Abstract
DNA methylation, the only known covalent modification of mammalian DNA, occurs primarily in CpG dinucleotides. 51% of CpGs in the human genome reside within repeats, and 25% within Alu elements. Despite that, no method has been reported for large-scale ascertainment of CpG methylation in repeats. Here we describe a sequencing-based strategy for parallel determination of the CpG-methylation status of thousands of Alu repeats, and a computation algorithm to design primers that enable their specific amplification from bisulfite converted genomic DNA. Using a single primer pair, we generated amplicons of high sequence complexity, and derived CpG-methylation data from 31 178 Alu elements and their 5′ flanking sequences, altogether representing over 4 Mb of a human cerebellum epigenome. The analysis of the Alu methylome revealed that the methylation level of Alu elements is high in the intronic and intergenic regions, but low in the regions close to transcription start sites. Several hypomethylated Alu elements were identified and their hypomethylated status verified by pyrosequencing. Interestingly, some Alu elements exhibited a strikingly tissue-specific pattern of methylation. We anticipate the amplicons herein described to prove invaluable as epigenome representations, to monitor epigenomic alterations during normal development, in aging and in diseases such as cancer.
INTRODUCTION
DNA methylation has been associated with the control of gene expression, genomic imprinting and the maintenance of genome integrity (1,2). Dynamic changes in genome-wide methylation occur during development, aging and cancer progression (3). For many tissues, the percentage of 5-methylcytosine in the genome, and in repetitive DNA particularly, decreases over time. Such loss of DNA methylation was shown to be age-related (4,5). Although the functional aspects of age-related genome demethylation are still largely unclear, such a decrease in the genome-wide level of DNA methylation has been associated with genomic instability (5–7). In addition, genome-wide hypomethylation has been recognized as a hallmark in many tumors (8,9). Recently, hypomethylation of repetitive elements was demonstrated to be highly associated with cancer progression and poor clinical outcome (10–12). Rather than being a passive bystander, repetitive elements may play a critical role in the establishment of genome-wide methylation patterns. In the past decade, homology dependent methylation has been found to be a mechanism that initiates de novo DNA methylation and transmits methylation patterns. DNA methylation has been proposed to result from homologous DNA:DNA or DNA:RNA pairings (13,14). Both types of pairings have been observed with repetitive elements, which may serve as methylation ‘way stations’ (15–17). Recently, AluY/S elements were shown to be enriched at the junction between hypermethylated and hypomethylated genomic regions (18). Thus, epigenetic analyses of repetitive elements will contribute significantly to our understanding of the dynamics of DNA methylation in the human genome.
Numerous approaches have been developed for genome-wide methylation analysis (19). These approaches can be classified into three major categories: restriction enzyme-based, microarray-based and bisulfite sequencing-based (18–21). Data generated with methylation-sensitive restriction enzymes are limited to the recognition sites of the enzymes used. Microarray-based methods for methylation profiling have limitations, not the least of which is the fact that they cannot distinguish among members of repetitive DNA families. Sequencing of bisulfite-treated DNA is thus still considered as the gold standard for high-resolution DNA-methylation profiling. By bisulfite-sequencing specific loci, Beck and colleagues determined the methylation profile of over 0.1% of the human epigenome in normal and in disease tissues (20). Although this provided a great deal of information, it cannot be conveniently scaled up. Recently, shotgun sequencing of bisulfite-converted genomic DNA has been exploited to generate an epigenomic map for Arabidopsis (22,23). Furthermore, ‘reduced representation bisulfite sequencing’ has been proposed for large-scale analysis of epigenomes of greater complexity (24,25). However, mapping of short sequence reads obtained from bisulfite converted genomic DNA remains challenging, most especially when derived from repetitive sequences. Besides a recent effort to track hypomethylated Alu elements in normal and in cancer cells (26), no high-throughput method has been reported to date for genome-wide ascertainment of the status of CpG methylation of repetitive elements and their flanking sequences.
Here we report a strategy to amplify and sequence large sets of repetitive elements and their flanking sequences, from bisulfite converted genomic DNA. Using this approach, we generated a methylation map of Alu elements in normal human cerebellum.
METHODS
The generation of a nucleotide position weight matrix for Alu elements
The human genome sequence (build36/hg18, March 2006) and the annotated Alu repetitive elements were obtained from the UCSC Genome Database (27). Considering genomic coordinates provided by UCSC database, 1 180 972 Alu sequences were extracted. Consensus sequences of 32 Alu subfamilies were downloaded from RepBase (28). For each Alu element and the consensus sequences, in silico bisulfite treatment was performed by converting CpG dinucleotides in consensus sequences to YG dinucleotides, and all other Cs—not in CpGs—to Ts. Together with 32 consensus sequences, multiple sequencing alignments were conducted for each Alu element using CLUSTALW (29). To keep track of over one million alignment results, the consensus sequence derived from multiple sequence alignment of the 32 Alu subfamilies was used as reference. In other words, the consensus sequence was used as a standard to provide coordinates for each base along the Alu sequences. Primers with specificity to a particular subset of Alu repeats were identified using this matrix, upon comparison of nucleotide occurrences in the given subset of Alu sequences with those in the remainder Alu elements.
