Abstract
FRAXA is one of a number of fragile sites in human chromosomes that are induced by agents like fluorodeoxyuridine (FdU) that affect intracellular thymidylate levels. FRAXA coincides with a >200 CGG•CCG repeat tract in the 5′ UTR of the FMR1 gene, and alleles prone to fragility are associated with Fragile X (FX) syndrome, one of the leading genetic causes of intellectual disability. Using siRNA depletion, we show that ATR is involved in protecting the genome against FdU-induced chromosome fragility. We also show that FdU increases the number of γ-H2AX foci seen in both normal and patient cells and increases the frequency with which the FMR1 gene colocalizes with these foci in patient cells. In the presence of FdU and KU55933, an ATM inhibitor, the incidence of chromosome fragility is reduced, suggesting that ATM contributes to FdU-induced chromosome fragility. Since both ATR and ATM are involved in preventing aphidicolin-sensitive fragile sites, our data suggest that the lesions responsible for aphidicolin-induced and FdU-induced fragile sites differ. FRAXA also displays a second form of chromosome fragility in absence of FdU, which our data suggest is normally prevented by an ATM-dependent process.
INTRODUCTION
Fragile X syndrome (FXS) is the most common heritable cause of intellectual disability. The disorder is named for a fragile site on the X chromosome that is seen when patient cells are treated with agents like folate or 5-fluorodeoxyuridine (FdU) that affect a key enzyme in the pyrimidine biosynthetic pathway, thymidylate synthase (1,2). This causes a nucleotide pool imbalance thereby slowing replication and perhaps requiring repair to remove misincorporated bases. The fragile site appears as a gap, constriction or break in metaphase chromosomes. The Fragile X fragile site is also known as FRAXA (Fragile site on X chromosome, site A). FRAXA is coincident with a stretch of >200 CGG•CCG-repeats in the 5′ UTR of the FMR1 gene that is responsible for FXS (3,4). Alleles with this number of repeats arise from an increase in the number of repeats on maternal transmission of an allele with 55–200 repeats. The relationship between this repeat expansion and chromosome fragility is unknown.
The sequence basis of six other folate-sensitive fragile sites in the human genome, FRAXE, FRAXF, FRA10A, FRA11A, FRA11B, FRA12A and FRA16A, have so far been determined. In all instances, the responsible sequence is also a long CGG•CCG-repeat tract (5–10). FRAXA is a frequent translocation breakpoint in rodent-human somatic hybrids (4,11). FRA11B, is associated with deletion of the telomeric end of long arm of chromosome 11 in a number of cases of Jacobsen Syndrome (12,13).
More than 110 other fragile sites are found in human genomes, many of which are common translocation breakpoints associated with different forms of cancer. Many of these sites are induced by aphidicolin (APC) and 13 such sites have been studied so far at the sequence level. They are all associated with hundreds of kilobases or even megabases of DNA with no particularly distinctive sequences (5,14–27). In the case of FRA3B, the most active APC-sensitive fragile site, breakage is seen in multiple places throughout a 4-Mb region and it has been suggested that no single sequence is responsible (28). Agents that induce chromosome fragility all share with folate-stress the potential to interfere with DNA replication. APC, for example, is an inhibitor of DNA polymerase α, δ and ε (29,30). At high concentrations these compounds may also affect DNA repair.
ATR and ATM are kinases responsible for the initiation and management of the DNA damage response (DDR) in mammals (31,32). Depending on the type of DNA damage, one or both of these enzymes are activated. The activated protein then initiates a kinase cascade that activates key downstream effectors of the DDR. The ATR-pathway is thought to be involved primarily in the cellular response to stalled replication forks and bulky DNA lesions that block DNA synthesis, while the ATM pathway is thought to respond primarily to double-strand breaks although some cross-talk between these pathways is apparent. Both ATR and ATM have been shown to protect the genome against APC-sensitive chromosome fragility (19,33–38).
Here we describe our analysis of FRAXA. Our data show that, as with the APC-sensitive sites, the ATR-pathway is involved in preventing FdU-sensitive chromosome fragility at FRAXA. KU55933, an inhibitor specific for ATM, reduces FdU-induced chromosome fragility. This suggests that a KU55933-sensitive enzyme, most likely ATM, is actually responsible for the generation of the fragile site. In addition, we show here that the Fragile X alleles exhibit a second form of fragility that is FdU-independent and that ATM is most likely responsible for preventing this form of chromosome fragility. This suggests that FRAXA differs in important ways from the APC-sensitive fragile sites and that FXS alleles form at least two different kinds of DNA lesions that gives rise to chromosome fragility, one that is normally repaired by ATR but not by ATM, and one that is repaired by ATM.
