dinucleotides. We conclude that the biochemical properties of ROS1 are consistent with its proposed role in protecting the plant genome from excess methylation.
INTRODUCTION
Methylation of cytosine bases is a post-synthetic modification of DNA found in the genome of many eukaryotic organisms. DNA methylation is established and perpetuated by DNA methyltransferases, which catalyse the transfer of a methyl group to the carbon 5 of cytosine to generate 5-methylcytosine (1). In animals methylation is mostly restricted to CG dinucleotides, whereas plant genomes also display substantial methylation levels in CHG and CHH contexts (where H = A, C or T) (2,3). DNA methylation is an epigenetic mark that promotes gene silencing and helps to preserve stable patterns of gene expression throughout cell divisions (4). It plays essential roles in tissue-specific gene expression, genomic imprinting, X-chromosome inactivation and genome defence against parasitic mobile elements (5,6). Furthermore, distortion of DNA methylation patterns is a central component in many forms of human disease, including cancer (7,8). The dynamic control of methylation requires DNA demethylation, which may take place as a passive process due to lack of maintenance of methylation during several cycles of DNA replication or as an active mechanism in the absence of replication. The enzymatic basis of active DNA demethylation in animal cells remains controversial (9). However, in plants there is convincing genetic and biochemical evidence that proteins from a family of DNA glycosylases typified by Arabidopsis ROS1 (REPRESSOR OF SILENCING 1) and DME (DEMETER) initiate 5-meC DNA demethylation through a base excision repair process (10–14).
ROS1 was identified in a screen for mutants with deregulated expression of the repetitive RD29A-LUC transgene (10). DME is expressed primarily in the central cell of the female gametophyte, where it is required for the expression of the maternal alleles of the imprinted genes MEA, FWA and FIS2 (11,15,16). In addition to ROS1 and DME, the genome of Arabidopsis encodes two additional paralogs, referred to as DEMETER-LIKE proteins DML2 and DML3 (11). All four proteins are large polypeptides containing a DNA glycosylase domain with significant sequence similarity to base excision DNA repair proteins in the HhH-GPD superfamily (17). The HhH-GPD superfamily of DNA glycosylases is widespread in all three domains of life (bacteria, archaea and eukaryotes) and its members are typically 200–400 amino acids long (18). However, proteins of the ROS1/DME family are unusually large (1100–2000 amino acids) compared with typical DNA glycosylases. Furthermore, they appear to be unique to plants, with putative orthologs present in mosses and unicellular green algae. This suggests that active demethylation through excision of 5-meC may have appeared early during plant evolution (9).
ROS1 and DME are the best in vitro-characterized members of this family of atypical DNA glycosylases (12–14). Both remove 5-meC as a free base from DNA using a glycosylase/lyase mechanism (12) and cleave the phosphodiester backbone at the 5-meC removal site by successive β,δ-elimination, leaving a gap that has to be further processed to generate a 3′-OH terminus suitable for polymerization and ligation (12,13). Excision of 5-meC in vitro is more efficient in sequences that are more likely to be methylated in vivo. Thus ROS1 and DME erase 5-meC at CG, CHG and CHH sequences, with a preference for CG sites (12,13), which matches the pattern of DNA methylation in plants. Furthermore, both proteins remove 5-meC more efficiently from a CAG context than when located in the outer position of a CCG context (12), consistent with the fact that CCG is the sequence with the lowest methylation level among the CHG sites (19). DML2 and DML3 are also 5-meC DNA glycosylases/lyases (20,21). While DML2 activity is very weak, at least in vitro, DML3 has an enzymatic activity and substrate specificity comparable to those of DME and ROS1 (20,21).
The precise in vivo roles of plant 5-meC DNA glycosylases are not fully understood. DME is required to demethylate MEA, FWA, FIS2 and perhaps other unidentified imprinted loci in female gametes before fertilization (13,15,16). ROS1 prevents transcriptional gene silencing and hypermethylation of a repetitive transgene (10) but also regulates endogenous loci that show reduced expression and hypermethylation in ros1 plants (22). Furthermore, genome-wide analyses of DNA methylation patterns have identified hundreds of regions that become hypermethylated in a ros1 dml2 dml3 triple mutant (20,23). Taken together, these results suggest that an important in vivo function of ROS1, DML2 and DML3 is to protect the genome from excess methylation. On the other hand, there is some evidence to suggest that these proteins may be needed not only to counteract deleterious methylation, but also to maintain high methylation levels at properly targeted sites (21,23). The prevailing view is that demethylation initiated by the ROS1/DME family of DNA glycosylases contributes to the stability and flexibility of the plant epigenome. In addition to 5-meC paired with guanine, DME, ROS1 and DML3 also remove thymine from a T:G mismatch located at CG, CHG and CHH sequences (12,13,21). Therefore, the possibility cannot be ruled out that DME, ROS1 and/or DML3 also play a role in repairing T:G mismatches arising from spontaneous deamination of 5-meC to thymine.
Discovering the molecular details of events involved in processing target bases by enzymes of the ROS1/DME family will be important to our understanding of the biological functions and relevance of these proteins. It remains unknown how the enzymes of the ROS1/DME family specifically recognize 5-meC in DNA and distinguish it from unmethylated C. A methylated cytosine is not a damaged base, but its effects on DNA structure are not yet completely understood. Some of them may involve subtle conformational changes, since it has been reported that the methyl group at C5 of cytosine induces a slight displacement of the surrounding bases to the minor groove of the helix, making it shallower (24,25). A complete understanding of how plant 5-meC DNA glycosylases recognize and excise target bases will require a thorough examination of their biochemical characteristics. In this study we have analysed the substrate specificity and functional properties of ROS1. We report that the nature of the substituent group at C5 of the target base has a major impact on defining the substrate specificity of ROS1 and that the identity of the opposite base affects 5-meC and T excision differently. We also found that ROS1 binds to the abasic (apurinic/apyrimidinic, AP) site intermediate generated during the reaction. This binding leads to a highly distributive behaviour on DNA substrates containing several 5-meC residues, and may help to avoid generation of double-strand breaks as bimethylated CG sites are processed. We discuss these functional properties in the light of the possible in vivo functions of ROS1.