DNA samples
Human genomic DNA samples for fetal adrenal gland tissues were purchased (BioChain Inc., Hayward, CA). Human snap-frozen cerebellum and cortex tissues were obtained from the tissue bank of the Department of Pathology, Children's Memorial Hospital at Chicago under an IRB-approved protocol. Genomic DNA was extracted with Qiagen DNeasy tissue kit (Qiagen, Valencia, CA).
Construction and sequencing of repeat amplicon library
Genomic DNA digestion, dephosphoration and ligation
One microgram of genomic DNA from cerebellum derived from an individual was digested with AluI restriction enzyme at 37°C for 2 h, followed by calf intestinal alkaline phosphatase (CIP) incubation at 37°C for 1 h. The dephosphorylated genomic DNA was mixed with 100-fold molar excess adaptors in the presence of T4 DNA ligase at 14°C for 12 h. The AluI restriction enzyme, alkaline phosphatase and T4 DNA ligase were purchased from New England BioLabs, Inc., Beverly, MA. The adaptors that we utilized were: 5′PO4-GAA GAT GAG TCA GGT CGG CAT CG (3′ddC)-3OH′ and 5′OH-GCG ATG CCG ACC TGA CTC ATC TTC-3′OH. To remove free adaptors, the ligation product was purified with PureLink PCR Purification kit (Invitrogen, Carlsbad, CA).
Bisulfite treatment and PCR
Bisulfite modification of genomic DNA was performed with EZ DNA Methylation Gold kit (Zymo Research, Orange, CA) according to the manufacturer's instructions. Briefly, 0.5 µg genomic DNA was bisulfite-treated, eluted with 10 µl elution solution, and stored at –20°C until ready for use. A 50 µl PCR mixture was prepared with 2 μl (100 ng) bisulfite treated DNA, 50 pmol each forward and reverse primers. The sequence of the primer utilized for annealing to the bisulfite converted Alu elements was: 5′-GCC TCC CTC GCG CCA TCA GTG TTA GTT AGG ATG GTT T-3′. The sequence of the primer utilized for annealing to the bisulfite converted adaptor was: 5′-GCC TTG CCA GCC CGC TCA GCC AAC CTA ACT CAT CTT C-3′. Both PCR primers were fusion primers containing sequences required for 454 sequencing (in bold print). The PCR reactions were performed with the high fidelity PCR master system (Roche Molecular Systems, Inc., Indianapolis, IN). After denaturation at 94°C for 3 min, PCR cycles were performed under the following conditions: 94°C for 15 s, 60°C for 30 s and 72°C for 1 min for three cycles, followed by 94°C for 15 s and 72°C for 1 min for 25 additional cycles. PCR products were first purified with PureLink PCR Purification kit (Invitrogen, Carlsbad, CA) and AMPure beads (Agencourt Bioscience Corporation, Beverly, MA) to remove short PCR products. The purified PCR products were then stored at 4°C until ready for 454 sequencing.
454 SEQUENCING
Total 454 sequencing was conducted with the Roche Genome Sequencer FLX System. Briefly, after size selection and quantification, PCR products were annealed to an excess of DNA capture beads and used as templates for bead-based emulsion PCR. The emulsion PCR was performed with emPCR Kit II (Amplicon A) to ensure that subsequent sequencing would be initiated at the end with the Alu specific primer. The DNA-containing capture beads and PCR reagents were emulsified in water-in-oil microreactors for clonal amplification. PCR reactions were performed according to manufacturer's instructions. After PCR, the microreactors were then broken to recover the beads. The beads with PCR products were further enriched and subjected to sequencing. The amplicon library was sequenced at the Sequencing Core Facility of the Children's Memorial Research Center of Northwestern University's Feinberg School of Medicine.
Processing the sequence reads
Sequence mapping
Raw sequencing images were first analyzed with the full data analysis pipeline provided by the Genome Sequencer FLX System. To identify the genomic origin of the sequence reads, a multistep mapping procedure was implemented with perl scripts. Since the sequences were derived from bisulfite converted DNA, an artificial reference genome database was developed to host the human genome sequence after in silico bisulfite conversion. More specifically, the cytosines were converted to thymidines in both strands of each of the chromosomes. In addition, we applied the same conversion to all sequence reads that we generated.
After removal of primer and adaptor sequences, sequences ≥40 bp were aligned to the reference genome using MegaBLAST. To facilitate the alignment procedure and to increase alignment stringency, multiple cycles of MegaBLAST were employed. The word size used in Megablast was set to 100 for the first cycle, it was decreased by 20 for every cycle thereafter till the last, for which the minimum length of best perfect match was set to 40. In addition, the identity percentage cutoff for a significant alignment was set to be 100 for the last cycle and 95 for all other cycles of Megablast. To ensure mapping accuracy, sequences that mapped to more than one position with similar scores (the differences in E-value are typically within 100-fold) were removed.
