Abstract
DNA interstrand crosslinks (ICLs) are among the most cytotoxic types of DNA damage, thus ICL-inducing agents such as psoralen, are clinically useful chemotherapeutics. Psoralen-modified triplex-forming oligonucleotides (TFOs) have been used to target ICLs to specific genomic sites to increase the selectivity of these agents. However, how TFO-directed psoralen ICLs (Tdp-ICLs) are recognized and processed in human cells is unclear. Previously, we reported that two essential nucleotide excision repair (NER) protein complexes, XPA–RPA and XPC–RAD23B, recognized ICLs in vitro, and that cells deficient in the DNA mismatch repair (MMR) complex MutSβ were sensitive to psoralen ICLs. To further investigate the role of MutSβ in ICL repair and the potential interaction between proteins from the MMR and NER pathways on these lesions, we performed electrophoretic mobility-shift assays and chromatin immunoprecipitation analysis of MutSβ and NER proteins with Tdp-ICLs. We found that MutSβ bound to Tdp-ICLs with high affinity and specificity in vitro and in vivo, and that MutSβ interacted with XPA–RPA or XPC–RAD23B in recognizing Tdp-ICLs. These data suggest that proteins from the MMR and NER pathways interact in the recognition of ICLs, and provide a mechanistic link by which proteins from multiple repair pathways contribute to ICL repair.
INTRODUCTION
DNA interstrand crosslinks (ICLs) can cause a block to DNA metabolic processes, such as replication and transcription, and are highly cytotoxic (1,2). Thus, ICL-forming compounds have been utilized in chemotherapeutic regimens to promote cancer cell death (3,4). Triplex-forming oligonucleotides (TFOs) bind to sites in duplex DNA in a sequence-specific fashion to form triple-helical structures, and when conjugated to a DNA damaging agent, can be used to direct site-specific DNA damage (5). By targeting DNA damaging agents to specific sites, drug-conjugated TFOs provide a potential mechanism to increase specificity for tumor cells and reduce cytotoxicity to normal cells. For example, Christensen et al. (6) used c-MYC-specific psoralen-modified TFOs to increase the incorporation of the anticancer nucleoside analogue gemcitabine into the targeted DNA in human breast cancer cells, which led to decreased anchorage-independent growth and increased cytotoxicity. Therefore, the use of combinations of TFOs with chemotherapeutic crosslinking agents may offer a new strategy for cancer treatment. However, the molecular mechanism(s) by which cells process TFO-directed psoralen ICLs (Tdp-ICLs) remains unclear. In addition, the repair of ICLs alone (in the absence of the triplex structure) in mammalian cells is still poorly understood.
In Escherichia coli, both the nucleotide excision repair (NER) pathway and homologous recombination (HR) pathway are involved in ICL repair (7,8). Escherichia coli UvrABC proteins, which function in the NER pathway are involved in resolving the ICLs (9). After recognition of the ICLs, the endonuclease complex UvrA2B recognizes and binds to the ICLs, and then UvrC and UvrB make 5′ and 3′ incisions flanking the ICLs, to release one of the DNA strands with the crosslinking agent still covalently linked to the other strand, a process called ‘unhooking’ (7,10). DNA polymerase IV binds to the excised ends and synthesizes the gap without the template, which is considered an error-prone type of repair. Then, DNA ligase I joins the end of the synthesized fragment to the excision. Finally, the strand with the unhooked fragment is excised by UvrABC and replicated using the repaired strand as a template, with ligase filling in the gap (9).