MATERIALS AND METHODS
DNA substrates
Oligonucleotides used as DNA substrates (see Supplementary Table 1) were synthesized by Operon and were purified by PAGE before use. Double-stranded DNA substrates were prepared by mixing a 5 μM solution of a 5′-fluorescein-labelled oligonucleotide (upper-strand) with a 10 μM solution of an unlabelled oligomer (lower-strand), heating to 95°C for 5 min and slowly cooling to room temperature.
DNA containing a stabilized AP site opposite a guanine was prepared by incubating a DNA duplex containing a U:G mispair (400 nM) with Escherichia coli uracil DNA glycosylase (Ung) at 30°C for 5 min. The reaction mixture was then incubated with 1 M NaBH4 at 25°C for 2 h, neutralized with 100 mM Tris–HCl, pH 7.4, and filtered through a Sephadex G-25 micro column (GE Healthcare). A DNA duplex containing a C:G pair was processed in parallel and used as a control.
Expression and purification of ROS1
The full-length ROS1 cDNA was inserted into the pET28a expression vector (Novagen) to add a polyhistidine (His6) Tag at the N-terminus of ROS1 protein. Expression of recombinant ROS1 was carried out in E. coli BL21(DE3) dcm− Codon Plus cells (Stratagene). A fresh single transformant colony was inoculated into 10 mL of LB medium containing kanamycin (30 µg/ml) and chloramphenicol (34 µg/ml) and the culture was incubated at 37°C overnight with shaking. A 2.5 ml aliquot of the overnight culture was inoculated into 250 ml of LB medium containing kanamycin (30 µg/ml) and chloramphenicol (34 µg/ml), and incubated at 37°C, 250 rpm, until the A600 was 0.1. The culture was then placed at 23°C, and incubation continued at 250 r.p.m. for 90 min before adding 5 mM betaine, 5 mM Na-glutamate and 0.5 M NaCl. When the A600 reached 0.7, expression was induced by adding isopropyl-1-thio-β-d-galactopyranoside (IPTG) to 1 mM and incubating for 2 h. After induction, cells were collected by centrifugation at 13 000 g for 30 min and the pellet frozen at –80°C. The stored pellet was thawed and resuspended in 3.5 ml of Sonication Buffer (SB: 20 mM Tris–HCl pH 8.0, 500 mM NaCl, 20% glycerol, 15 mM β-mercaptoethanol, 1% Tween-20) supplemented with 5 mM imidazole. Cells were disrupted by sonication and the lysate was clarified by centrifugation. The supernatant was loaded onto a Ni2+-sepharose column (GE Healthcare) preequilibrated with SB buffer supplemented with 5 mM imidazole. The column was washed with 10 ml of SB supplemented with 5 mM imidazole, followed by 10 ml of SB supplemented with 100 mM imidazole. Proteins were eluted with a 30 ml gradient of imidazole (100 mM to 1 M) in SB and collected in 0.5 ml fractions. An aliquot of each fraction was analysed by SDS–PAGE and those containing a single band of the overexpressed protein were pooled and dialysed against Dialysis Buffer (DB: 50 mM Tris–HCl pH 8.0, 500 mM NaCl, 1 mM DTT, 50% glycerol). The protein preparation was divided into aliquots, and stored at –80°C. All steps were carried out at 4°C or on ice. Protein concentrations were determined by the Bradford assay (26). Denatured proteins were analysed by SDS–PAGE (10%) using broad-range molecular weight standards (Bio-Rad) (Supplementary Figure 1). The identity of the purified protein was confirmed by MALDI-TOF mass spectrometry.
Enzyme activity assays
Double-stranded oligodeoxynucleotides (20 nM, unless otherwise stated) were incubated at 30°C for the indicated times in a reaction mixture containing 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 0.1 mg/ml BSA, and the corresponding amount of ROS1 protein in a total volume of 50 μl. Reactions were stopped by adding 20 mM EDTA, 0.6% sodium dodecyl sulphate, and 0.5 mg/ml proteinase K, and the mixtures were incubated at 37°C for 30 min. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol precipitated at –20°C in the presence of 0.3 mM NaCl and 16 μg/ml glycogen. Samples were resuspended in 10 μl 90% formamide and heated at 95°C for 5 min. Reaction products were separated in a 12% denaturing polyacrylamide gel containing 7 M urea. Fluorescein-labelled DNA was visualized using the blue fluorescence mode of the FLA-5100 imager and analysed using Multigauge software (Fujifilm). When measuring DNA glycosylase activity, samples were treated with NaOH 100 mM, and immediately transferred to 90°C for 10 min. An equal volume of 90% formamide was added and heated at 95°C for 5 min. Products were resolved and analysed as described above.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using a fluorescein-labelled duplex oligonucleotide containing either a synthetic AP site opposite guanine (AP:G) or a C:G pair, prepared as described above. DNA-binding reaction mixtures (10 μl) contained 100 nM of labelled duplex substrate, 60 ng of unlabelled pBluescript as competing nucleic acid, and different amounts of ROS1 (45, 90 and 135 nM) in 10 nM Tris–HCl pH 8.0, 1 mM DTT, 10 μg/ml BSA, 1 mM EDTA. After 60 min incubation at 25°C, reactions were immediately loaded onto 0.2% agarose gels in 1× TAE. Electrophoresis was carried out in 1× TAE for 40 min at 80 V at room temperature. Fluorescein-labelled DNA was visualized using the blue fluorescence mode of the FLA-5100 imager and analysed using Multigauge software (Fujifilm).
