Abstract
Cytokines secreted by infiltrating immune cells during atherogenesis modulate vascular remodeling. One exemplary event is the antagonism between transformed growth factor (TGF-β) and interferon gamma (IFN-γ) on the transcriptional control of type I collagen gene (COL1A2). Previously we have reported that IFN-γ up-regulates regulatory factor for X-box B (RFXB) to repress collagen transcription while down-regulates the expression of RFXBSV, a splice variant of RFXB that blocks collagen repression in fibroblasts. Here we demonstrate that TGF-β abrogated COL1A2 repression by IFN-γ through altering the relative expression of RFXB and RFXBSV. Unlike RFXB, RFXBSV did not bind to the collagen promoter and competed with RFXB for the co-repressor histone deacetylase 2 (HDAC2), limiting HDAC2 recruitment to the collagen transcription start site as evidenced by chromatin immunoprecipitation assays. Over-expression of RFXB by lentiviral infection in HASMCs enhanced HDAC2 enlistment, promoted histone deacetylation surrounding the collagen site by IFN-γ, and blocked the TGF-β antagonism, a pattern reversed by RFXBSV infection. On the contrary, silencing of RFXB, but not both RFXB and RFXBSV, expression promoted the TGF-β antagonism. Thus, we have identified a novel mechanism whereby TGF-β antagonizes the IFN-γ repression of collagen transcription in HASMCs and as such provided new insights into antiatherogenic strategies.
INTRODUCTION
Atherosclerosis is a multi-factorial process highlighted by a panel of inter-connected pathophysiological events. The aggregation and activation of immune cells initiate the formation in the arteries of an atherosclerotic lesion (1). Later, medial smooth muscle cells (SMCs) encroach into the intima synthesizing and depositing extracellular matrix (ECM) proteins en route to reshape the vessel wall (2), which ultimately leads to the maturation of the atherosclerotic plaque (atheroma). As the vasculature undergoes active remodeling, infiltrating immune cells secret cytokines to challenge SMCs and alter the synthesis of ECM proteins. Over-production of ECM content thickens the vessel wall and constrains the lumen, which impedes the blood flow resulting in cardiac ischemia. Atheroma with reduced ECM content, on the other hand, is prone to acute rupture causing myocardiac infarction. Therefore, the interplay between various cytokines and SMCs leads to deregulated ECM synthesis and largely dictates the clinical complications associated with arteriosclerosis.
Collagen type I is the most enriched ECM component in the vasculature and is found in abundance in the plaque, the levels of which affect the vulnerability of the plaque and correlate with the mortality and morbidity of atherosclerosis (3,4). Collagen type I is a heterotrimer of two alpha1 chains (COL1A1) and one alpha2 chain (COL1A2), whose transcription is coordinately regulated by various humoral factors in the plaque, among which are interferon gamma (IFN-γ) and transforming growth factor (TGF-β). IFN-γ and TGF-β, both of which are primarily secreted by T lymphocytes and macrophages in the plaque, often exhibit opposite characteristics during atherogenesis (5,6). For instance, IFN-γ promotes T-cell activation and exacerbates inflammation, whereas TGF-β is known to suppress the expression of a range of pro-inflammatory molecules associated with atherogenesis (7–9). Interestingly, although both cytokines tend to inhibit the proliferation of vascular SMCs, it is evident that TGF-β strongly activates collagen transcription (10), whereas IFN-γ is a potent repressor of collagen synthesis (11). Thus, IFN-γ and TGF-β differentially modulate the fibrous content of the plaque, creating a converging point where the stability of atheroma is regulated. The mechanism underlying the antagonism between IFN-γ and TGF-β, however, is still a matter of controversy (12–16).
Transcription of collagen type I is also fine-tuned by a group of proteins that ‘write’ the epigenetic information, namely DNA and histone modifying enzymes (17,18). Our earlier investigations have revealed that in fibroblast cells, IFN-γ mediated repression of COL1A2 transcription is accompanied by a decrease in histone acetylation surrounding the transcription start site, occupied by the trimeric regulatory factor for X-box (RFX) complex consisting of RFX5, RFXB and RFXAP (19). The RFX complex helps recruit a histone deacetylases 2 (HDAC2)-containing co-repressor complex to the COL1A2 transcription start site (20). Here we report that TGF-β differentially regulates the expression of RFXB, which interacts with HDAC2 and binds to the COL1A2 promoter, and its splice variant RFXBSV, which interacts with HDAC2 but does not bind to the COL1A2 promoter. In doing, TGF-β interferes with RFX-dependent recruitment of HDAC2 to and restores histone acetylation around the collagen transcription start site. Thus, we have identified a novel mechanism whereby TGF-β antagonizes the repression of COL1A2 transcription by IFN-γ.
MATERIALS AND METHODS
Cell culture and treatment
Human aortic smooth muscle cells (HASMCs, Lonza) were maintained in SMBM with supplements supplied by the vendor. Human embryonic kidney cells (293FT, Invitrogen) were grown in DMEM (Invitrogen) supplemented with 10% FBS (Hyclone). In certain experiments, cells were treated with cytokines. Cells were switched to 0.4% media 16–24 h prior to treatment. IFN-γ (100 U/ml, Roche) and/or TGF-β (2 ng/ml, R&D) were added and maintained in 0.4% media for indicated time points.
Plasmids, transfection and viral infection
The col1a2-luciferase constructs pH20 (−220/+54) and pGL3-COL-LUC (−357/+55) as well as the COL1A1-luciferase construct (−331/+114) were as previously described (21–23). The FLAG-tagged expression constructs pcDNA3/FLAG-RFXB and pcDNA3/FLAG-RFX5 were gifts from Dr Jenny Ting (24). pcDNA3/FLAG-HDAC2 was kindly provided by Dr Edward Seto (25). FLAG-RFXBSV was constructed by amplifying the RFXBSV cDNA from pRFXBΔ5 (26) and re-ligating the insert into pcDNA3/FLAG. The lentiviral constructs pLenti6/V5-RFXBSV and pLenti6/V5-RFXB were constructed similarly. All plasmids were verified by restriction digestion and direct sequencing. Transfections were performed with Lipofectamine 2000 (Invitrogen). Luciferase activities were measured using a luciferase reporter assay system (Promega). Experiments were routinely performed in triplicate wells and repeated three times. Lentivirus carrying the expression constructs pLenti6/V5-RFXB and pLenti6/V5-RFXBSV was generated in 293FT cells as previously described (27). Cells were harvested 48 h after infection.
Whole-cell protein extraction and co-immunoprecipitation
Whole-cell lysates were obtained as previously described (20). FLAG-conjugated beads (M2, Sigma) were added to and incubated with 500 μg protein lysates overnight. Precipitated immune complex was eluted with 3X FLAG peptide (Sigma). Western blot analyses were performed with monoclonal anti-FLAG (Sigma), polyclonal anti-RFX5 (Rockland), polyclonal anti-HDAC2 (Santa Cruz), monoclonal anti-β-actin (Sigma), monoclonal anti-V5 (Sigma) and polyclonal anti-collagen type I (Rockland) antibodies.