MATERIALS AND METHODS
Cell lines and reagents
Two male FX lymphoblastoid cells lines (GM03200B and GM07294), two normal lymphoblastoid cell lines (GM06895 and GM06865), an Ataxia Telangiectasia cell line (GM00719), a Seckel disease cell line (GM09703) and FXS fibroblasts (GM05848) were obtained from the Coriell Institute for Medical Research (Camden, NJ). The male FX lymphoblastoid cell line LL4118 was established by Dr Esterina Pascale (CNR, Rome, Italy; personal communication). The repeat numbers in all four FX patient cell lines was ∼500–700 (4,39). Lymphoblastoid cells were grown in RPMI 1640 supplemented with 10% FBS and 1× antibiotic–antimycotic liquid consisting of penicillin, streptomycin and fungisone (Invitrogen, Carlsbad, CA). Fibroblasts were grown in DMEM (Invitrogen) supplemented with 10% FBS and 1× antibiotic-antimycotic liquid. The BAC clones RP11-80F18 containing the Xq27.1 region centromeric to the FMR1 gene at Xq27.3, RP11-383P16 containing the Xq27–28 region and RP11-402H20 which contains Xq28 derived sequences were obtained from the Children's Hospital Oakland Research Institute (CHORI; Oakland, CA). A probe corresponding to chromosome 4q11-12 (RP11-535c7) was used as a negative control (CHORI). Previously described siRNA to ATR and their controls (34) were purchased from the Dharmacon subsidiary of Thermo Fisher Scientific (Lafayette, CO). Antibody to CHK2 phosphorylated at T68 (sc-16297-R) was purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA), antibodies to CHK1 phosphorylated at S317 (#2346) or S345 (#2341) and p53 phosphorylated at S15 (#9286) were purchased from Cell Signalling (Danvers, MA), antibody to RPA2/p34 (MS-691) was from Thermo Scientific (Waltham, MA), γ-H2AX (ab18311) was from Abcam (Cambridge, MA). UCN-01 (7-hydroxystaurosporine) was a kind gift of Dr Kondapaka (NCI, NIH). The ATM inhibitor KU55933 was from Calbiochem (San Diego, CA). Remaining reagents were purchased from Sigma-Aldrich (St Louis, MO) unless otherwise specified.
Fragile site analysis
Transfection of siRNAs was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Fibroblasts were grown for several hours in a six-well plate with 2 ml of medium without antibiotics. The cells were harvested after a total of 96 h. Fragile X expression was examined in cells treated with 0.1 µM or 1 µM FdU for 16 h. The depletion of ATR was confirmed by western blotting. KU55933 and UCN-01 were used where indicated at 10 µM and 100 nM respectively. Cells were then treated for 1–6 h with 50 ng/ml colcemid. Metaphase chromosome spreads were prepared and the fragile sites visualized either on Giemsa stained chromosomes or by FISH using standard procedures. FISH probes were prepared from the BAC clones by labeling using a nick-translation kit from Roche (Basel, Switzerland) and tetramethyl-Rhodamine-5-dUTP. BAC clones RP11-383P16 and RP11-402H20, containing sequences telomeric to FRAXA, were used as probes for FRAXA fragility. The BAC RP11-535c7 was used as a negative control. In the FISH experiments, chromosomes were counterstained with 4,6-diamidino-2-phenylindole-dihydrochroride (DAPI) and the spreads visualized by epifluorescence microscopy. At least 50 metaphases were examined for each experiment and each experiment was done in triplicate.
Western blotting
Analysis of the activation status of various proteins involved in the cellular response to DNA damage were examined in total protein extracts prepared from cells grown under the same conditions as those used to examine the fragile site. Preparation of extracts, and the SDS–gel electrophoresis and western blotting of these extracts was carried out using standard procedures. Detection was done using an ECL™ kit (GE Healthcare, Piscataway, NJ), according to the manufacturer's instructions.
Immunocytochemistry and interphase FISH
Lymphoblastoid cells were grown with or without 1 µM FdU for 18 h. After treatment, 2 × 105 cells were deposited on positively charged glass slides by spinning at 80 g for 5 min at RT in a cytospin centrifuge. Slides were fixed with methanol at −20°C for 30 min and then rinsed with cold acetone. Slides were stained with mouse monoclonal anti-γ-H2AX antibody (1/400 dilution) followed by Alexa-488-conjugated anti-mouse IgG (1/400 dilution, Molecular probes, Eugene, OR). Cells were then fixed with 50 mM ethylene glycol-bis(succinic acid N-hydroxy-succinimide ester). Probe hybridization was carried out on fixed cells as previously described (40). Briefly, DNA was denatured using 0.07 M NaOH at room temperature for 3 min. The biotin-labeled FMR1 probe was hybridized at 37°C overnight, followed by washing in pre-warmed 50% formamide/2× SSC and pre-warmed 0.1× SSC. The FMR1 probe was detected by Alexa-555-conjugated anti-streptavidin antibody (1/400 dilution, Molecular probes, Eugene, OR). DNA was counterstained with DAPI. The slides were then visualized using an Olympus fluorescent microscope (Olympus America Inc. Melville, NY). One hundred cells were counted for each experiment for a total of 200 cells/treatment.
Detection of γ-H2AX without FMR1 colocalization was done by fixing cells with 2% paraformaldehyde for 20 min at RT and depositing 2 × 105 cells of these fixed cells on positively charged glass slides by spinning at 80 g for 5 min at RT in a cytospin centrifuge. Cells were permeabilized with 70% ethanol at 4°C for 30 min and treated with mouse monoclonal anti-γ-H2AX (1/400 dilution) as described above. The DNA was counterstained with propidium iodide (PI). Laser scanning confocal microscopy was performed with a Nikon PCM 2000 (Nikon, Inc., Augusta, GA). Two hundreds cells were counted for each experiment using previously published criteria (41).