RESULTS
Substitutions at C5 of the target base and the nature of the opposite base exert an important effect on ROS1 efficiency
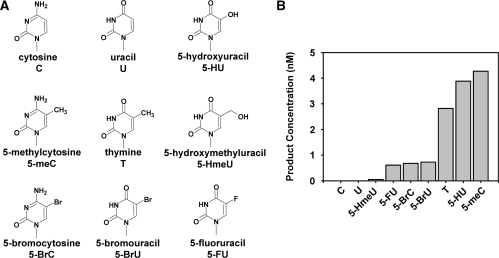
ROS1 is a 5-meC DNA glycosylase that also excises T (= 5-meU) from T:G mispairs, but does not show detectable activity on either C:G pairs or U:G mispairs (12). This discrimination suggests that the nature of the substituent group at C5 of the target base has an important role in determining the substrate specificity of the enzyme. To address this hypothesis, we first examined the activity of ROS1 on 51-mer duplex oligo substrates that contained different 5-substituted cytosine and uracil derivatives paired to G at position 29 in a CG context (Figure 1). We found that ROS1 incision activity was significant on 5-meC, T and 5-HU, with preferences ordered thus: 5-meC > 5-HU > T. In contrast, the activity with 5-substituted halogen derivatives of C (5-BrC) or U (5-BrU and 5-FU) was drastically lower than with their 5-methyl counterparts. On the other hand, substituting the methyl group by a hydroxymethyl group at C5 effectively abolished the capacity of ROS1 to excise the target base. These results indicate that the identity of the substituent group at C5 of the target has a major impact on the substrate specificity of ROS1.
Figure 1.
ROS1 incision activity on duplex DNA substrates containing different modified bases paired with G. (A) Chemical structures of substrate DNA bases tested in this study. (B) Purified ROS1 (22.5 nM) was incubated at 30°C for 2 h with 51-mer double-stranded oligonucleotide substrates (20 nM) containing at position 29 of the labelled upper-strand different target DNA bases paired with G. Products were separated in a 12% denaturing polyacrylamide gel and the amount of incised oligonucleotide was quantified by fluorescent scanning.
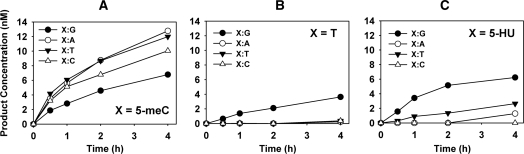
We next investigated the effect of changing the opposite base on ROS1 activity when acting on DNA substrates containing 5-meC, T or 5-HU (Figure 2). We found that 5-meC was less efficiently excised when paired with G than when mispaired with either C, T or A (Figure 2A). A contrasting effect was observed for DNA substrates containing T, which was excised only when mispaired with G (Figure 2B), but not when paired with A or mispaired with T or C (Figure 2B). Excision of 5-HU followed a pattern qualitatively similar to that of T, since it was preferentially excised when mispaired with G, and much less efficiently when opposite to either T, A or C (Figure 2C). We also examined ROS1 activity on DNA duplexes containing C mispaired either with C, A, G or T, but no detectable incision was observed in any case (data not shown).
Figure 2.
Effect of base-pairing partner on ROS1 activity for 5-meC, T and 5-HU. The time-dependent generation of incision products was measured by incubating purified ROS1 (22.5 nM) at 30°C with double-stranded oligonucleotide substrates (20 nM) containing 5-meC (A), T (B) or 5-HU (C) opposite G (filled circles), A (open circles), T (filled triangles) or C (open triangles) on the complementary strand. Reactions were stopped at the indicated times, products were separated in a 12% denaturing polyacrylamide gel, and the amount of incised oligonucleotide was quantified by fluorescence scanning.
Taken together, these results indicate that the substituent group at C5 of the target and the identity of the base at the complementary strand are key factors in determining the substrate specificity of ROS1.
Base excision and strand incision are coordinated
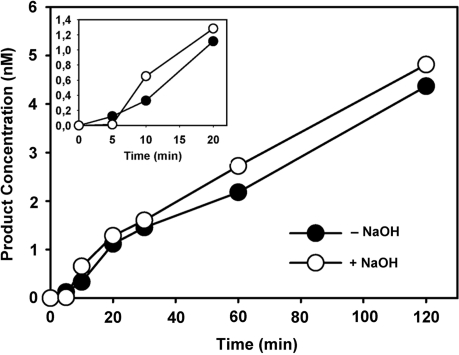
Unlike monofunctional glycosylases, bifunctional DNA glycosylases/lyases are endowed with an AP lyase activity that cleaves the phosphodiester backbone after glycosyl bond scission. Earlier studies with some bifunctional enzymes found that glycosylase action and AP lyase catalysis occur concomitantly (27), and this finding is consistent with a unified mechanism of DNA glycosylases/AP lyases that postulates coordinated base excision and strand incision as a result of Schiff base formation (28). However, other studies have reported that in some cases the two activities are uncoupled, since the rate of AP lyase cleavage is significantly slower than that of base excision (29,30). We therefore decided to examine whether the AP lyase activity of ROS1 is coupled to its 5-meC DNA glycosylase activity (Figure 3). To differentiate 5-meC excision and strand incision the reaction products were analysed with or without alkaline treatment with NaOH, which cleaves all AP sites generated by the enzyme. Strand-nicking detected after treating the reaction products with NaOH reflects the glycosylase activity of ROS1, while cleavage in the absence of NaOH treatment reflects the combined DNA glycosylase/AP lyase action of the enzyme. Quantification of the reaction products generated by ROS1 revealed that the amounts of strand incision with and without NaOH treatment were not significantly different (Figure 3). Therefore, no glycosyl bond scission was detected without an accompanying phosphodiester bond incision. We conclude that the 5-meC excision by ROS1 is coupled to the AP lyase step.
Figure 3.