Nuclear protein extraction and DNA affinity pull-down
Nuclear protein extracts were obtained essentially as previously described (20). A piece of double-stranded collagen DNA oligos was used as the probe (COL1A2 − 25/+30, Genbank accession number AF004877) (28). In vitro DNA affinity pull-down assays were performed as previously described (20). Briefly, pre-cleared nuclear proteins were incubated with biotin-labeled DNA probe. DNA–protein complex formed was then captured by incubating with the streptavidin beads on a shaking platform. Ternary complex (biotin-labeled DNA–protein–streptavidin) was washed extensively and eluted with 1× SDS electrophoresis sample buffer and analyzed by Western blot analysis. Competition of binding was performed with specific as well as non-specific DNA oligos as previously described (20).
RNA extraction and real-time PCR
RNA was extracted using an RNeasy RNA isolation kit (Qiagen). Reverse transcriptase reactions were performed using a SuperScript First-strand synthesis system (Invitrogen). Real-time PCR reactions were performed on an ABI Prism 7500 (Applied Biosystems). The oligonucleotide forward and reverse PCR primers and fluorescent probes for RFXB, RFXBSV, collagen type I and HLA-DRα were as previously described (20). SM22α primer/probe set was purchased from Applied Biosystems.
Chromatin immunoprecipitation (ChIP)
ChIP analyses were performed essentially as described before (19). Aliquots of cell lysates containing 200 μg of protein were used for each immunoprecipitation reaction with anti-RFX5 (Rockland), anti-HDAC2 (Santa Cruz), anti-acetylated H3 (Millipore), anti-V5 (Sigma) and anti-FLAG (Sigma) antibodies. Precipitated DNA was dissolved in 50 μl deionized distilled water and 10 μl was used for each real-time PCR reaction. The primers surrounding the collagen start site for real time PCR have been described previously (19). Serial dilutions of genomic DNA extracted from normal cells were included with ChIP samples as standards to determine the amount of DNA being precipitated by a particular antibody. Ten percent of the starting material was also included as the input. Data were normalized to input and expressed as pg of DNA precipitated per ng of input DNA.
Short hairpin RNA (shRNA)-mediated silencing
The procedure for the construction of RFXB shRNA plasmid (designated shB herein) and RFXB/RFXBSV shRNA plasmid (designated shB/SV herein) was described before (27). Briefly, designing of reverse primers (Supplementary Data Table I) was facilitated by the Cold Spring Harbor RNAi oligo retriever program (katahdin.cshl.org:9331/RNAi/html/rnai.html). PCR products were purified and ligated into pLenti6-V5-TOPO vector using a Viralpower directional cloning kit (Invitrogen). Positive clones were verified by direct sequencing. Silencing was performed by infecting HASMCs with different viral plasmids (shB or shB/SV) as described earlier (27).
Statistical analysis
One-way ANOVA with post-hoc Scheffe analyses were performed using an SPSS package. P-values <0.05 were considered statistically significant.
RESULTS
TGF-β blocks the repression of COL1A2 transcription by IFN-γ while altering the relative expression of RFXB/RFXBSV in HASMCs
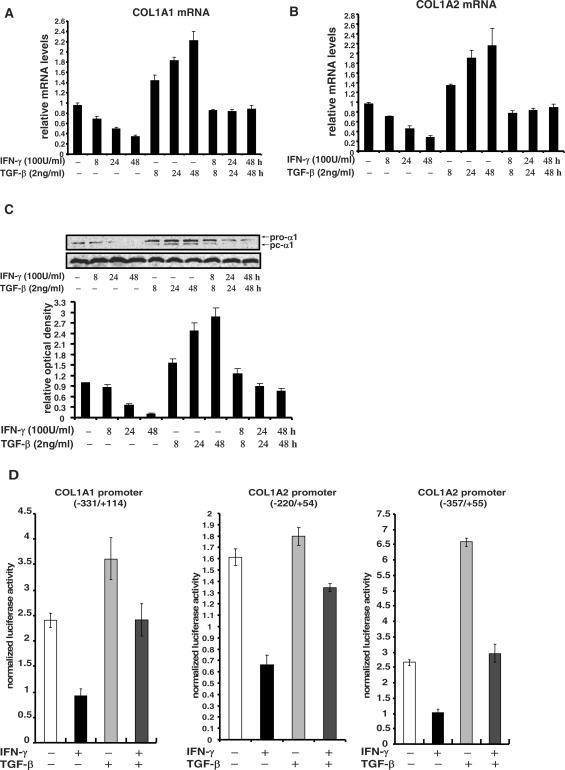
We first examined the expression of collagen type I in human primary aortic smooth muscle cells (HASMCs) with IFN-γ and/or TGF-β treatment. IFN-γ potently down-regulated both mRNA levels (Figure 1A and B) and protein levels (Figure 1C) of collagen type I, which was completely abrogated by TGF-β treatment, in line with findings observed in other cell types (15,29,30). Antagonism between IFN-γ and TGF-β on collagen synthesis primarily occurred at the transcriptional level as judged by promoter-reporter assays. IFN-γ repressed the activities of two COL1A2 promoter-reporter constructs as well as one COL1A1 construct in HASMCs (Figure 1D). Although TGF-β up-regulated the activities of three promoter constructs with various magnitude, it abolished the repression by IFN-γ similarly and comparable to changes in message as well as protein levels of type I collagen.
Figure 1.
TGF-β blocks the repression of COL1A2 transcription by IFN-γ in HASMCs. (A, B) HASMCs were treated with IFN-γ and/or TGF-β for various time points as indicated. Collagen type I message levels were measured by real-time PCR. Data are expressed as relative RNA levels compared to control levels, normalized to 18S RNA and presented as average ± SD. (C) HASMCs were treated with IFN-γ and/or TGF-β, and measured for collagen protein levels by Western blot. A representative picture is shown. Normalized collagen levels were compared to the control group and expressed as relative densitometric values. Shown in the graph are data collected from three independent experiments and expressed as average ± SD. (D) HASMCs were transfected with different collagen type I promoter constructs along with GFP for normalization. Cells were treated with IFN-γ and/or TGF-β 24 h after transfection. Luciferase activities were assayed 48 h after transfection, normalized by both protein concentration and GFP fluorescence, and presented as average ± SD.