RESULTS
FdU and APC-treatment have similar effects on the activation of DDR proteins
Activation of the ATM and ATR DDR pathways leads to phosphorylation of a number of proteins downstream of these kinases. In order to test which of these proteins are activated in conditions that result in FRAXA fragility, we carried out western blotting on protein extracts prepared from FdU-treated cells from both affected and unaffected individuals using antibodies for the phosphorylated forms of various DDR proteins. Activation with FdU was compared to that seen with APC, etoposide, which induces DNA double strand breaks, and hydroxyurea (HU), an inhibitor of ribonucleotide reductase that results in replication fork stalling.
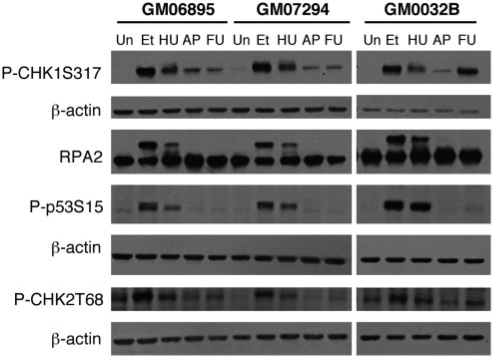
The results for two-patient cell lines and an age-matched control are shown in Figure 1. FdU-treatment caused increased phosphorylation of CHK1 at S317 and S345 even at concentrations 100-fold lower than that used to visualize the FX fragile site (Figure 1 and data not shown). Agents like etoposide and HU increased phosphorylation of CHK2 at threonine 68 and p53 at serine 15 (Figure 1). However, little or no phosphorylation of CHK2 or RPA2 was seen with either APC or FdU. No phosphorylation of p53 was seen in GM07294 although a small amount was seen in GM0032B (Figure 1), and two other cell lines in the presence of FdU (data not shown). GM07294 also shows less CHK1 phosphorylation than GM0032B. It is thus possible that the DDR is less efficiently induced in this cell line. We thus cannot exclude the possibility that some phosphorylation of p53 occurs in this cell line that is below the sensitivity of our assay. Our results differ somewhat from previously published data where APC was shown to cause significant phosphorylation of Chk2, p53 and RPA2 (36). These differences could be due to differences in the growth conditions or to genetic variation in the DDR in the human population. Nevertheless, our results demonstrate that in our cell lines, the DDR response to FdU and APC are similar and that the CHK1 pathway, rather than the typical DSB-response pathway that involves CHK2, is the major pathway activated in response to both APC and FdU.
Figure 1.
Activation of DNA damage signaling proteins by FdU treatment. Western blots of extracts from an unaffected cell line, GM06895 and 2 FX cell lines, GM07924 and GM03200B, treated with various DNA damaging agents were probed with the indicated antibodies. Etoposide (Et) was used at 68 µM for 2 h. Hydroxyurea (HU) was used at a concentration of 2 mM for 24 h. Fluorodeoxyuridine, abbreviated in this figure as FU for reasons of space, was used at 1 µM for 18 h. Aphidicolin, indicated here by the abbreviation AP, was used at 0.4 µM for 24 h. Note that the same gel was used for the detection of RPA2 and CHK1 and so the same β-actin control applies. The β-actin data for both experiments is shown beneath the CHK1 data.
Knockdown of ATR or UCN-01 treatment increases FRAXA expression
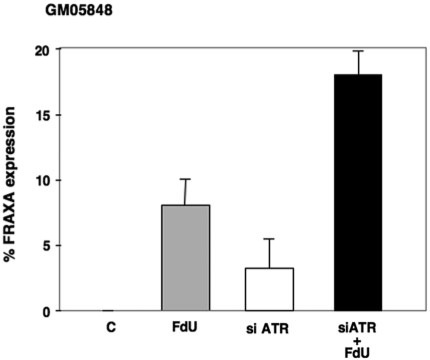
In order to assess the role of different DDR pathways on expression of the FX fragile site more directly, we first treated FX fibroblasts with siRNAs targeted to ATR and examined the frequency of chromosome breaks and rearrangements at the FMR1 locus. Fibroblasts were used rather than lymphoblastoid cells because of their higher transfection efficiency. The incidence of breaks and rearrangements was compared to that seen without either FdU or siRNAs to ATR, with FdU alone and when ATR siRNAs were used in combination with FdU. No chromosome fragility was seen in normal cells under any of these conditions or in untreated patient cells. In contrast, ATR knockdown, with or without FdU treatment, resulted in chromosome fragility in patient cells that was detected using probes containing sequences telomeric to FMR1 (Figure 2). Similar results were obtained with both RP11-383P16 and RP11-402H20, which contain sequences from Xq27-28 and Xq28, respectively. No fragility was seen when the BAC, RP11-80F18, which contains sequences centromeric to FRAXA, was used a probe (data not shown).
Figure 2.
Effect of siRNA knockdown of ATR on fragile site expression at FRAXA in FX fibroblasts. The level of FRAXA expression in FX fibroblasts (GM05848) with and without ATR depletion in the presence and absence of FdU. The data shows the average of three independent experiments. C: control.