DNA glycosylase and AP lyase activities of ROS1. The time-dependent generation of incision products was measured by incubating purified ROS1 (22.5 nM) at 30°C with double-stranded oligonucleotide substrates (20 nM) containing a single 5-meC:G pair. Reactions were stopped at the indicated times and products were separated in a 12% denaturing polyacrylamide gel. Filled circles: nick incisions detected without alkaline treatment. Open circles: nick incisions detected after incubation with 100 mM NaOH at 90°C for 10 min, in order to cleave AP sites. The amount of incised oligonucleotide was quantified by fluorescence scanning.
ROS1 is a slow-turnover catalyst
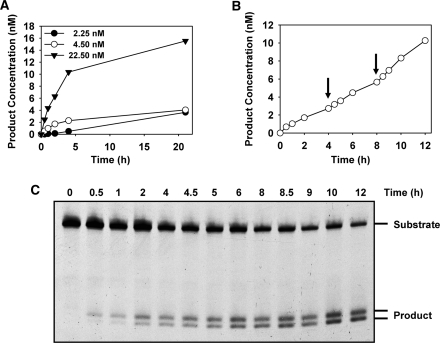
We next examined the kinetics of ROS1 activity on a 5-meC:G pair, incubating 20 nM substrate DNA with either 2.25, 4.5 or 22.5 nM ROS1 (Figure 4A). The reaction curves showed an initial burst of product accumulation followed by a slower phase. Most of the final product concentration was generated during the initial burst, which lasted about 4 h in the three curves. Over the following 17 h the rate of formation of additional product was much lower. Importantly, the amplitude of the initial burst was correlated with the enzyme concentration used (Figure 4A). This suggests that product concentration is limited by the concentration of ROS1 DNA glycosylase.
Figure 4.
Time-dependent product accumulation in reactions with different enzyme concentrations. (A) Different amounts of purified ROS1 (2.25 nM, filled circles; 4.5 nM, open circles; 22.5 nM, filled triangles), were incubated at 30°C with a double-stranded oligonucleotide substrate (20 nM) containing a 5-meC:G pair. (B and C) Reaction was started by addition of ROS1 (4.5 nM) and additional enzyme aliquots (4.5 nM) were added at 4 and 8 h (arrows). Reactions were stopped at the indicated times, products were separated in a 12% denaturing polyacrylamide gel (C), and the amount of incised oligonucleotide was quantified by fluorescence scanning (B).
To confirm that the fraction of DNA substrate converted into product was proportional to the enzyme concentration we incubated 20 nM substrate with 4.5 nM ROS1 and added additional enzyme aliquots once the reaction approached its plateau (Figure 4B and C). The results show that additional and approximately equivalent amounts of substrate were converted to product after supplementing with equal molar amounts of enzyme. We conclude that ROS1 does not exhibit turnover in vitro and removes a near-stoichiometric amount of 5-meC.
ROS1 binds to DNA containing an AP site
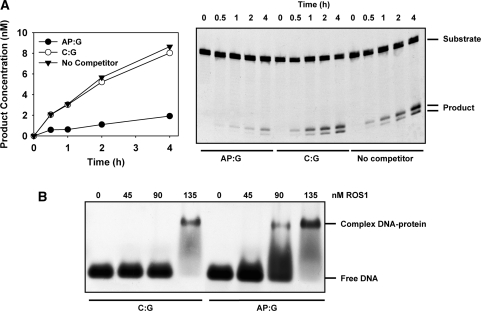
We next investigated whether this behaviour arises because the enzyme is inactivated during the reaction (i.e. it acts as a ‘suicide’ enzyme) or rather because it does not efficiently release its reaction product, as previously reported for other DNA glycosylases (31–34). We preincubated ROS1 with an unlabelled double-stranded oligonucleotide containing a synthetic AP site opposite guanine (AP:G), before measuring the activity on an equivalent labelled 5-meC:G substrate. As control reactions, ROS1 was also preincubated with unlabelled DNA containing a C:G pair or with reaction buffer (Figure 5A). The preincubation with unlabeled AP:G DNA strongly inhibited the processing of the 5-meC:G pair, which nevertheless was not significantly altered when unlabelled C:G DNA was used as competitor (Figure 5A). We performed an analogous experiment using a reduced AP site as competitor, and the results were equivalent (Supplementary Figure 2). We also confirmed that both the synthetic and the reduced AP site used in the preincubation experiments are refractory to incision by ROS1 (data not shown), as previously reported for other bifunctional glycosylases (35,36).
Figure 5.
ROS1 binding to DNA containing an AP site. (A) Effect of preincubation with DNA containing an AP site on ROS1 activity. Purified ROS1 (22.5 nM) was preincubated for 30 min with reaction buffer (filled triangles) or with 40 nM unlabelled duplex oligonucleotide containing either a synthetic AP site opposite guanine (AP:G, filled circles) or a C:G pair (open circles). Then, the fluorescein-labelled 5-meC:G substrate (40 nM) was added and the reactions were monitored for 4 h. Products were separated in a 12% denaturing polyacrylamide gel, and the relative amount of incised oligonucleotide was quantified by fluorescence scanning. (B) EMSA. Increasing amounts of ROS1 were incubated with a labelled duplex oligonucleotide containing either a synthetic AP site opposite guanine (AP:G) or a C:G pair. After nondenaturing gel electrophoresis, protein–DNA complexes were identified by their retarded mobility compared with that of free DNA, as indicated.
To test whether ROS1 inhibition results from binding to the AP site, we performed a gel-mobility shift assay incubating the enzyme with a labelled duplex oligonucleotide containing either a synthetic AP site opposite guanine (AP:G) or a C:G pair (Figure 5B). At increasing ROS1 concentrations we detected a retarded band indicating the formation of a complex with the DNA duplex containing the AP site. ROS1 also bound to the homoduplex containing a C:G pair, but only at the highest protein concentration. These results indicate that the protein can bind non-specifically to DNA, but with a much lower affinity than it binds to DNA containing an abasic site. Therefore, we conclude that ROS1 inhibition by preincubation with an AP site is due to strong binding of the enzyme to this reaction intermediate.