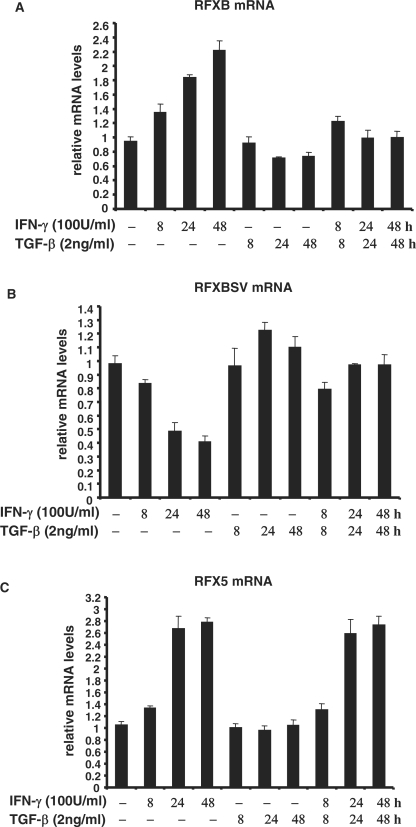
Previous investigations indicate that IFN-γ promotes RFXB expression while suppressing RFXBSV expression in HEK293 cells (31) and lung fibroblast cells (19). In SMCs, IFN-γ augmented RFXB expression and reduced RFXBSV expression simultaneously in a time-dependent manner, in agreement with previous reports (Figure 2A and B). TGF-β alone marginally altered the expression of both RFXB and RFXBSV without statistical significance. However, TGF-β abolished the induction of RFXB expression and repression of RFXBSV expression by IFN-γ, suggesting that differential expression of RFXB isoforms may play a role in the antagonism between IFN-γ and TGF-β. As a control, message levels of RFX5, another component of the RFX complex, were stimulated by IFN-γ but unopposed by TGF-β under the same condition (Figure 2C).
Figure 2.
TGF-β alters the relative expression of RFXB splice variants in response to IFN-γ in HASMCs. HASMCs were treated with IFN-γ and/or TGF-β. Message levels of RFXB (A), RFXBSV (B) and RFX5 (C) were measured by real-time PCR.
RFXB alleviates the antagonism of IFN-γ mediated collagen transcription repression by TGF-β
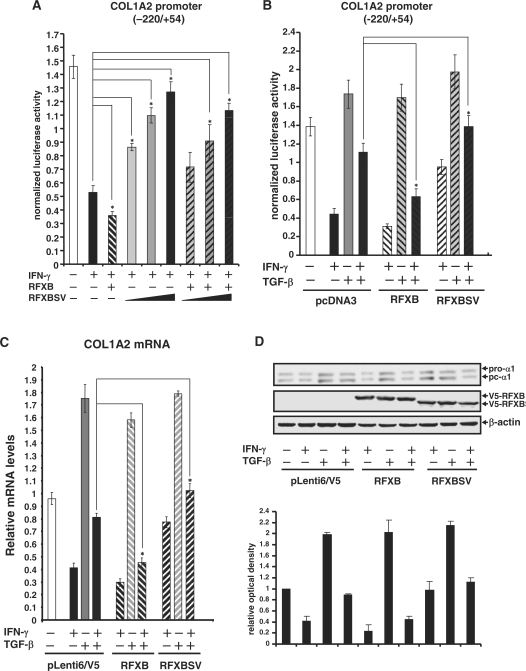
Since we observed an alteration in the relative expression of RFXB versus RFXBSV during the TGF-β antagonism of IFN-γ mediated repression of collagen synthesis, the individual roles of RFXB and RFXBSV were probed in HASMCs. First, consonant with our previous report, over-expression of RFXB enhanced IFN-γ repression of COL1A2 promoter activity. Increasing concentrations of RFXBSV, however, abrogated the IFN-γ repression and antagonized the enhancement by RFXB in SMCs (Figure 3A). Next, the antagonism between TGF-β and IFN-γ on COL1A2 transcription was examined in the presence of ectopically expressed RFXB or RFXBSV. When RFXB was over-expressed, the reversion of IFN-γ repression by TGF-β was substantially inhibited (Figure 3B). On the other hand, RFXBSV significantly promoted the TGF-β antagonism when it was present in excess. Finally, SMCs were infected with viral particles carrying either RFXB or RFXBSV and endogenous COL1A2 message and protein levels were examined. RFXB restored the repression of endogenous collagen expression by IFN-γ in the presence of TGF-β (Figure 3C and D). RFXBSV, however, not only suppressed the repression of collagen synthesis by IFN-γ, but further reinforced the antagonism by TGF-β. To rule out the possibility that these observations were artifact caused by RFXB/RFXBSV over-expression, the transcription of HLA-DRα, which faces the same competition between IFN-γ and TGF-β 32, was examined by promoter-luciferase and quantitative PCR assays. Unlike collagen type I gene, the antagonism between IFN-γ and TGF-β on HLA-DRα transcription was not affected by infection of either RFXB or RFXBSV (Supplementary Data Figure 1). Together, these data suggest that RFXB and RXBSV had opposite roles in the regulation of collagen synthesis. Moreover, relative expression of RFXB and RFXBSV also determined the balance of antagonism between IFN-γ and TGF-β.
Figure 3.
RFXB alleviates the antagonism of IFN-γ mediated collagen transcription repression by TGF-β. (A) A collagen type I promoter construct (pH 20) was co-transfected with expression constructs for RFXB and/or RFXBSV into HASMCs. IFN-γ was added 24 h after transfection and incubated with cells for additional 24 h. Luciferase activities were normalized by both protein concentration and GFP fluorescence, and presented as average ± SD. (B) A collagen type I promoter construct (pH 20) was co-transfected with expression constructs for RFXB and/or RFXBSV into HASMCs as indicated. Cells were treated with IFN-γ and/or TGF-β for 24 h. Data were obtained and analyzed as described under (A). (C, D) HASMCs were infected with lentiviral stocks carrying an empty vector (pLenti6/V5), RFXB or RFXBSV as indicated. Cells were treated with IFN-γ and/or TGF-β 24 h after infection. RNA or protein was extracted 24 h after treatment. Message (C) and protein (D) levels of collagen type I were assessed by real-time PCR and Western blot, respectively. Data were processed and presented as described in Figure 1.
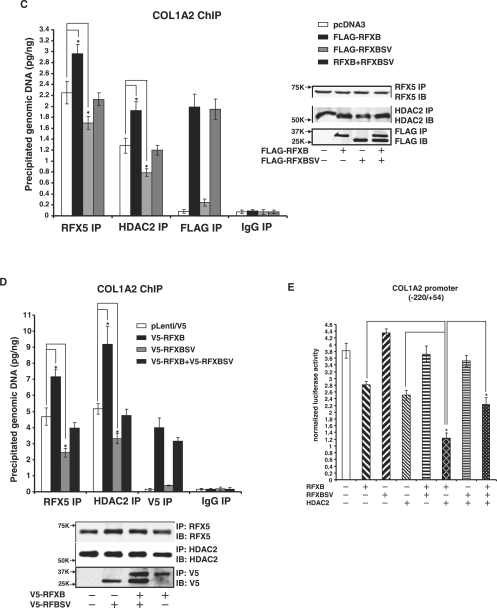
RFXB and RFXBSV compete for the binding of HDAC2 thus limiting its recruitment to the collagen transcription start site
Our earlier findings suggest that RFXB forms a complex with RFX5 and recruits HDAC2 to the collagen promoter, which in turn deacetylates core histones and represses transcription of collagen type I in response to IFN-γ (19,20). Therefore, we examined whether RXBSV existed in a similar complex with RFX5 and HDAC2.