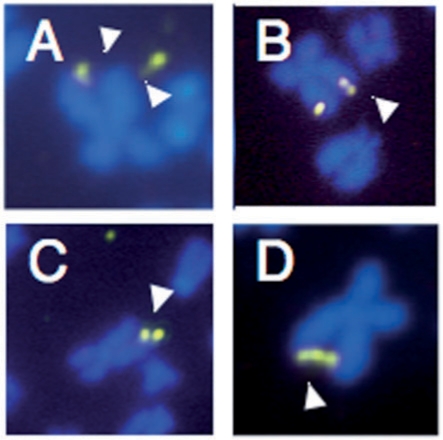
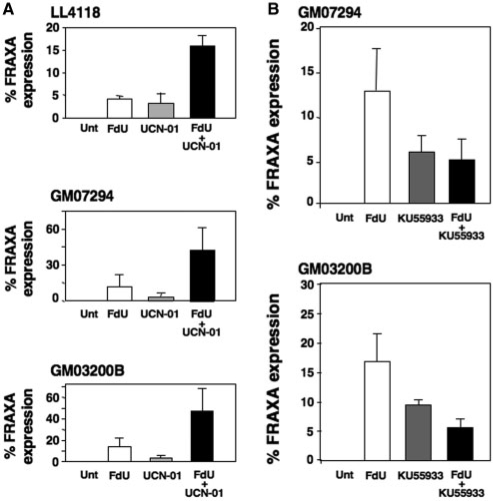
In addition to more typical fragile sites (Figure 3A), a small number of unusual derivative chromosomes were seen, including those containing duplications of the Xq27–28 region or two sister chromatids joined by three copies of the Xq27–28 region (Figure 3B–D). These chromosomes, which constituted <10% of the derivative chromosomes, are reminiscent of the products of breakage-fusion-bridge (BFB) cycles first described by Barbara McClintock (42), and subsequently demonstrated for a number of APC-inducible fragile sites (43,44). A similar increase in chromosome fragility and chromosome abnormalities was seen when patient cells were treated with UCN-01, a strong inhibitor of CHK1 (Figure 4A).
Figure 3.
Representative metaphases seen in FX lymphoblasts after treatment with 1 µM FdU and/or 100 nM UCN-01. (A) X chromosome showing breakage of both sister chromatids. (B) FISH showing X chromosome in which one chromatid shows what appears to be a partial duplication of part of the FRAXA region. (C) X chromosome in which one chromatid has lost the FRAXA region and the other has two copies of that region. (D) X chromosome in which the end of the long arm of the sister chromatids are linked by three copies of the FRAXA region.
Figure 4.
Effect of CHK1 and ATM inhibition on fragile site expression at FRAXA in FX lymphoblastoid cells. (A) FX lymphoblastoid cells treated with UCN-01 with and without FdU. (B) FX lymphoblastoid cells treated with KU55933 with and without FdU. The data shows the average of three independent experiments for each cell line. Unt: untreated or vehicle only.
KU55933 increases chromosome breakage at FRAXA but reduces the incidence of FdU-dependent FRAXA fragility
We also examined the effect of the ATM inhibitor KU55933 on chromosome fragility at FRAXA. Unlike many commonly used inhibitors of the phosphatidylinositol 3′-kinase-related kinase (PIKK) family to which ATM and ATR belong, KU55933 is considered a relatively specific inhibitor of ATM (45). It is 100 times more active against ATM than other PIKK family members, and does not inhibit ATR, the kinase to which ATM is most similar, even when used at 10 times the concentration we used here (45). Normal cells show no increase in chromosome fragility at FMR1 in the presence of KU55933 (data not shown). In the absence of FdU, KU55933 caused an increase in fragile site expression in FX cells. However, unlike what was seen with ATR depletion or UCN-01 treatment, KU55933 did not exacerbate the effect of FdU. In fact, the fraction of cells showing fragility at FRAXA when treated with both FdU and KU55933 was much lower than what was seen in FdU alone (Figure 4B). The fact that ATR depletion and KU55933 treatment have such different effects rules out an off-target effect of KU55933 on ATR and lends support to the idea that ATM is in fact the kinase most likely to be affected by the inhibitor. Our data would be consistent with the idea that some ATR-independent DDR pathway, most likely one mediated by ATM, contributes to FdU-sensitive chromosome fragility but protects against fragility arising during cell growth in the absence of FdU. Activation of ATM as well as ATR by FdU would be consistent with previous reports of activation of both kinases by thymidine-stress, which like FdU causes a nucleotide pool imbalance (46) and also induces FRAXA chromosome fragility (47).