Determination of CpG methylation
To obtain methylation information and estimate bisulfite conversion rate, comparisons between sequences generated and corresponding genomic sequences were conducted. Before sequence alignment with CLUSTALW, all cytosines in corresponding genomic sequences were converted to thymidines in silico, except for the ones in CpGs. However, no conversion was applied to the sequence reads that we generated. The final methylation data were extracted by perl scripts that we wrote, and stored in two files of Generic Feature Format (GFF). One corresponds to the methylation status of each CpG dinucleotide in the reads, and the other to the methylation level of each read.
Data integration and visualization
Human genome sequence data and annotations for repetitive elements and known genes were obtained from the UCSC Genome Database (30). The coordinates of putative transcription start sites for known genes were extracted from the UCSC annotation database. A MySOL database was designed and implemented to host the methylation data. At the same time, the database is also hosting publicly available datasets from several genome-wide methylation studies, gene expression studies and annotations for SNPs, genes, CpG islands and Alu elements. The structure of this database allows integration of these different types of data. On top of the database, a genome browser was developed to allow synchronic view of genetic/epigenetic features (http://cmbteg.childrensmemorial.org/cgi-bin/gbrowse/btech/). With the genome browser, users can navigate genomic regions and search for data and annotations available for these regions. All the data that are populated in the database can be displayed on the genome browser. The genome browser installed locally was derived from the building blocks provided by The Generic Model Organism System Database Project (31). All perl scripts implementing primer design, sequence processing and mapping in this study are available upon request.
Pyrosequencing verification
Nested PCR reactions were adopted to amplify the target regions from bisulfite modified genomic DNA. Two runs of PCR reactions were carried out using the Hotstart Taq polymerase kit (Qiagen, Valencia, CA) in 25 µl total volume and with 50 pmol each of forward and reverse primers. In the first PCR reaction, 50 ng of the bisulfite converted DNA was used as template. After 5 min of initial denaturation at 95°C, the cycling conditions of 40 cycles consisted of denaturation at 95°C for 15 s, annealing at 55°C for 30 s and elongation at 72°C for 30 s. One microliter of PCR product from the first run was used as template for the second PCR reaction, which after initial denaturation at 95°C for 5 min comprised 40 cycles with denaturation at 95°C for 15 s, annealing at 55°C for 30 s and elongation at 72°C for 15 s. The PCR products were stored at 4°C until ready for pyrosequencing.
Pyrosequencing was performed using the PyroMark MD Pyrosequencing System (Biotage, Charlottesville, VA). The final PCR product was purified using streptavidin-Sepharose HP beads (GE Healthcare, Uppsala, Sweden) and processed to yield single-stranded DNA. The single-stranded DNA was prepared for pyrosequencing using the PyroMark Vacuum Prep Tool (Biotage, Charlottesville, VA). In brief, the PCR product was bound onto Sepharose beads. Beads containing the immobilized PCR product were washed, denatured using a 0.2 M NaOH solution, washed again and neutralized. Pyrosequencing primer at a concentration of 0.3 µM was annealed to the purified single-stranded PCR products at 28°C. Methylation quantification was performed using the manufacturer-provided software. The primers used in the PCR runs and pyrosequencing reactions are shown in Supplementary Table 2.
RESULTS
Sequencing strategy for methylation analysis of bisulfite converted repetitive DNA elements
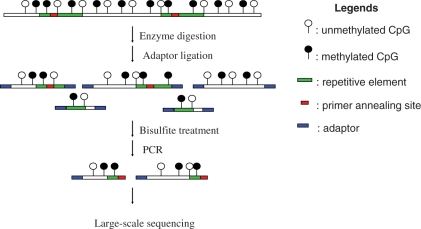
Based on the reference genome sequence (hg18, http://genome.ucsc.edu/), there are over 28 million CpG dinucleotides in the human genome. Repeat elements contain more than half of these CpG dinucleotides. The main repeat families, SINEs, LINEs and LTRs, contribute 27%, 12% and 7% of CpG dinucleotides in the human genome, respectively. In order to obtain a detailed methylation profile of repetitive elements, we devised and implemented the strategy shown in Figure 1. In this strategy, one needs to design primers within the repetitive elements and one has to identify a methylation insensitive restriction enzyme that does not have recognition site(s) between the primer annealing region and the end of repeats. After digestion with the selected restriction endonuclease, genomic DNA is ligated to an alien adaptor and treated with bisulfite. Using primers specific for a targeted group of repeats, a complex amplicon library is generated and sequenced. It should be emphasized that each read encompasses repetitive as well as (most often) unique flanking sequences. Unequivocal mapping of repetitive elements is therefore enabled by their unique flanking sequences.
Figure 1.