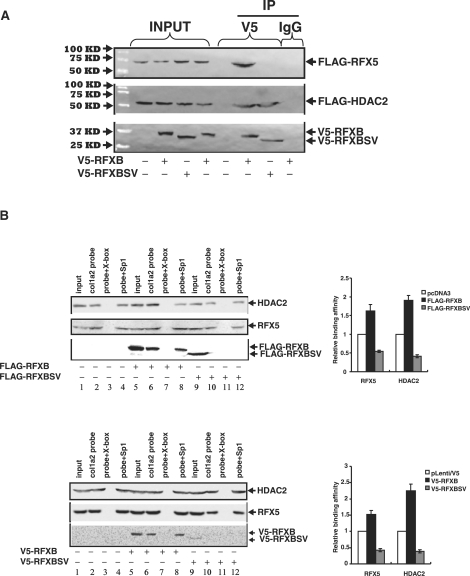
First, V5-tagged RFXB or RFXBSV was co-transfected with FLAG-tagged RFX5 or HDAC2 into HEK293 cells followed by immunoprecipitation with an anti-V5 antibody to examine the interactions. As shown in Figure 4A, both RFXB and RFXBSV came down along with HDAC2 (bottom and middle panels). Interestingly, unlike RFXB, RFXBSV failed to interact with RFX5 (top panel).
Figure 4.
RFXB and RFXBSV compete for the binding of HDAC2, thus limiting its recruitment to the collagen transcription start site. (A) 293FT cells were transfected with V5-tagged RFXB or V5-tagged RFXBSV along with either FLAG-tagged RFX5 or FLAG-tagged HDAC2. Immunoprecipitations were performed as described under ‘Materials and Methods’. Ten percent of the starting material was also loaded as input. (B) 293FT cells were transfected with FLAG-tagged RFXB, FLAG-tagged RFXBSV or an empty vector (top panel). Alternatively, HASMCs were infected with pLenti/V5-RFXB, pLenti/V5-RFXBSV or an empty vector (bottom panel). DNA affinity pull-down assays were performed as described. Binding of RFX5 and HDAC2 to the collagen probe in the presence of RFXB and RFXBSV was normalized to the control group and expressed as relative binding affinity. Shown in the graph are data collected from three independent experiments and expressed as average ± SD. (C, D) 293FT cells were transfected with FLAG-tagged RFXB and/or FLAG-tagged RFXBSV (C). Alternatively, HASMCs were infected with pLenti/V5-RFXB and/or pLenti/V5-RFXBSV (D). Chromatin immunoprecipitation assays were performed with anti-RFX5, anti-HDAC2, anti-FLAG, anti-V5 or pre-immune IgG as indicated. Precipitated DNA was amplified by real-time PCR with primers spanning the COL1A2 transcription start site (*P < 0.05). (inset) 10% of the IP samples were separated by SDS-PAGE gels and examined for precipitation efficiency with Western blot. (E) A collagen type I promoter construct (pH 20) was co-transfected with expression constructs for RFXB, RFXBSV or HDAC2 into HASMCs. Luciferase activities were normalized by both protein concentration and GFP fluorescence, and presented as average ± SD (*P < 0.05).
Next, in order to determine whether RFXB and RFXBSV could bind to COL1A2 transcription start site and recruit HDAC2, nuclear proteins extracted from HEK293 cells transfected with either FLAG-tagged RFXB or RFXBSV were incubated with a biotin-labeled collagen probe. Both RFX5 and HDAC2 were present on the collagen site without ectopically expressed RFXB or RFXBSV (Figure 4B, lane 2). The bindings were eliminated by a specific competitor for RFX5 (X-box, lane 3) but not by a non-specific competitor (Sp1, lane 4), suggesting that recruitment of HDAC2 relied on RFX5. Over-expressed exogenous RFXB was able to bind to the collagen site (lane 6) and slightly increased the bindings of both RFX5 and HDAC2 (compare lane 6 to lane 2). On the other hand, RFXBSV was not associated with the collagen probe. Moreover, over-expression of RFXBSV down-regulated the binding of RFX5 and HDAC2 (compare lane 10 to lane 2). Similar results were also observed in physiologically relevant HASMCs infected with viral clones carrying either V5-RFXB or V5-RFXBSV (Figure 4B, bottom panel).
Next, in order to evaluate the binding capacity of RFXB and RFXBSV as well as their ability to promote the recruitment of HDAC2 to the endogenous COL1A2 start site, ChIP analyses were performed. Consistent with the in vitro DNA affinity pull-down assay, RFXB but not RFXBSV was bound to the endogenous collagen promoter (Figure 4C). More importantly, more HDAC2 (1.5X) and RFX5 (1.3X) were found to be associated with the same site when there was an over-expression of RFXB. On the contrary, when RFXBSV was over-expressed, there was a significant decrease in the binding of HDAC2 and RFX5. In addition, stimulation of recruitment of HDAC2 as well as RFX5 by RFXB was blunted by RFXBSV. Similarly, over-expression of RFXBSV by viral infection in HASMCs dampened the recruitment of RFX5 and HDAC2 to the collagen promoter, whereas over-expression of RFXB promoted the binding of RFX5 and HDAC2, and relieved the antagonism by RFXBSV (Figure 4D). To verify whether altered HDAC2 recruitment by RFXB/RFXBSV was specific to the collagen promoter, the binding of HDAC2 to the promoter of SM22α, an SMC marker gene whose transcription is regulated by HDAC2 (33), was examined. Indeed, HDAC2 was associated with the SM22α promoter in ChIP assays (Supplementary Data Figure 2). Neither RFXB nor RFXBSV over-expression impacted the binding of HDAC2 or SM22α mRNA levels.
Finally, we examined the functional relevance of the interactions among RFXB, RFXBSV and HDAC2 on collagen transcription. As shown in Figure 4E, RFXB and HDAC2 repressed collagen promoter activity both singularly and synergistically when added together, whereas RFXBSV by itself activated the promoter slightly. More significantly, RFXBSV counteracted the repression of the collagen promoter activity by RFXB and HDAC2. Combined, these data suggest that RFXBSV interacted with HDAC2 but not with RFX5, thus competing for the cellular pool of HDAC2 that could be recruited to the collagen promoter by the RFX5/RFXB complex to repress collagen transcription.