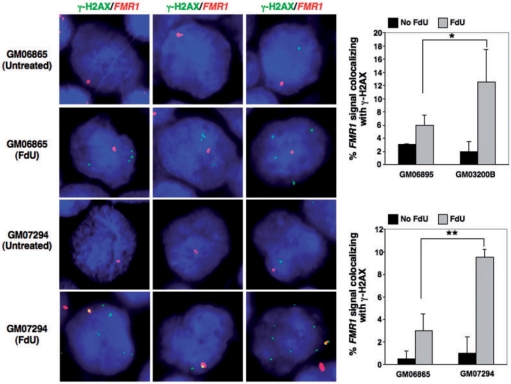
γ-H2AX colocalizes with the FMR1 locus in response to FdU treatment
APC-inducible fragile sites colocalize with the γ-H2AX foci that normally form at double strand breaks (DSBs) and double-strand ends found at stalled replication forks (41). In order to test if the same is true of FRAXA, we carried out in situ hybridization with a FMR1 BAC clone in combination with immunofluorescence with an antibody to γ-H2AX. In the absence of FdU, an average of ∼1 γ-H2AX focus/cell was seen in both normal and FXS cells (Figure 5). Colocalization of these foci with the FMR1 locus probe was seen in a small percentage of untreated normal and FX cells (Figure 5). This may be related to the presence of the common fragile site, FRAXD, in this region (48) or to some inherent problem present at this locus even in normal alleles. FdU-treatment increased the number of γ-H2AX foci in both normal and FXS cells to ∼4–5/cell. This contrasts with the much larger number of foci seen when cells are treated with enough APC to induce fragility at common fragile sites (37). Thus, despite the prolonged treatment with this FdU dosage, large-scale DNA damage is not apparent. However, this treatment did increase the number of cells in which γ-H2AX foci colocalized with probes containing FRAXA (Figure 5). In particular, the number of cells in which colocalization was observed was significantly higher in patient cells than in cells from unaffected individuals and is of the same order of magnitude as the number of fragile sites seen under the same conditions (Figure 5). This lends support to the idea that the FX allele is associated with the formation of DSBs or related structures in response to FdU. Since thymidine-stress does not produce DSBs (46), a structure other than a frank DSB is perhaps more likely.
Figure 5.
Colocalization of FX alleles with γ-H2AX foci in the presence of FdU. Immunocytochemistry combined with FISH was used to examine the colocalization of γ-H2AX foci and the FMR1gene in normal (GM06865 and GM06895) and FXS lymphoblastoid cells (GM07294 and GM03200B) treated with or without 1 µM FdU for 18 h. (A) Representative images of γ-H2AX (green) and FMR1 (red) stained cells. (B) Proportion of cells in which γ-H2AX and FMR1 colocalize. The data shows the average of two independent experiments in which a 100 FMR1 signals were counted for each experiment. Error bars signify standard deviation. Statistical analysis was carried out using Fishers exact test. A single asterisk indicates a P-value <0.01, while a double asterisk indicates a P-value <0.005. Note that the X chromosome tends to be located at the nuclear periphery (54). This can make it seem in some cases as if the FMR1 signal is located outside of the nucleus.
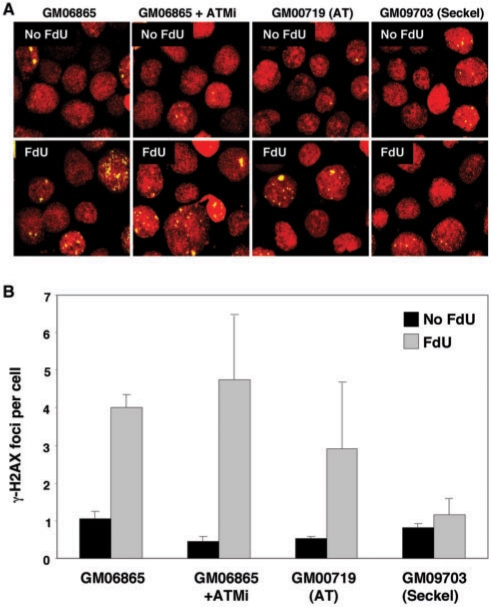
The basis of the FdU-induced foci seen in both normal and patient cells is currently unknown. The simplest explanation is that these foci represent regions of the human genome that, like FX alleles, contain long CGG•CCG-repeat tracts or related sequences with similar biological properties. In a cell line, GM09703, that has reduced levels of ATR and is deficient in the ATR pathway (49), no increase in the number of γ-H2AX foci was seen in response to FdU (Figure 6). This suggests that the ATR-pathway is responsible for the generation of the common FdU-induced γ-H2AX foci. It remains to be seen whether ATR is also responsible for the foci that colocalize with FRAXA. No change in the number of FdU-induced foci was seen in AT-cells or in normal cells treated with KU55933 (Figure 6). The latter result would be consistent with the effect of KU55933 being ATM-specific and these data taken together suggest that the common FdU-induced γ-H2AX foci are ATM-independent.
Figure 6.
The FdU-induced common γ-H2AX foci are dependent on ATR not ATM. GM06865 with and without ATM inhibitor (ATMi), an AT cell line, GM00719, and an ATR-deficient cell line, GM09703 were examined for the presence of FdU-induced γ-H2AX foci after growth with or without FdU as indicated. (A) shows representative fields obtained for each condition. (B) shows the combined data for two independent experiments in which a total of 400 nuclei were counted for each condition. The error bars indicate standard deviations.