Diagram of experimental design. Genomic DNA is first digested with a methylation insensitive enzyme, ligated to adaptors and then subjected to bisulfite treatment. Bisulfite treated DNA is amplified with adaptor and primer specific for the targeted repeat elements. PCR products contain repeat sequence and flanking unique genomic sequence. Lastly, the repeat-containing amplicon library thus constructed is subjected to large-scale sequencing. The methylated cytosines are indicated with the filled circles while the unmethylated cytosines are indicated with the open circles. The green and red segments represent the repeat elements with the red segment indicating the region to which the primer anneals. The adaptors are indicated by blue boxes.
In the human genome, ∼1.2 million Alu elements belong to the SINE superfamily altogether encompassing in excess of 7.1 million CpG dinucleotides, which correspond to over 25% of all CpGs in the genome. Therefore, Alu elements were prioritized for epigenomic analysis in this study. The consensus sequences of Alu subfamilies indicate that all Alu subfamilies are rich in CpG dinucleotides (28). However, due to frequent deamination of methylated cytosines in CpG dinucleotides within Alu elements, the oldest Alu subfamilies exhibit a significant depletion of CpG dinucleotides (32). We investigated the preservation of CpG dinucleotides among all Alu subfamilies. AluYa-h, the most recent members of the Alu superfamily, were found to have 28 CpG dinucleotides on average. In contrast, less than three CpG sites are present in AluJ elements, the oldest Alu subfamily. It was also found that approximately half of all Alu elements have two or fewer CpGs in their 5′ most 80 nt.
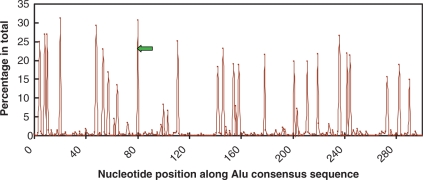
We identified 27 059 Alu elements that contain seven or more CpG dinucleotides within their 5′ most 80 bases. In order to target the specific amplification of this subset of Alu elements, a primer design algorithm was developed and implemented. We determined the nucleotide position weight matrices for all Alu elements and for the aforementioned selected subset of Alu elements, as described in the Methods section. The 5′ region of Alu elements contains the A box and the B box promoter regions of RNA Polymerase III. It is noteworthy that the 5′-terminal region of Alu repeats contains a greater number of CpG dinucleotides than the 3′-end (Figure 2). Hence, it was targeted for primer design. Comparison of the two matrices revealed the region from position 82 to 100 in the Alu consensus sequence as the most discriminating of the selected Alu subset, hence targeted for primer design (Supplementary Table 1). The lack of CpG dinucleotides in the primer annealing region ensures that both methylated and unmethylated Alu elements (Figure 2) can be amplified following bisulfite treatment.
Figure 2.
The preservation of CpG dinucleotides along Alu consensus sequences. X-axis represents the nucleotide position within the consensus Alu sequence. The Y-axis represents the percentage of all Alus that contain CpG dinucleotides for each given position. The green arrow represents the PCR primer designed in this study.
To select the best suited restriction endonuclease, we performed in silico genomic DNA digestions of all restriction enzymes with recognition sequences of 4 bp and 5 bp, singly and in all combinations. As a result of this analysis, the restriction enzyme AluI was chosen for this study. The length of the PCR products derived from the targeted Alu elements, i.e. those specifically amplified with the designed primer, after digestion of human genomic DNA with the AluI restriction endonuclease, was in the range of 150–500 bp. A critical issue in our approach is the specificity of PCR amplification. Due to the high sequence similarity among Alu elements, a low annealing temperature during PCR would lead to non-specific amplification of undesired Alu elements. In contrast to ‘degenerate’ PCR, we used the designed primer and conducted PCR under very stringent conditions. Accordingly, a series of PCR amplifications was performed to determine the optimal annealing temperature, i.e. the highest temperature to still yield products, which was found to be 60°C.
Statistics of the reads and CpG sites sequenced
An amplicon library was constructed from bisulfite treated human cerebellum DNA derived from a single individual, using the primer, restriction enzyme and annealing temperature discussed above. A total of 48 839 943 nt sequences were generated from the amplicon library using the Roche Genome Sequencer FLX™ 454 system. After removal of primer and adaptor sequences, a total of 370 612 sequence reads comprising 37 011 779 bases, with an average of 100 bp per read, were obtained. Sequence mapping was conducted as described in the ‘Methods’ section (Supplementary Figure 1). Prior to sequence mapping, all cytosines in the sequence reads and in the reference human genome sequence were converted to thymidines in silico. Thus, the methylation variability and incomplete bisulfite conversion would not affect the accuracy of sequence mapping.
A total of 245 825 sequence reads encompassing a total of 28 650 038 bases were mapped to 31 871 non-overlapping genomic regions. These non-overlapping genomic regions cover 4 223 824 bases in the human genome. The overall mapping efficiency was 66% (245 825/370 612) and it increases for the longer sequence reads (Supplementary Figure 2). Based on the comparisons of mapped sequence reads and the corresponding genomic sequences, the bisulfite conversion rate in this study was estimated to be 99.1%. The average coverage of each targeted region was found to be ∼7-fold (28 650 038/4 223 824). The cluster size distribution of sequences that could be unequivocally mapped to the genome is shown in Supplementary Figure 3. Of the 31 871 sequence clusters, 38% contained only one sequence read and 95% of all clusters had fewer than 32 sequence reads. This indicated that the amplicon library obtained from a single amplification was of high complexity. In addition, we also examined the specificity of amplification. Of the 245 825 sequences that were successfully mapped to the genome, 94% were derived from 31 178 distinct Alu elements (Table 1). Over 86% of all sequences generated were mapped to genomic loci with sequence, after bisulfite conversion, 100% identical to the PCR primer designed.