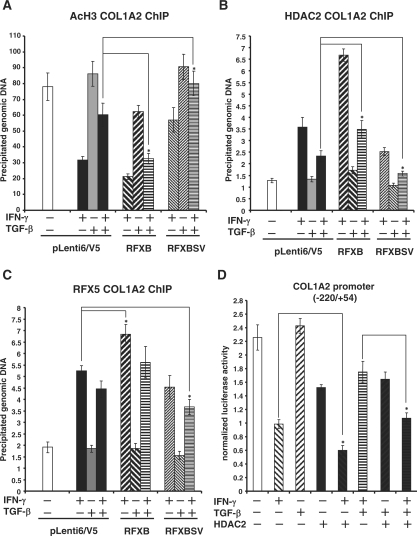
TGF-β blocks the recruitment of HDAC2 by IFN-γ and restores acetylation of core histones at the collagen transcription start site
Previously we have reported that IFN-γ represses collagen transcription by down-regulating levels of histone acetylation around the COL1A2 transcription start site through the recruitment of HDAC2 in lung fibroblast cells (19,20). Since we observed that the association of HDAC2 with the collagen promoter was regulated by the relative expression of RFXB and RFXBSV, we decided to assess the possibility that TGF-β opposed the IFN-γ repression by blocking the recruitment of HDAC2, thus up-regulating histone acetylation. As expected, IFN-γ treatment led to a decrease in the acetylation of core histones (Figure 5A and data not shown) accompanied by a concurrent increase of HDAC2 recruitment (Figure 5B) in HASMCs. TGF-β alone did not have any significant impact on the histone acetylation or HDAC2 binding. Nor did it affect the histone acetylation by a large margin, suggesting that TGF-β may activate collagen synthesis through mechanisms other than blocking HDAC2-mediated histone deacetylation. It did, however, ameliorate the inhibition of histone acetylation and prevent the induction of HDAC2 enlistment by IFN-γIntroduction into HASMCs exogenous RFXB led to further decrease in histone acetylation and increased HDAC2 recruitment by IFN-γ. It also completely blocked the TGF-β induced antagonism of HDAC2 binding and histone deacetylation (Figure 5A and B). RFXBSV, on the other hand, disrupted the change in histone acetylation and HDAC2 binding initiated by IFN-γ and substantiated the TGF-β antagonism. As a control, we also examined the binding of RFX5 to the collagen transcription start site under the same conditions. IFN-γ markedly increased the level of RFX5 associated with the collagen site, which was only slightly decreased by TGF-β. RFXB infection led to additional RFX5 binding, whereas RFXBSV infection resulted in a decrease in RFX5 binding, both of which relied on the presence of IFN-γ. Nonetheless, the changes in RFX5 binding were much milder than those of either HDAC2 binding or histone acetylation. More importantly, when HDAC2 was over-expressed in HASMCs, the TGF-β antagonism of collagen transcriptional repression by IFN-γ was greatly alleviated (Figure 5D). On the contrary, knock-down of HDAC2 expression significantly enhanced the antagonism of TGF-β on the down-regulation of histone acetylation around COL1A2 transcription start site as well as COL1A2 repression by IFN-γ (Supplementary Data Figure 3). Collectively, these data indicate that the antagonism between IFN-γ and TGF-β on collagen transcription manifests, at the chromatin level, as alterations in HDAC2 binding and histone acetylation around the collagen transcription start site, which are mediated by the relative expression of RFXB and RFXBSV.
Figure 5.
TGF-β blocks the recruitment of HDAC2 and restores the acetylation of core histones at the collagen transcription start site. (A–C) HASMCs were infected with V5-tagged RFXB or V5-tagged RFXBSV or an empty vector followed by treatment with IFN- γ and/or TGF-β as indicated. Chromatin immunoprecipitation assays were performed as described under Materials and Methods with either anti-acetylated histone H3 (A), or anti-HDAC2 (B), or anti-RFX5 (C) antibodies as indicated. Precipitated DNA was amplified by real-time PCR with primers spanning the COL1A2 transcription start site (*P < 0.05). (D) A collagen type I promoter construct (pH 20) was co-transfected with expression constructs for HDAC2 into HASMCs as indicated. Cells were treated with IFN-γ and/or TGF-β for 24 h. Luciferase activities were normalized by both protein concentration and GFP fluorescence, and presented as average ± SD (*P < 0.05).
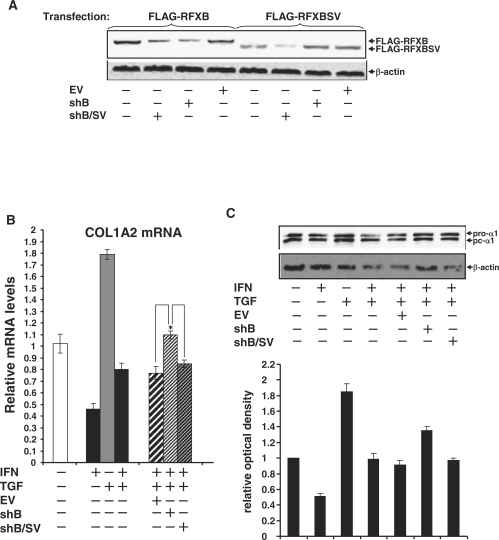
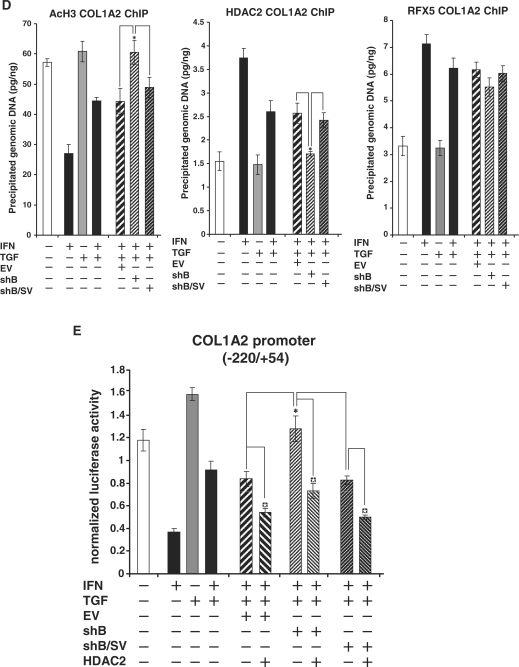
Silencing of RFXB expression favors the TGF-β antagonism of IFN-γ induced collagen repression
To further probe the role of RFXB and its splice variant in mediating the antagonism between IFN-γ and TGF-β, the following strategy was employed. Because RFXB and RFXBSV share substantial sequence identity, our attempt to silence them individually was not successful. Through extensive screening, we obtained two plasmids expressing shRNA: one targets RFXB only (shB), whereas the other targets both RFXB and RFXBSV (shB/SV). Specificity and efficiency of these two plasmids were verified by western blot (Figure 6A) and quantitative PCR (Supplementary Data Figure 4). Infection of HASMCs with viral stocks carrying shB led to further antagonism of IFN-γ induced collagen repression by TGF-β (Figure 6B and C), indicating that the balance between IFN-γ and TGF-β relies on the relative expression of RFXB and RFXBSV. Infection with shRNA targeting both RFXB and RFXBSV, however, had no such effect, suggesting that a new balance could be established between IFN-γ and TGF-β once both RFXB and RFXBSV were removed. Similar changes were observed on the collagen chromatin where RFXB silencing resulted in additional decrease in HDAC2 binding and increase in histone H3 acetylation compared to control or silencing of both RFXB and RFXBSV (Figure 6D). Finally, enhanced TGF-β antagonism, brought by RFXB silencing, of IFN-γ induced repression of collagen promoter was alleviated by the presence of HDAC2 (Figure 6E), again indicating the competition converges on HDAC2 that is available inside the cell.