DISCUSSION
To assess the similarities between APC-inducible and FdU inducible fragile sites, we have examined the activation of different DNA damage response pathways and chromosome fragility at FRAXA after FdU treatment. Our data demonstrate that as with APC treatment, FdU causes phosphorylation of CHK1 but, little, if any CHK2, RPA2 or p53 activation. FdU also increases the colocalization of the FMR1 locus with γ-H2AX foci (Figure 5), analogous to what is seen with APC-sensitive fragile sites (37). Thus, in some ways APC and FdU have grossly similar effects on human cells and their respective fragile sites.
However, important differences between the APC and FdU response are also seen. ATR knockdown and the use of KU55933, an ATM inhibitor, have opposite effects on FdU-induced FRAXA chromosome fragility, increasing and decreasing FRAXA expression respectively (Figures 2 and 4B). Since ATM and ATR are both involved in protecting the genome from APC-sensitive chromosome fragility (37), our data suggest that the basis of the FdU-sensitive FX fragile site is different from that of the APC-sensitive fragile sites.
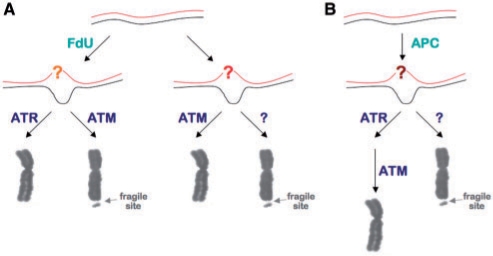
Since ATR is activated primarily in response to stalled replication forks and bulky DNA lesions that block DNA polymerase, and CGG•CCG-repeats form non-canonical structures like hairpins and tetraplexes that block DNA synthesis in vitro (50) and are thought to cause replication fork stalling in bacteria (51) and yeast (52), it is tempting to think that these sorts of structures may be responsible for the problems that give rise to the FdU-induced FRAXA chromosome fragility. In this view, illustrated in Figure 7A, the nucleotide pool imbalance resulting from FdU treatment slows replication through the repeat region, perhaps causing uncoupling of leading and lagging strand DNA synthesis. This may create a greater opportunity for structures like hairpins and tetraplexes to form that block DNA synthesis. The nucleotide pool imbalance may also lead to the misincorporation of bases that would then need to be excised. Since ATR depletion exacerbates FdU-sensitive chromosome fragility while the ATM inhibitor reduces it, it may be that ATM and ATR normally compete for the repair of the underlying lesion. When ATR is recruited to the lesion, it is able to prevent the generation of a fragile site. However, when ATM is involved, attempts to repair the lesion lead to chromosome fragility. ATM inhibition may reduce chromosome fragility by allowing more ATR-dependent repair to take place.
Figure 7.
Comparison of models for FRAXA and APC-inducible chromosome fragility. (A) In the presence of FdU, a lesion arises in the FMR1 gene in FXS cells that result in activation of ATM and ATR. ATR activation results in the lesion being successfully repaired, but ATM activation can lead to chromosome fragility. In the absence of FdU, a second type of chromosome fragility is seen, one that is normally resolved by ATM. In the absence of ATM, an as yet unidentified DNA damage response pathway is used instead and this leads to the production of a fragile site. (B) A model for APC-inducible fragile sites based on the data of Ozeri-Galai et al. (37). In the case of these fragile sites, a lesion of some sort arises in the presence of APC. ATR is primarily responsible for dealing with this lesion, but in the absence of ATR, ATM can substitute for ATR. However, when both ATR and ATM are depleted, an error-prone mechanism is used to deal with the problem resulting in the formation of a fragile site. It is unclear how these three types of fragile site differ at the molecular level. For this reason the responsible lesion is indicated by different colored question marks.
Since the ATM inhibitor does increase chromosome fragility, albeit of a type that is not exacerbated by FdU, there is apparently also a previously unappreciated FdU-independent form of chromosome fragility at FRAXA. The basis of this fragility is unknown. However, the presence at FRAXA of both ATR-sensitive and lesions that may be sensitive to ATM is of interest given the other ATR and ATM-sensitive events that occur in long CGG•CCG-repeat tracts. We have shown that a mouse model with a long CGG•CCG-repeat tract in the murine Fmr1 gene shows two types of repeat expansion, one that is ATR-sensitive and one that is ATM-sensitive [(53) and manuscript in preparation]. These data would be consistent with the idea that long CGG•CCG-repeats cause at least two different kinds of DNA lesions and that failure to resolve or repair them appropriately can lead to chromosome fragility and/or repeat expansion.