Table 1.
Statistics of Alu elements sequenced
| Alu sub family | Number of Alu elements in genome | Number of distinct Alu elements sequenced | Number of sequences generated | Precentage of sequences generateda |
|---|---|---|---|---|
| AluY | 137 925 | 23 857 | 207 602 | 84.5 |
| AluSc | 49 325 | 1725 | 7365 | 3.0 |
| AluSx | 339 002 | 1438 | 2744 | 1.1 |
| AluSg | 81 915 | 1304 | 4381 | 1.8 |
| AluYa_hb | 9646 | 672 | 3494 | 1.4 |
| AluJb | 128 539 | 539 | 1044 | 0.4 |
| AluJo | 141 841 | 510 | 806 | 0.3 |
| AluSq | 94 487 | 450 | 926 | 0.4 |
| AluSp | 50 917 | 231 | 467 | 0.2 |
| Others | 147 375 | 452 | 1218 | 0.5 |
| Total | 1 180 972 | 31 178 | 230 047 | 93.6 |
aThe percentage was calculated as the number of sequences from Alu subfamily divided by the total number of sequences mapped (245 825 sequence reads).
bAluYa-h denotes the young AluY families: AluYa to AluYh.
Once mapped, sequences were compared to the reference human genomic sequence. With respect to methylation, 1 252 562 methylation data points were obtained from 159 225 distinct CpG sites. On average, five CpG dinucleotides were present per sequence read. In total, the methylation statuses of 0.6% of all CpG dinucleotides found in the human genome were determined in this study. The genomic distribution of the CpG dinucleotides sequenced in this study was further examined. For each chromosome, the ratio of the number of CpG dinucleotides sequenced to the total number of CpG dinucleotides in the chromosome was calculated (Supplementary Figure 4A). This ratio varied in the 0.5–0.8% range across chromosomes. Since Alu elements are known to be co-localized with genes (33), the chromosomes rich in genes, such as chr19, are the ones with the highest percentage of CpG dinucleotides sequenced. The CpG dinucleotides sequenced in this study were widely distributed within each chromosome (Chr1 is shown as example in Supplementary Figure 4B). This demonstrated that, with a single primer, we could target thousands of CpG dinucleotides simultaneously in a genome-wide manner.
Alu methylation profile in human normal cerebellum
It has been well known that human Alu elements are heavily methylated in normal tissues, including brain (34). However, the genome-wide pattern of Alu methylation remains unknown due to the lack of an efficient methodology. To obtain a comprehensive understanding of the methylation data generated in this study, we assessed the methylation data at four levels: sequence reads, sequence read clusters, all methylation values obtained and the methylation of CpG sites (the clusters of methylation values). We found that 75.1% of all reads were found to be completely methylated and less than 2% of reads were completely unmethylated. After the reads were clustered, 43.5% of the 31 871 read clusters were completely methylated and 3.6% were completely unmethylated. In terms of individual methylation data points, 93.5% of the 1.2 million methylation data points generated were found to be methylated. Once these methylation values were assigned to individual CpG sites, 76.2% of 159 225 distinct CpG sites sequenced were always methylated and 7.4% were always unmethylated while the remainder showed a mixed methylation pattern.
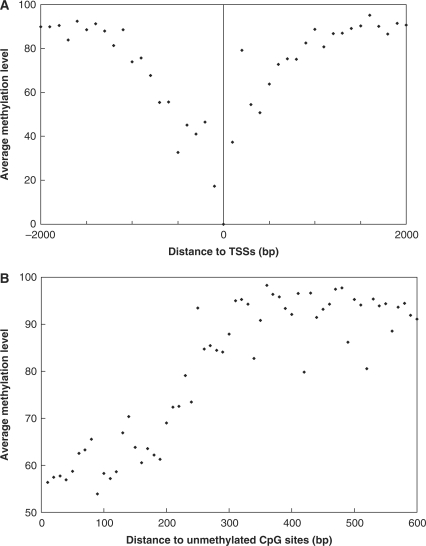
To further understand the underlying mechanisms controlling the methylation of the 5′-end and flanking sequences of Alu elements, we examined the methylation levels of CpG sites with respect to their genomic localization (Table 2). The methylation levels of CpG sites in introns, 3′ UTRs and in intergenic regions were ∼94%. The methylation levels of CpG sites in coding regions, in 5′UTRs and within 1 kb from transcription start sites (as indicated in the UCSC Genome Database) were 79.2%, 82.8% and 84.2%, respectively. We further examined the methylation statuses of 6477 CpG sites that mapped within ±2 kb of the TSSs of 1081 known genes. We found that methylation levels decreased the closer the CpG dinucleotides were to the TSSs, and that they in turn increased symmetrically the farther they were to the TSSs (Figure 3A). In addition, we observed that the unmethylated CpG sites identified in our study tended to be in clusters. We then examined the methylation levels of CpG sites adjacent to 11 740 unmethylated CpG dinucleotides. Significantly less methylation was observed within 200–300 bp from an unmethylated CpG. Furthermore, we found that such a correlation deteriorates rapidly after 300 bp (Figure 3B).