Figure 6.
RFXB silencing favors the TGF-β antagonism of IFN-γ induced collagen repression. (A) HEK293 cells were transfected with FLAG-tagged RFXB or RFXBSV plasmids in the presence of shB, shB/SV or an empty vector (EV). Silencing efficiency was measured by Western blot. (B, C, D) HASMCs were infected with viral stocks carrying shB, shB/SV or an EV followed by treatment with IFN-γ and/or TGF-β as indicated. Message (B) and protein (C) levels of collagen type I were assessed by real-time PCR and Western blot, respectively as described in Figure 1. ChIP assays (D) with anti-AcH3, -HDAC2 and –RFX5 antibodies were performed as described in Figure 5. (E) A collagen type I promoter construct (pH 20) was co-transfected with expression constructs for HDAC2 into HASMCs along with shB, shB/SV or an EV as indicated, followed by treatment with IFN-γ and/or TGF-β. Luciferase activities were normalized by both protein concentration and GFP fluorescence, and presented as average ± SD (*P < 0.05).
DISCUSSION
It has long been documented that collagenous content in atheroma contributes to the overall susceptibility of atherosclerosis, highlighting the importance of regulation of collagen levels during atherogenesis (34). Here we examined the possible mechanism underlying the antagonism between IFN-γ and TGF-β on the transcriptional regulation of collagen type I (COL1A2) in human aortic smooth muscle cells. Our data suggest that TGF-β interferes with the differential expression of RFXB and its splice variant RFXBSV induced by IFN-γ, thus abrogating HDAC2-dependent histone deacetylation surrounding the collagen transcription start site.
IFN-γ and TGF-β are two major cytokines found in the atherosclerotic plaque that modulate the expression of collagen. Whereas IFN-γ suppresses collagen production and renders the plaque prone to rupture, TGF-β stimulates collagen synthesis and stabilizes the plaque (35). The mechanism underneath the antagonism between IFN-γ and TGF-β on collagen production has been tackled on by several independent groups under different circumstances, mostly in fibroblast cells to study fibrosis. For instance, it has been proposed that IFN-γ-induced Stat-1 sequesters an essential transcription activator, either AP-1 (14) or CBP/p300 (15), that is required for TGF-β dependent transcriptional activation of collagen type I. Alternatively, YB-1, another protein up-regulated by IFN-γ, either competes for the binding of CBP/p300 (12) or stimulates the expression of the inhibitory TGF-β mediator Smad7 (13), thus dampening the fibrogenic effect of TGF-β Another possible scenario involves direct inhibition of TGF-β synthesis by IFN-γ in human primary skin fibroblast cells (16). Notably, these studies all concentrate on the more distal promoter region where TGF-β has a much stronger response of activation (presumably through stimulating Smad binding). It is evident, however, that even on shorter promoter constructs, where TGF-β only marginally activates collagen transcription, TGF-β is still capable of blanking the repression by IFN-γ (Figure 1D, compare the three collagen promoter constructs of different sizes), indicating that at least part of the antagonism may be mediated by factors that bind to the proximal and/or start site of the collagen promoter. Indeed, TGF-β blocks the up-regulation of RFXB (Figure 2A), which binds to the collagen transcription start site and recruits HDAC2 when complexed with RFX5 and RFXAP to mediate the IFN-γ effect (19,20) Furthermore, TGF-β also restores the expression of RFXBSV (Figure 2B), a naturally occurring splice variant of RFXB (26). Thus, TGF-β antagonizes the repression by IFN-γ through manipulating the molecular events surrounding the COL1A2 transcription start site.
Previously, we have demonstrated that RFX5 interacts with HDAC2 and enlists it to the collagen transcription start site in response to IFN-γ (20). Our new data summarized here suggest that both RFXB and RFXBSV are associated with HDAC2 (Figure 4A). RFXBSV, however, does not interact with RFX5 (Figure 4A) and fails to bind to the COL1A2 transcription start site (Figure 4B–D). Furthermore, in the presence of excessive RFXBSV the binding of HDAC2 as well as RFX5 to the collagen site is hindered (Figure 4B–D). Thus, it appears that there are at least two reservoirs of HDAC2 complex inside the cell. One contains RFXB that binds to the collagen site to promote histone deacetylation and hence transcriptional repression by IFN-γ (Figures 5 and 6). The other contains RFXBSV that ‘stays clear’ the collagen site and impedes the formation and/or promoter recruitment of the alternative complex, abrogating the repression of collagen transcription by IFN-γ (Figures 5 and 6).
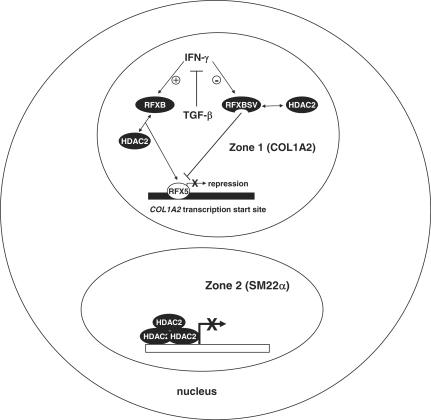
One major pitfall with the current model of HDAC2-squelching presented here (Figure 7) is how the ratio of RFXB versus RFXBSV, by itself, could effectively control the nuclear pool of HDAC2, a common co-factor with seemingly unlimited availability. This discrepancy can be reconciled by several possible explanations. Type I collagen gene has been mapped to discrete sites in the nucleus by fluorescent immuno-in-situ hybridization (FISH) (36). Indeed, it has been demonstrated that individual transcriptional events do not occur at random sub-nuclear compartments. Instead, actively transcribed genes are found at various highly organized foci in the nucleus (37). Presumably, RFXBSV and RFXB (and a portion of all the available HDAC2 in the nucleus) are confined to a specific ‘zone’, where collagen transcription is taking place. Therefore, within the speckle, HDAC2 availability becomes a rate-limiting factor that can be modulated by the differential expression of RFXB and RFXBSV. Alternatively, since recruitment of HDAC2 is only observed on the collagen promoter, but not on the SM22α promoter, we propose that the ability of HDAC2 to be sequestered is reliant on its affinity for different promoters. It is possible that HDAC2 binds with higher affinity to some promoters (e.g. SM22α, likely through stronger protein–protein interactions with SM22α-specific transcription factors) than to others (e.g. COL1A2), rendering it less susceptible to squelching by increasing levels of RFXBSV. Yet a third possibility exists that the impact exerted by the decrease in the recruitment of HDAC2 to the collagen promoter is multifold. In addition to affecting the chromatin environment surrounding the COL1A2 transcription start site, HDAC2 could fine-tune the post-translational modifications (PTMs) of key protein(s) involved the repression of collagen transcription. In fact, our previous publication suggests that the acetylation level of RFX5 is regulated by HDAC2, which directly contributes to the COL1A2 repression (3) (It also needs to be pointed out that target proteins for HDAC2-mediated deacetylation that are involved in the COL1A2 repression may not be limited to RFX5). Deacetylatoin of RFX5 by HDAC2, in turn, may result in an increase of HDAC2 recruitment via enhanced protein–protein interactions. Consequently, HDAC2 may facilitate the formation of a multi-protein repression complex on the collagen promoter, by modifying the overall chromatin structure and acetylation status of individual transcription factors, fulfilling its role in the IFN-γ induced repression of COL1A2 transcription. Thus, although RFXBSV-mediated down-regulation of HDAC2 recruitment may be inadequate by itself to induce significant changes in collagen transcripion, later events (e.g. up-regulation of RFX5 acetylation and release of HDAC2 from the COL1A2 promoter due to reduced affinity) nonetheless could operate in a self-sustainable manner to amplify the original signal such that de-repression of COL1A2 transcription can be achieved. Of note, these scenarios are not necessarily exclusive. For instance, the distinct affinity of HDAC2 for different promoters may well determine its sub-nuclear localizations and ultimately, its availability. Further investigations are warranted to explore these possibilities.