FUNDING
Funding for open access charge: Intramural program of the NIDDK, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Howard-Peebles PN, Stoddard GR. X-linked mental retardation with macro-orchidism and marker X chromosomes. Hum. Genet. 1979;50:247–251. doi: 10.1007/BF00399389. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland GR, Ashforth PL. X-linked mental retardation with macro-orchidism and the fragile site at Xq 27 or 28. Hum. Genet. 1979;48:117–120. doi: 10.1007/BF00273283. [DOI] [PubMed] [Google Scholar]
- 3.Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Jr, Warren ST, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 4.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 5.Debacker K, Winnepenninckx B, Longman C, Colgan J, Tolmie J, Murray R, van Luijk R, Scheers S, Fitzpatrick D, Kooy F. The molecular basis of the folate-sensitive fragile site FRA11A at 11q13. Cytogenet Genome Res. 2007;119:9–14. doi: 10.1159/000109612. [DOI] [PubMed] [Google Scholar]
- 6.Gecz J, Gedeon AK, Sutherland GR, Mulley JC. Identification of the gene FMR2, associated with FRAXE mental retardation. Nat. Genet. 1996;13:105–108. doi: 10.1038/ng0596-105. [DOI] [PubMed] [Google Scholar]
- 7.Gu Y, Shen Y, Gibbs RA, Nelson DL. Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nat. Genet. 1996;13:109–113. doi: 10.1038/ng0596-109. [DOI] [PubMed] [Google Scholar]
- 8.Parrish JE, Oostra BA, Verkerk AJ, Richards CS, Reynolds J, Spikes AS, Shaffer LG, Nelson DL. Isolation of a GCC repeat showing expansion in FRAXF, a fragile site distal to FRAXA and FRAXE. Nat. Genet. 1994;8:229–235. doi: 10.1038/ng1194-229. [DOI] [PubMed] [Google Scholar]
- 9.Sarafidou T, Kahl C, Martinez-Garay I, Mangelsdorf M, Gesk S, Baker E, Kokkinaki M, Talley P, Maltby EL, French L, et al. Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics. 2004;84:69–81. doi: 10.1016/j.ygeno.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Winnepenninckx B, Debacker K, Ramsay J, Smeets D, Smits A, FitzPatrick DR, Kooy RF. CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am. J. Hum. Genet. 2007;80:221–231. doi: 10.1086/510800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren ST, Knight SJ, Peters JF, Stayton CL, Consalez GG, Zhang FP. Isolation of the human chromosomal band Xq28 within somatic cell hybrids by fragile X site breakage. Proc. Natl Acad. Sci. USA. 1990;87:3856–3860. doi: 10.1073/pnas.87.10.3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones C, Penny L, Mattina T, Yu S, Baker E, Voullaire L, Langdon WY, Sutherland GR, Richards RI, Tunnacliffe A. Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature. 1995;376:145–149. doi: 10.1038/376145a0. [DOI] [PubMed] [Google Scholar]
- 13.Jones C, Slijepcevic P, Marsh S, Baker E, Langdon WY, Richards RI, Tunnacliffe A. Physical linkage of the fragile site FRA11B and a Jacobsen syndrome chromosome deletion breakpoint in 11q23.3. Hum. Mol. Genet. 1994;3:2123–2130. doi: 10.1093/hmg/3.12.2123. [DOI] [PubMed] [Google Scholar]
- 14.Callahan G, Denison SR, Phillips LA, Shridhar V, Smith DI. Characterization of the common fragile site FRA9E and its potential role in ovarian cancer. Oncogene. 2003;22:590–601. doi: 10.1038/sj.onc.1206171. [DOI] [PubMed] [Google Scholar]
- 15.Curatolo A, Limongi ZM, Pelliccia F, Rocchi A. Molecular characterization of the human common fragile site FRA1H. Genes Chromosomes Cancer. 2007;46:487–493. doi: 10.1002/gcc.20432. [DOI] [PubMed] [Google Scholar]
- 16.Denison SR, Callahan G, Becker NA, Phillips LA, Smith DI. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer. 2003;38:40–52. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- 17.Huang H, Qian J, Proffit J, Wilber K, Jenkins R, Smith DI. FRA7G extends over a broad region: coincidence of human endogenous retroviral sequences (HERV-H) and small polydispersed circular DNAs (spcDNA) and fragile sites. Oncogene. 1998;16:2311–2319. doi: 10.1038/sj.onc.1200202. [DOI] [PubMed] [Google Scholar]
- 18.Mangelsdorf M, Ried K, Woollatt E, Dayan S, Eyre H, Finnis M, Hobson L, Nancarrow J, Venter D, Baker E, et al. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 2000;60:1683–1689. [PubMed] [Google Scholar]
- 19.Mishmar D, Rahat A, Scherer SW, Nyakatura G, Hinzmann B, Kohwi Y, Mandel-Gutfroind Y, Lee JR, Drescher B, Sas DE, et al. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl Acad. Sci. USA. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paradee W, Wilke CM, Wang L, Shridhar R, Mullins CM, Hoge A, Glover TW, Smith DI. A 350-kb cosmid contig in 3p14.2 that crosses the t(3;8) hereditary renal cell carcinoma translocation breakpoint and 17 aphidicolin-induced FRA3B breakpoints. Genomics. 1996;35:87–93. doi: 10.1006/geno.1996.0326. [DOI] [PubMed] [Google Scholar]
- 21.Rassool FV, Le Beau MM, Shen ML, Neilly ME, Espinosa R, 3rd, Ong ST, Boldog F, Drabkin H, McCarroll R, McKeithan TW. Direct cloning of DNA sequences from the common fragile site region at chromosome band 3p14.2. Genomics. 1996;35:109–117. doi: 10.1006/geno.1996.0329. [DOI] [PubMed] [Google Scholar]