ROS1 activity on bimethylated and hemimethylated CG sites
In plants, as in animals, methylation is most frequent in symmetrical CG contexts (37). We and others have previously shown that ROS1 and its paralog DME are more active with 5-meC located at CG sequences (12,13). Since symmetrical sequences may be present in either the hemimethylated or fully methylated state, it is reasonable to ask whether 5-meC excision at a CG site in one strand is affected by the methylation status of the C in the complementary strand. It has been previously reported that DME is more active on a 5-meC located at a CG sequence when the C in the complementary strand is not methylated, thus suggesting that this enzyme has a preference for hemimethylated over fully methylated CG sequences (13).
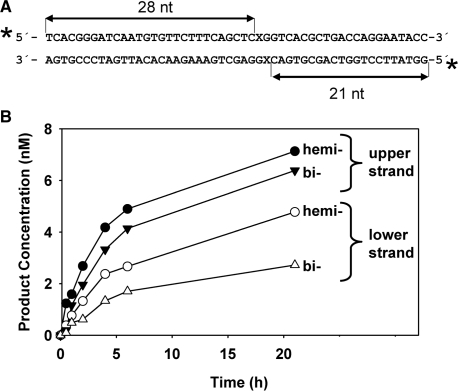
We therefore decided to examine whether the methylation status of the CG sequence has any effect on ROS1 activity on 5-meC residues. We reasoned that the use of DNA duplexes labelled only on the upper strand, such as those used in the experiments describe above, might lead to erroneous conclusions due to inhibition by AP sites generated in the lower, unlabelled strand. To compare rigorously ROS1 activity in hemimethylated versus bimethylated DNA we decided to measure simultaneously the enzymatic activity in both DNA strands. We incubated ROS1 with double-stranded oligonucleotides labelled at both 5′-ends, which contained a single hemimethylated or a fully methylated CG site (Figure 6). In these substrates, incisions in the upper or lower strand are distinguished as 28-mer or 21-mer labelled oligonucleotides, respectively.
Figure 6.
ROS1 activity on hemimethylated and bimethylated DNA. (A) Structure and length of the DNA substrates and products. X indicates target base. (B) Purified ROS1 (22.5 nM) was incubated with 20 nM duplex DNA labelled on both strands that contained a single hemimethylated or a bimethylated CG site. Reactions were stopped at the indicated times and products were separated in a 12% denaturing polyacrylamide gel. The number of incisions on the upper (filled symbols) and/or lower (open symbols) strand on the hemimethylated (circles) and bimethylated (triangle) DNA substrate was quantified by fluorescence scanning.
As shown in Figure 6, incisions in the lower strand accumulated at a slower rate than in the upper strand, in both the hemimethylated and bimethylated substrates. This suggests that ROS1 activity is modulated by the specific sequence context around the CG site, which is different in each strand. In fact, we have some evidence indicating that the sequence surrounding a CG site influences 5-meC excision rate (our unpublished observations). In addition, incision activity in each strand was affected by the methylation status of the CG site. Thus, for the upper and lower strands the rate of incision was higher in the hemimethylated DNA than the bimethylated substrate. We also tested whether both strands could be incised simultaneously at a bimethylated CG site, thus leading to double-strand breaks (DSBs). ROS1 was incubated with hemimethylated or bimethylated DNA and the products were separated by electrophoresis on agarose gels (Supplementary Figure 3). Even after 21 h incubation, no DSBs were detected in a bimethylated DNA substrate, indicating that incision in one 5-meC residue prevents processing of the 5-meC located in the other strand.
Taken together, these results suggest that ROS1 activity at symmetrical CG sites depends on the specific sequence around the target 5-meC and the methylation status of the C residue in the complementary strand.
ROS1 exhibits distributive behaviour on DNA substrates containing multiple 5-meC residues
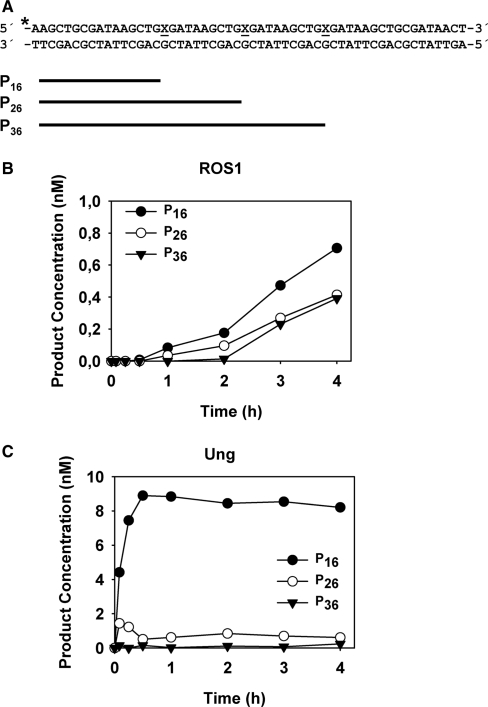
Loci subjected to ROS1 demethylation in vivo contain multiple 5-meC residues, many of them separated by short distances (10,22). We therefore decided to examine the effect of the slow-turnover of ROS1 on its ability to excise several 5-meC residues in close proximity. We specifically aimed to test whether ROS1 acts processively (excising several 5-meC residues without releasing the DNA substrate) or exhibits a distributive behaviour (releasing DNA after each catalytic event). To determine whether ROS1 behaves in a processive or distributive fashion, the enzyme was incubated with a double-stranded oligonucleotide substrate containing three 5-meC residues in the upper strand separated by 9 nt and located in the same sequence context (Figure 7) (see Supplementary Table 1). In a control reaction, we used E. coli Uracil DNA glycosylase (Ung) and an equivalent DNA substrate containing three uracil residues (Figure 7). We reasoned that a processive mechanism would convert the substrate to a final reaction product represented by a 16-nt labelled fragment, whereas a distributive mechanism would lead to the accumulation of partially processed reaction intermediates represented by 26- and 36-nt labelled fragments. As shown in Figure 7A, ROS1 processed 5-meC in a highly distributive fashion: there was a steady accumulation of a 16-nt fragment but also of significant amounts of reaction intermediates. In contrast, a processive enzyme such as Ung led to a rapid accumulation of the final reaction product with a negligible amount of reaction intermediates being detected (Figure 7B). We conclude that ROS1 does not show significant processivity in vitro.