Figure 7.
A schematic representation of proposed model for RFXB variants mediated antagonism between IFN-γ and TGF-β in collagen type I transcription. IFN-γ stimulates the production of RFXB, which interacts with RFX5 and recruits HDAC2 to the COL1A2 start site (in a specialized sub-nuclear ‘zone’) to repress transcription. Meanwhile, IFN-γ down-regulates the levels of RFXBSV, which does not interact with RFX5 and fails to bind to the COL1A2 start site, thus squelching HDAC2 away and alleviating the repression. TGF-β antagonizes IFN-γ induced COL1A2 repression by reshuffling the balance between RFXB and RFXBSV through differential regulation of their expression. In a separate ‘zone’ where SM22α gene is located, HDAC2 binds to the promoter more strongly and is immune from the squelching by RFXBSV.
In conclusion, we have identified here a novel mechanism whereby TGF-β antagonizes the transcriptional repression of collagen type I gene (COL1A2) by IFN-γ in human aortic smooth muscle cells (Figure 7). Since TGF-β is also known to attenuate the up-regulation of MHC II expression and antigen-dependent T-cell activation by IFN-γ (38–40), it is possible that a similar mechanism involving a different set of co-factors (instead of HDACs) applies. Intriguingly, a recent study by Cunha et al. reports that alternative splicing of the human ANK2 gene, closely related to RFXB, is implicated in cardiac function (41). Thus, post-transcriptional processing could be a shared feature among ankyrin proteins, alluding to the importance of this regulatory dimension in maintaining the physiological integrity of the cardiovascular system. Therefore, modulation of alternative splicing and differential expression of RFXB isoforms may present as a potential target for therapeutics against atherosclerosis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (30800436); Jiangsu Provincial Natural Science Foundation (BK2008443); NJMU Natural (all to Y.X.). X.Y.W. was supported by a research grant from NJMU (08NMUM004). Funding for open access charge: 30800436 and BK2008443.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors are thankful to Nanjing Medical University for providing a start-up fund.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat. Rev. Immunol. 2006;6:508–519. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- 2.Katsuda S, Kaji T. Atherosclerosis and extracellular matrix. J. Atheroscler. Thromb. 2003;10:267–274. doi: 10.5551/jat.10.267. [DOI] [PubMed] [Google Scholar]
- 3.Libby P. Changing concepts of atherogenesis. J. Intern. Med. 2000;247:349–358. doi: 10.1046/j.1365-2796.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- 4.Rekhter MD, Zhang K, Narayanan AS, Phan S, Schork MA, Gordon D. Type I collagen gene expression in human atherosclerosis. Localization to specific plaque regions. Am. J. Pathol. 1993;143:1634–1648. [PMC free article] [PubMed] [Google Scholar]
- 5.Warner SJ, Friedman GB, Libby P. Immune interferon inhibits proliferation and induces 2′-5′-oligoadenylate synthetase gene expression in human vascular smooth muscle cells. J. Clin. Invest. 1989;83:1174–1182. doi: 10.1172/JCI113998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frostegard J, Ulfgren AK, Nyberg P, Hedin U, Swedenborg J, Andersson U, Hansson GK. Cytokine expression in advanced human atherosclerotic plaques: dominance of pro-inflammatory (Th1) and macrophage-stimulating cytokines. Atherosclerosis. 1999;145:33–43. doi: 10.1016/s0021-9150(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 7.Harvey EJ, Ramji DP. Interferon-gamma and atherosclerosis: pro- or anti-atherogenic? Cardiovasc. Res. 2005;67:11–20. doi: 10.1016/j.cardiores.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg MW, Jain MK. Role of transforming growth factor-beta1/Smads in regulating vascular inflammation and atherogenesis. Panminerva Med. 2005;47:169–186. [PubMed] [Google Scholar]
- 9.Robertson AK, Rudling M, Zhou X, Gorelik L, Flavell RA, Hansson GK. Disruption of TGF-beta signaling in T cells accelerates atherosclerosis. J. Clin. Invest. 2003;112:1342–1350. doi: 10.1172/JCI18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, et al. Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc. Natl Acad. Sci. USA. 1986;83:4167–4171. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenbloom J, Feldman G, Freundlich B, Jimenez SA. Transcriptional control of human diploid fibroblast collagen synthesis by gamma-interferon. Biochem. Biophys. Res. Commun. 1984;123:365–372. doi: 10.1016/0006-291x(84)90422-4. [DOI] [PubMed] [Google Scholar]
- 12.Higashi K, Inagaki Y, Fujimori K, Nakao A, Kaneko H, Nakatsuka I. Interferon-gamma interferes with transforming growth factor-beta signaling through direct interaction of YB-1 with Smad3. J. Biol. Chem. 2003;278:43470–43479. doi: 10.1074/jbc.M302339200. [DOI] [PubMed] [Google Scholar]
- 13.Dooley S, Said HM, Gressner AM, Floege J, En-Nia A, Mertens PR. Y-box protein-1 is the crucial mediator of antifibrotic interferon-gamma effects. J. Biol. Chem. 2006;281:1784–1795. doi: 10.1074/jbc.M510215200. [DOI] [PubMed] [Google Scholar]
- 14.Eickelberg O, Pansky A, Koehler E, Bihl M, Tamm M, Hildebrand P, Perruchoud AP, Kashgarian M, Roth M. Molecular mechanisms of TGF-(beta) antagonism by interferon (gamma) and cyclosporine A in lung fibroblasts. FASEB J. 2001;15:797–806. doi: 10.1096/fj.00-0233com. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh AK, Yuan W, Mori Y, Chen S, Varga J. Antagonistic regulation of type I collagen gene expression by interferon-gamma and transforming growth factor-beta. Integration at the level of p300/CBP transcriptional coactivators. J. Biol. Chem. 2001;276:11041–11048. doi: 10.1074/jbc.M004709200. [DOI] [PubMed] [Google Scholar]