Table 2.
The average methylation level of CpG clusters with different genomic localization
| Number of methylation value | Number of distinct CpG dinucleotides | Methylation level | |
|---|---|---|---|
| Within 1 kb upstream from TSSsa | 11 185 | 1437 | 84.2 |
| 5′UTR | 586 | 135 | 82.8 |
| CDS | 857 | 187 | 79.2 |
| 3′UTR | 5082 | 687 | 93.3 |
| Intron | 443 817 | 58 718 | 93.5 |
| Intergenicb | 791 035 | 98 061 | 93.6 |
| SUM | 1 252 562 | 159 225 |
aTSSs denotes Transcription Start Sites.
bThe genomic regions within 1-kb upstream from TSSs were excluded from intergenic regions.
Figure 3.
The methylation profile of CpG sites sequenced. (A) CpG methylation near transcription start sites (TSSs). The X-axis represents the distance of CpG sites sequenced to the TSSs. The Y-axis represents the average methylation level. The average methylation levels were calculated for CpG sites adjacent to TSSs in 100-bp increments. (B) Spatial methylation correlation surrounding unmethylated CpG sites. The X-axis represents the distance to the nearest unmethylated CpG site. The Y-axis represents the average methylation level. The average methylation level of CpG dinucleotides were determined and plotted against the spatial distance, in 10-bp increments, to the nearest unmethylated CpG site.
A recent study tracking hypomethylated Alus identified 59 hypomethylated Alu elements in normal colonic mucosa (26). In that study, based on a simulation, 4104 Alu elements (2.6% of all Alu elements) with sequence AACCCGGG were predicted to be unmethylated or partially unmethylated. In our study, excluding the sequence reads without CpG dinucleotides, 1690 clusters derived from Alu elements (∼5% of all sequenced) were found to contain at least one completely unmethylated read. To further understand the methylation pattern of Alu repeats, the family distribution of unmethylated Alu elements was examined (Supplementary Table 3). Only 1.6% of very recent Alu elements (AluYa-h) were found to have completely unmethylated reads, while over 20% of the oldest Alu subfamily (AluJb) were found to have completely unmethylated reads. Such significant enrichment for unmethylated CpGs in old Alu subfamilies suggested that the oldest Alu elements (AluJb and AluJo), with fewer CpG sites in avereage and mutated RNA Polymerase III promoter regions, are more likely to be completely unmethylated.
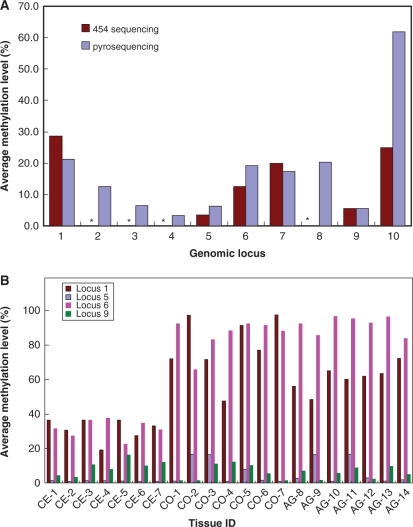
As an independent approach, pyrosequencing was used to verify the methylation data generated. Since most of Alu elements are heavily methylated, a successful verification for hypermethylated Alus could be achieved simply by chance. To maximize the power of validation, we focused on the Alu elements identified to be hypomethylated based on the 454 sequencing results. Ten different hypomethylated loci with various numbers of 454 sequence reads generated and different levels of methylation were selected (Supplementary Table 4). Of the ten loci examined, nine were confirmed to exhibit a low level of methylation, including two loci for which only two 454 sequence reads were available (Figure 4A). The remaining one locus showed a significant level of methylation (61.9). It is noteworthy, however, that there was a single 454 sequence read available for this locus. For all 54 CpG sites verified, the methylation levels varied from 1% to 72%, with an average of 15% (Supplementary Table 4). Considering that 93.5% of the methylation data points generated in this study was methylated, these pyrosequencing results strongly indicate that the 454 sequence data herein reported are reliable.
Figure 4.