- 22.Rozier L, El-Achkar E, Apiou F, Debatisse M. Characterization of a conserved aphidicolin-sensitive common fragile site at human 4q22 and mouse 6C1: possible association with an inherited disease and cancer. Oncogene. 2004;23:6872–6880. doi: 10.1038/sj.onc.1207809. [DOI] [PubMed] [Google Scholar]
- 23.Sawinska M, Schmitt JG, Sagulenko E, Westermann F, Schwab M, Savelyeva L. Novel aphidicolin-inducible common fragile site FRA9G maps to 9p22.2, within the C9orf39 gene. Genes Chromosomes Cancer. 2007;46:991–999. doi: 10.1002/gcc.20484. [DOI] [PubMed] [Google Scholar]
- 24.Helmrich A, Stout-Weider K, Matthaei A, Hermann K, Heiden T, Schrock E. Identification of the human/mouse syntenic common fragile site FRA7K/Fra12C1—relation of FRA7K and other human common fragile sites on chromosome 7 to evolutionary breakpoints. Int. J. Cancer. 2007;120:48–54. doi: 10.1002/ijc.22049. [DOI] [PubMed] [Google Scholar]
- 25.Limongi MZ, Pelliccia F, Rocchi A. Characterization of the human common fragile site FRA2G. Genomics. 2003;81:93–97. doi: 10.1016/s0888-7543(03)00007-7. [DOI] [PubMed] [Google Scholar]
- 26.Morelli C, Karayianni E, Magnanini C, Mungall AJ, Thorland E, Negrini M, Smith DI, Barbanti-Brodano G. Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene. 2002;21:7266–7276. doi: 10.1038/sj.onc.1205573. [DOI] [PubMed] [Google Scholar]
- 27.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 28.Corbin S, Neilly ME, Espinosa R, 3rd, Davis EM, McKeithan TW, Le Beau MM. Identification of unstable sequences within the common fragile site at 3p14.2: implications for the mechanism of deletions within fragile histidine triad gene/common fragile site at 3p14.2 in tumors. Cancer Res. 2002;62:3477–3484. [PubMed] [Google Scholar]
- 29.Cheng CH, Kuchta RD. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry. 1993;32:8568–8574. doi: 10.1021/bi00084a025. [DOI] [PubMed] [Google Scholar]
- 30.Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 31.Hurley PJ, Bunz F. ATM and ATR: components of an integrated circuit. Cell Cycle. 2007;6:414–417. doi: 10.4161/cc.6.4.3886. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, Xu ZP, Huang Y, Hamrick HE, Duerksen-Hughes PJ, Yu YN. ATM and ATR: sensing DNA damage. World J. Gastroenterol. 2004;10:155–160. doi: 10.3748/wjg.v10.i2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 35.Durkin SG, Arlt MF, Howlett NG, Glover TW. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene. 2006;25:4381–4388. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- 36.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 37.Ozeri-Galai E, Schwartz M, Rahat A, Kerem B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene. 2008;27:2109–2117. doi: 10.1038/sj.onc.1210849. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz M, Zlotorynski E, Goldberg M, Ozeri E, Rahat A, le Sage C, Chen BP, Chen DJ, Agami R, Kerem B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19:2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gray SJ, Gerhardt J, Doerfler W, Small LE, Fanning E. An origin of DNA replication in the promoter region of the human fragile X mental retardation (FMR1) gene. Mol. Cell Biol. 2007;27:426–437. doi: 10.1128/MCB.01382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClintock B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics. 1938;23:315–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, et al. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum. Mol. Genet. 2002;11:2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- 44.Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, Smith DI, Trakhtenbrot L, Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 45.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 46.Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M. ATM is required for the cellular response to thymidine induced replication fork stress. Hum. Mol. Genet. 2004;13:2937–2945. doi: 10.1093/hmg/ddh316. [DOI] [PubMed] [Google Scholar]
- 47.Howard-Peebles PN, Stoddard GR, Mims MG. Familial X-linked mental retardation, verbal disability, and marker X chromosomes. Am. J. Hum. Genet. 1979;31:214–222. [PMC free article] [PubMed] [Google Scholar]
- 48.Ledbetter SA, Ledbetter DH. A common fragile site at Xq27: theoretical and practical implications. Am. J. Hum. Genet. 1988;42:694–702. [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Matsuda A, Plunkett W. Ataxia-telangiectasia and Rad3-related and DNA-dependent protein kinase cooperate in G2 checkpoint activation by the DNA strand-breaking nucleoside analogue 2′-C-cyano-2′-deoxy-1-beta-D-arabino-pentofuranosylcytosine. Mol. Cancer Ther. 2008;7:133–142. doi: 10.1158/1535-7163.MCT-07-0416. [DOI] [PubMed] [Google Scholar]
- 50.Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 52.Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell Biol. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Entezam A, Usdin K. ATR protects the genome against CGG.CCG-repeat expansion in Fragile X premutation mice. Nucleic Acids Res. 2008;36:1050–1056. doi: 10.1093/nar/gkm1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dyer KA, Canfield TK, Gartler SM. Molecular cytological differentiation of active from inactive X domains in interphase: implications for X chromosome inactivation. Cytogenet. Cell Genet. 1989;50:116–120. doi: 10.1159/000132736. [DOI] [PubMed] [Google Scholar]