Figure 7.
Processivity analysis of ROS1 activity. (A) Structure and length of the DNA substrates and products. X indicates target base. (B) Purified ROS1 (4.5 nM) was incubated with a double-stranded oligonucleotide substrate (10 nM) containing three 5-meC:G pairs. Reactions were stopped at the indicated times and products were separated in a 12% denaturing polyacrylamide gel. Product concentration was quantified by fluorescence scanning: 16 nt (filled circles), 26 nt (open circles) and 36 nt (filled triangles). (C) A control reaction was performed incubating purified E. coli Ung (1 × 10–3 U) with an equivalent double-stranded oligonucleotide substrate (10 nM) containing three U:G pairs.
DISCUSSION
Factors determining ROS1 substrate specificity
A central issue in base excision repair is how DNA glycosylases recognize specific types of bases and discriminate against non-target substrates. ROS1 displays the remarkable capacity to excise 5-meC and T while retaining the ability to discriminate effectively against C and U (12). This suggests that the enzyme makes specific contacts with the methyl group at C5 of the target base. In this work we examined ROS1 activity on several 5-substituted derivatives of C and T. We found that substitution of the methyl group of thymine by OH actually increased enzymatic activity, while the introduction of a bulkier substituent, such as CH2OH, virtually abolished it. On the other hand, replacement of the methyl group by halogen substituents Br or F substantially decreased excision of the target base. It should be noted that this substrate preference contrasts strikingly with that of other thymine glycosylases, such as TDG (38–40) and MBD4 (41), which show higher activity with uracil and its halogenated derivatives. This preference has been explained in terms of substrate reactivity. Electron-withdrawing substituents, such as Br or F, stabilize the transition state and enhance the leaving ability of the base, while electron-donating groups, such as CH3, actually destabilize the transition state and may slow the reaction (39). At least in the case of TDG, this has led to the proposal that specificity depends on the stability of the scissile C–N bond rather than the selective recognition of substrates at the active site (39,40). In the case of ROS1, increasing leaving ability of the target base does not overcome the negative effect of the substituents on excision efficiency. Thus, our results suggest that ROS1 may rather specifically recognize 5-meC and T by selective steric and/or electrostatic interactions involving the methyl group at position C5.
We also examined the effect of pairing the target base with C, T or A, rather than G, and found entirely different effects on the capacity of ROS1 to excise 5-meC and T. 5-meC was excised less efficiently when correctly paired with G than when mispaired with any of the other three bases. It is possible that processing mismatched 5-meC is biologically relevant in vivo. It has been reported that DNA methyltransferases can efficiently catalyse methylation of mismatched C in vitro (42,43). A methylated C in a mismatch may also arise from the erroneous insertion of an incoming nucleotide opposite 5-meC during DNA replication. In both scenarios, ROS1 activity could lead to mutations if 5-meC is the correct base in the mispair, because it would initiate its incorrect removal. However, it is perhaps more likely that the efficient processing of mismatched 5-meC by ROS1 in vitro simply reflects the reduced thermodynamic stability of 5-meC mispairs compared with a correct 5-meC:G pair. In contrast to 5-meC excision specificity, we found that ROS1 only excised T when mispaired with G, whereas no detectable activity was observed with either T:A, T:T or T:C. This strict preference may be rationalized in terms of the possible consequences of each excision event in vivo. The failure to excise thymine when correctly paired with A suggests that the enzyme has an effective mechanism for avoiding activity on the huge excess of normal T:A pairs in DNA. On the other hand, the absence of activity with T:T or T:C mispairs may avoid potential fixation of DNA replication errors into mutations due to the presumed inability of ROS1 to distinguish the erroneous base. This extreme preference for G as a pairing partner for the substrate T is analogous to that observed for other thymine DNA glycosylases. Thus, the rate of T removal by TDG from T:G mismatches is 1–2 orders of magnitude higher than that from T:C or T:T mismatches, and no activity on T:A pairs has been detected (32,44). Similarly, no MBD4 activity has been detected on either T:T, T:C or T:A pairs (33).
In rationalizing the contrasting effects of opposite residues on 5-meC and T excision it must be remembered that the substrate specificity of DNA glycosylases is shaped by natural selection driven only by the base pairs and mispairs likely to be encountered in cells (45). ROS1 avoids mutagenic excision of T from normal T:A pairs and also from T:C and T:T mispairs. However, it is not precluded from excising 5-meC from thermodynamically unstable 5-meC mispairs, probably because they are very rarely encountered by the enzyme in vivo. Something similar has been observed with E. coli Fpg, which removes 8-oxoguanine residues from DNA. Excision of 8-oxoG from the uncommon 8-oxoG:G and 8-oxoG:T mispairs is 10–30 times faster than from 8-oxoG:C, but the enzyme effectively avoids excision from the frequent 8-oxoG:A pairs generated by erroneous 8-oxoG replication (46). Nevertheless, a comprehensive mechanistic explanation of ROS1 specificity will require detailed kinetic and structural information.
Coupling of glycosylase and lyase activities
We have examined the DNA glycosylase activity of ROS1 separately from its AP lyase activity, and found that 5-meC excision and AP incision are concurrent. The factors determining the coupling or dissociation of glycosylase and lyase activities are not yet understood. The unified mechanism proposed for bifunctional DNA glycosylases postulates coordination of base excision and beta-elimination as a result of Schiff base formation (28). We have previously reported that excision of 5-meC by both ROS1 and DME proceeds via a Schiff base (12). In other bifunctional DNA glycosylases, such as E. coli 8-oxoG DNA glycosylase Fpg, base excision is also tightly coupled with DNA strand cleavage (47). However, not all bifunctional DNA glycosylases involve coupling of excision and incision. Thus, human Endonuclease III (hNTH1) and mammalian OGG1 display a significant dissociation of the two activities, with the rate of AP lyase-mediated strand cleavage being significantly slower than the glycosylase-mediated base excision (29,30).