Pyrosequencing validation and tissue specific Alu methylation (A) Pyrosequencing validation for high-throughput sequencing results. Asterisks represent the genomic regions identified to be completely unmethylated by high-throughput sequencing. The X-axis represents the distinct genomic loci selected. The Y-axis represents the average methylation level determined by 454 sequencing or by pyrosequencing. (B) Methylation profiles of four loci adjacent to promoters in cerebellum, cortex and fetal adrenal gland. The X-axis represents the distinct tissue sample IDs. CE, CO and AG represent the cerebellum, cortex and fetal adrenal gland tissues, respectively. Paired cerebellum and cortex were shown with the same number. The Y-axis represents the average methylation level determined by pyrosequencing. Similar methylation profiles were observed in duplicate experiments.
Tissue-specific Alu methylation
Recently, Ladd-Acosta et al. (35) examined the methylation profiles of 1505 CpG sites representing 807 genes in different brain regions. At least 20 CpG sites were shown to be differentially methylated in matched cerebellum and cortex. To determine whether Alu elements might also exhibit a brain-region-specific methylation pattern, we investigated the methylation profiles of a few hypomethylated Alu elements, which are adjacent to promoters, in paired cerebellum and cortex tissues from seven individuals and in fetal adrenal gland tissues from seven other individuals. Interestingly, the Alu elements demonstrated very distinct methylation profiles (Figure 4B). Locus 1 and Locus 6 seem to be tissue-specifically methylated. Locus 1 localizes in the intron of the Fem-1 homolog b gene at a distance of 3 kb from the promoter. The average methylation levels of Locus 1 are 31% in cerebellum, 79% in cortex and 61% in fetal adrenal gland. Locus 6 is in the 3′-UTR of the telomeric repeat binding factor 2 gene, ∼5 kb away from the promoter of the downstream gene, which codes for the transmembrane emp24 protein. The average methylation levels of Locus 6 are 32% in cerebellum, 86% in cortex and 92% in fetal adrenal gland. Both locus 5 and locus 9 were found to exhibit a low level of methylation in all samples examined. Locus 5 localizes in the intron of the A2ML1 gene at a distance of 4 kb from an alternative promoter, while locus 9 is located 300 bp from a CpG island that overlaps with the promoter of the transmembrane protein 183A gene. Based on microarray, SAGE and EST data provided by GeneCards (http://www.genecards.org/), all the genes adjacent to the unmethylated Alu elements herein investigated are expressed in cerebellum. Additional studies will be required to investigate the significance of the unmethylated status of these Alu elements to the expression of the nearby genes.
To visualize the methylation data reported in this study, a genome browser has been developed and installed at http://cmbteg.childrensmemorial.org/cgi-bin/gbrowse/btech. This genome browser integrates the methylation data that we generated with publicly available data on repeat elements, CpG islands, gene structure and other genomic features. A representative screen of this browser is shown in Supplementary Figure 5. In addition, we have collected several sets of genome-scale DNA-methylation data, including the methylation data for over 2500 genomic loci determined by Human Epigenome Consortium (20). More detailed descriptions of all datasets were provided on the website. Although the direct comparison of these datasets could be challenging due to the differences in tissue samples studied and techniques used, the genome browser may enable the researchers to examine the details of methylation profiles for specific genomic loci.
DISCUSSION
The determination of a complete DNA-methylation profile of the human genome, the methylome, will further our understanding of the human epigenome and of its functional significance. Bisulfite sequencing is the best approach to generate methylome data at a single base resolution (20,22–24). However, limited bisulfite sequencing data have been generated for the human genome to date. We have developed a bisulfite-based sequencing strategy targeting the methylome of Alu repeats and their flanking sequences. Using a single Alu specific primer, we simultaneously amplified thousands of Alu elements and obtained over 1.2 million methylation data points for 31 871 genomic regions, 94% of which were derived from the 5′ end regions and 5′ flanking sequences of Alu elements. The high sequence complexity and target specificity of the amplicons thus produced make them invaluable as epigenome representations, in studies aimed at uncovering epigenomic alterations associated with normal development and disease. Accordingly, it is noteworthy that we have successfully utilized this approach to monitor epigenomic alterations in non-aggressive and aggressive pediatric ependymomas, based on the methylation statuses of a common set of ∼100 000 CpGs (Xie, H. and Soares, M.B., data not shown).
In our study, only a small subset of a million Alu elements was sampled. Since the primer used was designed for Alu elements rich in CpG dinucleotides, this study is biased for younger Alu elements. To scale up the coverage, primers targeting different sets of Alu elements could be designed with a similar approach. On the other hand, this study covered a significant number of genomic loci, although the number of sequence reads generated was not enough to provide deep read coverage for all loci. Thus, the result for individual CpG sites or locus without deep read coverage should be taken cautiously. To increase the read coverage and reduce the sequencing cost, this approach may be modified for pair-end sequencing or to target hypomethylated Alu elements alone. In addition, the digestion with AluI enzyme releases various lengths of fragments and the shorter ones were amplified in much higher efficiency. To reduce such PCR bias, linear amplification should be explored in the future.