- 16.Tredget EE, Wang R, Shen Q, Scott PG, Ghahary A. Transforming growth factor-beta mRNA and protein in hypertrophic scar tissues and fibroblasts: antagonism by IFN-alpha and IFN-gamma in vitro and in vivo. J. Interferon Cytokine Res. 2000;20:143–151. doi: 10.1089/107999000312540. [DOI] [PubMed] [Google Scholar]
- 17.Rishikof DC, Ricupero DA, Liu H, Goldstein RH. Phenylbutyrate decreases type I collagen production in human lung fibroblasts. J. Cell Biochem. 2004;91:740–748. doi: 10.1002/jcb.10742. [DOI] [PubMed] [Google Scholar]
- 18.Guenette DK, Ritzenthaler JD, Foley J, Jackson JD, Smith BD. DNA methylation inhibits transcription of procollagen alpha 2(I) promoters. Biochem. J. 1992;283(Pt 3):699–703. doi: 10.1042/bj2830699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Y, Wang L, Buttice G, Sengupta PK, Smith BD. Interferon gamma repression of collagen (COL1A2) transcription is mediated by the RFX5 complex. J. Biol. Chem. 2003;278:49134–49144. doi: 10.1074/jbc.M309003200. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Sengupta PK, Seto E, Smith BD. Regulatory factor for X-box family proteins differentially interact with histone deacetylases to repress collagen alpha2(I) gene (COL1A2) expression. J. Biol. Chem. 2006;281:9260–9270. doi: 10.1074/jbc.M511724200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg H, Helaakoski T, Garrett LA, Karsenty G, Pellegrino A, Lozano G, Maity S, de Crombrugghe B. Tissue-specific expression of the mouse alpha 2(I) collagen promoter. Studies in transgenic mice and in tissue culture cells. J. Biol. Chem. 1992;267:19622–19630. [PubMed] [Google Scholar]
- 22.Zhu XS, Ting JP. A 36-amino-acid region of CIITA is an effective inhibitor of CBP: novel mechanism of gamma interferon-mediated suppression of collagen alpha(2)(I) and other promoters. Mol. Cell Biol. 2001;21:7078–7088. doi: 10.1128/MCB.21.20.7078-7088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta P, Xu Y, Wang L, Widom R, Smith BD. Collagen alpha1(I) gene (COL1A1) is repressed by RFX family. J. Biol. Chem. 2005;280:21004–21014. doi: 10.1074/jbc.M413191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu XS, Linhoff MW, Li G, Chin KC, Maity SN, Ting JP. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai SC, Seto E. Regulation of histone deacetylase 2 by protein kinase CK2. J. Biol. Chem. 2002;277:31826–31833. doi: 10.1074/jbc.M204149200. [DOI] [PubMed] [Google Scholar]
- 26.Nagarajan UM, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss JM. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Wang L, Buttice G, Sengupta PK, Smith BD. Major histocompatibility class II transactivator (CIITA) mediates repression of collagen (COL1A2) transcription by interferon gamma (IFN-gamma) J. Biol. Chem. 2004;279:41319–41332. doi: 10.1074/jbc.M404174200. [DOI] [PubMed] [Google Scholar]
- 28.Sengupta PK, Fargo J, Smith BD. The RFX family interacts at the collagen (COL1A2) start site and represses transcription. J. Biol. Chem. 2002;277:24926–24937. doi: 10.1074/jbc.M111712200. [DOI] [PubMed] [Google Scholar]
- 29.Sobral LM, Montan PF, Martelli-Junior H, Graner E, Coletta RD. Opposite effects of TGF-beta1 and IFN-gamma on transdifferentiation of myofibroblast in human gingival cell cultures. J. Clin. Periodontol. 2007;34:397–406. doi: 10.1111/j.1600-051X.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- 30.Gu L, Zhu YJ, Guo ZJ, Xu XX, Xu WB. Effect of IFN-gamma and dexamethasone on TGF-beta1-induced human fetal lung fibroblast-myofibroblast differentiation. Acta. Pharmacol. Sin. 2004;25:1479–1488. [PubMed] [Google Scholar]
- 31.Das S, Lin JH, Papamatheakis J, Sykulev Y, Tsichlis PN. Differential splicing generates Tvl-1/RFXANK isoforms with different functions. J. Biol. Chem. 2002;277:45172–45180. doi: 10.1074/jbc.M204117200. [DOI] [PubMed] [Google Scholar]
- 32.Zika E, Greer SF, Zhu XS, Ting JP. Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation. Mol. Cell Biol. 2003;23:3091–3102. doi: 10.1128/MCB.23.9.3091-3102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Invest. 2003;112:863–871. doi: 10.1172/JCI19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velican C. Relationship between regional aortic susceptibility to atherosclerosis and macromolecular structural stability. J. Atheroscler. Res. 1969;9:193–201. doi: 10.1016/s0368-1319(69)80054-2. [DOI] [PubMed] [Google Scholar]
- 35.Amento EP, Ehsani N, Palmer H, Libby P. Cytokines and growth factors positively and negatively regulate interstitial collagen gene expression in human vascular smooth muscle cells. Arterioscler Thromb. 1991;11:1223–1230. doi: 10.1161/01.atv.11.5.1223. [DOI] [PubMed] [Google Scholar]
- 36.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 38.Bridoux F, Badou A, Saoudi A, Bernard I, Druet E, Pasquier R, Druet P, Pelletier L. Transforming growth factor beta (TGF-beta)-dependent inhibition of T helper cell 2 (Th2)-induced autoimmunity by self-major histocompatibility complex (MHC) class II-specific, regulatory CD4(+) T cell lines. J. Exp. Med. 1997;185:1769–1775. doi: 10.1084/jem.185.10.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nandan D, Reiner NE. TGF-beta attenuates the class II transactivator and reveals an accessory pathway of IFN-gamma action. J. Immunol. 1997;158:1095–1101. [PubMed] [Google Scholar]
- 40.Panek RB, Lee YJ, Benveniste EN. TGF-beta suppression of IFN-gamma-induced class II MHC gene expression does not involve inhibition of phosphorylation of JAK1, JAK2, or signal transducers and activators of transcription, or modification of IFN-gamma enhanced factor X expression. J. Immunol. 1995;154:610–619. [PubMed] [Google Scholar]
- 41.Cunha SR, Le Scouarnec S, Schott JJ, Mohler PJ. Exon organization and novel alternative splicing of the human ANK2 gene: implications for cardiac function and human cardiac disease. J. Mol. Cell Cardiol. 2008;45:724–734. doi: 10.1016/j.yjmcc.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.