ROS1 binds to the AP:G intermediate generated during reaction
We investigated the time-course of 5-meC excision and found that the product accumulated biphasically, with an initial burst whose amplitude was correlated with the amount of enzyme used. Furthermore, we confirmed that the amount of enzyme effectively limited the amount of product generated in the reaction, and that ROS1 removed a near-stoichiometric quantity of 5-meC. The reason for this behaviour is that the turnover of the enzyme was exceedingly low, since it binds strongly to the AP site generated after 5-meC excision.
These results suggest that, as previously described for other DNA glycosylases (31–34), the rate-limiting step in the action of ROS1 occurs after base excision. Given the coupling between base excision and strand incision, our findings imply that, in most cases, ROS1 does not dissociate from the AP site intermediate and engages in the AP lyase reaction. Such a restraint at the expense of turnover probably reflects an essential coordination with enzymes acting on subsequent steps in the base excision pathway, thus assuring protection of the potentially harmful AP site that arises during the reaction.
ROS1 activity on hemimethylated and bimethylated DNA
Avoiding hazardous reaction intermediates may be particularly important in the case of ROS1, because simultaneous excision on bimethylated CG sites could generate deleterious double-strand breaks. We found that, even after extended incubation, ROS1 did not produce detectable levels of DSB on a DNA substrate containing a bimethylated CG site. This suggests that 5-meC excision in one strand efficiently prevents processing of the methylated cytosine in the complementary strand. It has been reported that the activity of the ROS1-related enzyme DME on a hemimethylated CG site is inhibited by about 10-fold by the presence of an AP site on the opposite strand (13). Our results suggest that in the case of ROS1 it is unlikely that this circumstance actually occurs in vivo, and it is more probable that the glycosylase will prevent processing of the opposite strand by strongly binding to the AP site until the next step in the excision pathway. There are precedents for such protection lingering with its product in other DNA glycosylases. Thus, E. coli MutY removes A from 8-oxoG:A mispairs, but remains bound to its product in order to prevent double-strand breaks due to premature 8-oxoG excision by Fpg (48,49). The binding of ROS1 to the AP:G intermediate probably also explains the slower 5-meC excision rate observed on bimethylated DNA compared with hemimethylated DNA. It is predicted that product inhibition will be faster in the first case, since the 5-meC/enzyme ratio is 2-fold higher in bimethylated DNA compared to the hemimethylated substrate.
The action of ROS1 on symmetrical CG sites is further complicated by the fact that the enzyme certainly encounters different sequence contexts in the two strands. We found that 5-meC processing by ROS1 occurs at different rates on each strand of the same DNA molecule. This confounding factor should be taken into account when examining the substrate activity of 5-meC DNA glycosylases on bimethylated substrates.
ROS1 initiates demethylation in a distributive fashion
DNA glycosylase processivity has been defined as the ability to excise several close target bases without dissociating from DNA (45). We tested the processivity of 5-meC excision by ROS1 using a DNA duplex with three target residues separated from each other by 9 nt. We found that, under conditions where E. coli Ung displays processive behaviour, ROS1 excises 5-meC in a near-exclusively distributive fashion. Our experiments were performed at low salt concentrations (2.5 mM), using DNA substrates that contained target bases in close proximity (9 nt), therefore requiring very little translocation or short range hopping. Even under these favourable conditions, no evidence for significant ROS1 processivity was found.
Nevertheless, it should be emphasized that these results do not rule out the possibility that target search by ROS1 proceeds though one-dimensional diffusion by sliding and/or hopping (50). Processivity is a property arising from correlated cleavage, i.e. the probability of the enzyme resuming the walk after catalysis is complete (45). In contrast, one-dimensional diffusion applies to correlated searches, i.e. the probability that the enzyme remains bound to the DNA after one step of random walk (45). A case in point is hOGG1, which at physiological salt concentrations displays limited processivity (51,52) but undergoes rapid sliding while searching for target lesions (53). In this regard it should be noted that ROS1 displays some non-specific DNA binding, a property also found in other DNA glycosylases such as TDG (54).
The very low processivity of ROS1 is remarkable, even by comparison with enzymes that are catalytically less efficient than Ung. In addition to Ung (55,56), other DNA glycosylases such as hOGG1, Fpg, MutY, T4-pdg and human alkyladenine DNA glycosylase (AAG) (52,57–59) as well as human AP endonuclease (APE1) (60), exhibit some degree of significant processivity when tested at low salt concentration. A distributive initiation of demethylation events may be well suited for an enzyme possibly performing a protecting role in a plant genome containing significant amounts of 5-meC. Genome-wide methylation analyses have uncovered hundreds of discrete hypermethylated regions in ros1 dml2 dml3 triple-mutant plants, but overall DNA methylation levels are similar to those of wild-type plants (20,23). Hypermethylation primarily affects the 5′ and 3′ gene ends in genic regions (20,23), but also transposon sequences (22,23). Therefore, 5-meC DNA glycosylases seem to be active throughout the genome at a variety of loci. There is strong evidence suggesting that demethylation by ROS1 may be guided by small RNAs bound to ROS3 (61), but it is still an open question whether ROS1 and related enzymes are targeted to specific DNA sequences for unknown reasons or simply to genomic regions that are likely to suffer excessive methylation. The ROS3-dependent demethylation of a transgenic locus subjected to continuous short interfering RNA (siRNA)-directed DNA methylation (61) seems to argue in favor of the latter possibility. Overall, the available data suggest that one of the functions of plant 5-meC DNA glycosylases is to counteract excess DNA methylation, thus protecting the plant genome from a robust DNA modification machinery that evolved for defensive purposes (20). This idea is consistent with the hypothesis that plant 5-meC DNA glycosylases diverged from a common ancestor dedicated to DNA maintenance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Educación y Ciencia, Spain [grant number BFU2007-60956/BMC]; Junta de Andalucía, Spain [grant number P07-CVI-02770]. Ph.D. Fellowship from the Junta de Andalucía, Spain (to M.I.P.-M.). Funding for open access charge: Ministerio de Educación y Ciencia, Spain [grant number BFU2007-60956/BMC].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of our laboratory for helpful discussions.