Despite the limitations discussed above, some interesting observations were made in our study. By examining the methylation patterns of 17 different LINE-1 elements, Phokaew et al. (36) demonstrated that the LINE1 methylation can be influenced differentially depending on their genomic locations. We found that the methylation statuses of CpG dinucleotides that occur within the 5′ most 80 bases of Alu elements can also be entirely different even though their sequences may be nearly identical, thus indicating that the methylation levels of Alu elements are not simply determined by their sequences either. The CpG sites sequenced in our study were 10% more methylated in intergenic, intronic and 3′ untranslated regions than in promoters, 5′-UTRs and coding regions. Although our dataset is very different from the data generated by the Human Epigenome Consortium (20), we also found that the methylation level is low in the regions close to transcription start sites and that it increases symmetrically with distance from TSSs. In addition, less methylation was observed within 200–300 bp of an unmethylated CpG and such co-methylation effect deteriorated with distance. With pyrosequencing, we were able to verify the methylation patterns of a small set of hypomethylated Alu elements. More interestingly, our result suggested that some Alu elements might be tissue specifically methylated.
Dynamic DNA methylation, particularly in repetitive elements, has been shown during normal development and disease progression (2,37–39). As mentioned previously, AluY/S elements were shown to be enriched at the junction of hypermethylated and hypomethylated genomic regions (18). Hence, Alu elements may serve as sensors for DNA-methylation changes. Therefore, it would be interesting to use the approach outlined in this article to study the methylation differences not only among different tissues but also within the same tissue at different stages of development, as well as during disease progression. This approach will enable monitoring of the methylation status for a specific subset of Alu elements and will contribute significantly to the understanding of the dynamics of DNA methylation. In addition, the loss of DNA methylation in the genome of cancers was found to be associated with genome instability. Accordingly, it is noteworthy that the sequence-based strategy herein described, while producing methylation data, can also reveal associated structural alterations, such as insertions and deletions. Additionally, the strategy outlined in this study could be adapted to study the epigenome of other model organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Everett/O'Connor Charitable Trust; Dr Ralph & Marian C. Falk Medical Research Trust; Gus Foundation; The Maeve McNicholas Memorial Foundation; Medical Research Institute Council. Funding for open access charge: Dr. Ralph & Marian C. Falk Medical Research Trust.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kochanek S, Renz D, Doerfler W. Transcriptional silencing of human Alu sequences and inhibition of protein binding in the box B regulatory elements by 5'-CG-3' methylation. FEBS Lett. 1995;360:115–120. doi: 10.1016/0014-5793(95)00068-k. [DOI] [PubMed] [Google Scholar]
- 2.Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000116. e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J. Biol. Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- 5.Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 6.Krichevsky S, Pawelec G, Gural A, Effros RB, Globerson A, Yehuda DB, Yehuda AB. Age related microsatellite instability in T cells from healthy individuals. Exp. Gerontol. 2004;39:507–515. doi: 10.1016/j.exger.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Cadieux B, Ching TT, Vandenberg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 8.Riggs AD, Jones PA. 5-methylcytosine, gene regulation, and cancer. Adv. Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 10.Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM, Kim D, Kang GH. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J. Pathol. 2007;211:269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 11.Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, Issa JP. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, San Jose-Eneriz E, Garate L, Cordeu L, Cervantes F, Prosper F, et al. Repetitive DNA hypomethylation in the advanced phase of chronic myeloid leukemia. Leuk. Res. 2008;32:487–490. doi: 10.1016/j.leukres.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 13.Miao VP, Singer MJ, Rountree MR, Selker EU. A targeted-replacement system for identification of signals for de novo methylation in Neurospora crassa. Mol. Cell Biol. 1994;14:7059–7067. doi: 10.1128/mcb.14.11.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 15.Selker EU. Gene silencing: repeats that count. Cell. 1999;97:157–160. doi: 10.1016/s0092-8674(00)80725-4. [DOI] [PubMed] [Google Scholar]
- 16.Allen E, Horvath S, Tong F, Kraft P, Spiteri E, Riggs AD, Marahrens Y. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl Acad. Sci. USA. 2003;100:9940–9945. doi: 10.1073/pnas.1737401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 18.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–237. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez J, Vives L, Jorda M, Morales C, Munoz M, Vendrell E, Peinado MA. Genome-wide tracking of unmethylated DNA Alu repeats in normal and cancer cells. Nucleic Acids Res. 2008;36:770–784. doi: 10.1093/nar/gkm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 29.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing J, Hedges DJ, Han K, Wang H, Cordaux R, Batzer MA. Alu element mutation spectra: molecular clocks and the effect of DNA methylation. J. Mol. Biol. 2004;344:675–682. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]
- 33.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 34.Gama-Sosa MA, Wang RY, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983;11:3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, Callinan PA, Fan JB, Potash JB, Feinberg AP. DNA methylation signatures within the human brain. Am. J. Hum. Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res. 2008;36:5704–5712. doi: 10.1093/nar/gkn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hellmann-Blumberg U, Hintz MF, Gatewood JM, Schmid CW. Developmental differences in methylation of human Alu repeats. Mol. Cell Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: the time is now. Crit. Rev. Oncol. Hematol. 2008;68:1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.