REFERENCES
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 6.Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 7.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat. Rev. Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 9.Roldan-Arjona T, Ariza RR. DNA demethylation. In: Grosjean H, editor. DNA and RNA modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution. Austin, TX: Landes Bioscience; 2009. pp. 149–161. [Google Scholar]
- 10.Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 11.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 12.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl Acad. Sci. USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl Acad. Sci. USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 16.Jullien PE, Kinoshita T, Ohad N, Berger F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell. 2006;18:1360–1372. doi: 10.1105/tpc.106.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 18.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol. Biol. Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 19.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl Acad. Sci. USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 23.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinemann U, Hahn M. C-C-A-G-G-C-m5C-T-G-G. Helical fine structure, hydration, and comparison with C-C-A-G-G-C-C-T-G-G. J. Biol. Chem. 1992;267:7332–7341. [PubMed] [Google Scholar]
- 25.Marcourt L, Cordier C, Couesnon T, Dodin G. Impact of C5-cytosine methylation on the solution structure of d(GAAAACGTTTTC)2. An NMR and molecular modelling investigation. Eur. J. Biochem. 1999;265:1032–1042. doi: 10.1046/j.1432-1327.1999.00819.x. [DOI] [PubMed] [Google Scholar]
- 26.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Williams SD, David SS. Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nucleic Acids Res. 1998;26:5123–5133. doi: 10.1093/nar/26.22.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodson ML, Michaels ML, Lloyd RS. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994;269:32709–32712. [PubMed] [Google Scholar]
- 29.Marenstein DR, Ocampo MT, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J. Biol. Chem. 2001;276:21242–21249. doi: 10.1074/jbc.M101594200. [DOI] [PubMed] [Google Scholar]
- 30.Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 31.Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waters TR, Swann PF. Kinetics of the action of thymine DNA glycosylase. J. Biol. Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 33.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J. Biol. Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 34.Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Substrate specificity of human endonuclease III (hNth1): effect of human AP endonuclease 1 (APE1) on hNth1 activity. J. Biol. Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- 35.Castaing B, Boiteux S, Zelwer C. DNA containing a chemically reduced apurinic site is a high affinity ligand for the E. coli formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1992;20:389–394. doi: 10.1093/nar/20.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Handley S, Scholes CP, Cunningham RP. Endonuclease III interactions with DNA substrates. 1. Binding and footprinting studies with oligonucleotides containing a reduced apyrimidinic site. Biochemistry. 1995;34:2528–2536. doi: 10.1021/bi00008a017. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 38.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan MT, Bennett MT, Drohat AC. Excision of 5-halogenated uracils by human thymine DNA glycosylase. Robust activity for DNA contexts other than CpG. J. Biol. Chem. 2007;282:27578–27586. doi: 10.1074/jbc.M704253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner DP, Cortellino S, Schupp JE, Caretti E, Loh T, Kinsella TJ, Bellacosa A. The DNA N-glycosylase MED1 exhibits preference for halogenated pyrimidines and is involved in the cytotoxicity of 5-iododeoxyuridine. Cancer Res. 2006;66:7686–7693. doi: 10.1158/0008-5472.CAN-05-4488. [DOI] [PubMed] [Google Scholar]
- 42.Klimasauskas S, Roberts RJ. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res. 1995;23:1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith SS, Hardy TA, Baker DJ. Human DNA (cytosine-5)methyltransferase selectively methylates duplex DNA containing mispairs. Nucleic Acids Res. 1987;15:6899–6916. doi: 10.1093/nar/15.17.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neddermann P, Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 45.Zharkov DO, Grollman AP. The DNA trackwalkers: principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J. Biol. Chem. 1994;269:15318–15324. [PubMed] [Google Scholar]
- 47.Krishnamurthy N, Haraguchi K, Greenberg MM, David SS. Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008;47:1043–1050. doi: 10.1021/bi701919u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 49.Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 50.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 51.Sidorenko VS, Nevinsky GA, Zharkov DO. Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair. 2007;6:317–328. doi: 10.1016/j.dnarep.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Sidorenko VS, Zharkov DO. Correlated cleavage of damaged DNA by bacterial and human 8-oxoguanine-DNA glycosylases. Biochemistry. 2008;47:8970–8976. doi: 10.1021/bi800569e. [DOI] [PubMed] [Google Scholar]
- 53.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- 55.Sidorenko VS, Mechetin GV, Nevinsky GA, Zharkov DO. Correlated cleavage of single- and double-stranded substrates by uracil-DNA glycosylase. FEBS Lett. 2008;582:410–414. doi: 10.1016/j.febslet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 56.Bennett SE, Sanderson RJ, Mosbaugh DW. Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosylase is affected by NaCl concentration. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- 57.Francis AW, David SS. Escherichia coli MutY and Fpg utilize a processive mechanism for target location. Biochemistry. 2003;42:801–810. doi: 10.1021/bi026375+. [DOI] [PubMed] [Google Scholar]
- 58.Hedglin M, O'Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lloyd RS. Investigations of pyrimidine dimer glycosylases–a paradigm for DNA base excision repair enzymology. Mutat. Res. 2005;577:77–91. doi: 10.1016/j.mrfmmm.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 60.Carey DC, Strauss PR. Human apurinic/apyrimidinic endonuclease is processive. Biochemistry. 1999;38:16553–16560. doi: 10.1021/bi9907429. [DOI] [PubMed] [Google Scholar]
- 61.Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.