The NER mechanism in human cells is more complex than that in E. coli. Proteins involved in the mammalian NER pathway, such as XPA, RPA, XPC–RAD23B, ERCC1, XPF and XPG, have all been reported to function in ICL repair (11). Among these proteins, the XPC–RAD23B complex is commonly accepted as the DNA damage recognition factor (12,13). ERCC1–XPF and XPG are structure-specific endonucleases that generate 5′ and 3′ incisions, respectively, flanking bulky DNA damage, which is similar to the excision pattern of DNA damage processed by UvrABC in E. coli (14). In addition to the NER pathway, the mismatch repair (MMR) protein complex MutSβ (MSH2-MSH3 heterodimer) binds DNA ICLs in purified systems and in cell lysates (15). Using a cell-based assay, Zhang et al. (16) showed that ICLs could be processed by an error-free homology-dependent recombination repair pathway after introducing a DNA double-strand break in close proximity to the ICL, and that this repair was dependent on MSH2, ERCC1–XPF, REV3 and Fanconi anemia proteins.
The repair of Tdp-ICLs may be even more complicated than that of ICLs alone due to the triple-helical structure at the site of the ICL. Previously, we reported that two NER protein complexes (XPA–RPA and XPC–RAD23B) bind to Tdp-ICLs in vitro (17,18), and that the bacterial UvrABC nuclease can recognize and incise Tdp-ICLs in vitro (19). Similar to duplex ICLs, more than one repair pathway may be involved in the recognition and processing of Tdp-ICLs. We have shown that the repair efficiency of Tdp-ICLs was reduced in MSH2-deficient human cell-free extracts, suggesting that MSH2 is involved in their repair (20). In addition, MSH2-deficient cells were sensitive to psoralen ICLs, yet the ICL-induced mutagenesis was similar to that in MSH2-proficient cells, indicating that the MMR protein MSH2 is involved in an error-free repair of ICLs (20). Thus, proteins from both the MMR and NER pathways have been implicated in the recognition and/or processing of psoralen ICLs in mammalian cells. However, it is not known how these proteins interact in the first, and rate-limiting step of repair, i.e. DNA damage recognition.
In the present study, we investigated the recognition of Tdp-ICLs by the MMR protein complex MutSβ and the NER protein complexes, XPA–RPA and XPC–RAD23B, and discovered that MutSβ interacts with these two complexes on Tdp-ICLs. At low protein concentrations, MutSβ and XPC–RAD23B bound the psoralen ICLs independently. However, increasing the concentrations of MutSβ and XPC–RAD23B triggered the formation of a higher-order complex containing the Tdp-ICL bound to both protein complexes. In contrast, MutSβ formed higher-order complexes with XPA-RPA on psoralen ICLs even at low protein concentrations. Chromatin immunoprecipitation (ChIP) analysis revealed that MutSβ bound to Tdp-ICLs in human cells. Our findings suggest that proteins from more than one repair pathway are involved in the recognition of ICLs in vitro and in vivo, and that these proteins may participate cooperatively or independently in ICL repair, depending on their local concentrations. Understanding the mechanism(s) of ICL repair in mammalian cells is essential for improving the efficacy of ICL-inducing chemotherapeutic agents and preventing the resistance of cancer cells to such treatment.
MATERIALS AND METHODS
TFO-directed psoralen ICL (Tdp-ICL) formation
TFOs were synthesized with the 5′-psoralen derivative, HMT, (2-[4′(hydroxymethyl)-4,5′,8-trimethylpsoralen]-hexyl-1-O-(2-cyanoethyl)-N,N-diisopropyl)-phosphoramidite) by the Midland Certified Reagent Company, Inc. (Midland, TX, USA). Synthetic 57-bp oligonucleotides were annealed to form the target duplex (Figure 1A, complementary oligonucleotides 71 + 72), then were 5′-end-labeled with [γ-32P]dATP, and gel purified as described previously (17,21). The psoralen-modified TFO (pAG30, 30-mer) was incubated with duplex target substrate in a triplex binding buffer (10 mM Tris–HCl, pH 7.6, 10 mM MgCl2, 10% [v/v] glycerol) in the dark at 37°C overnight, and then irradiated with 1.8 J/cm2 ultraviolet A light at 365 nm for 20 min to induce the formation of TFO-directed psoralen ICLs (Tdp-ICLs). The efficiency of crosslinking of the psoralen to the duplex targets was up to 90% as determined by native 12% polyacrylamide gel electrophoresis (PAGE).
Figure 1.
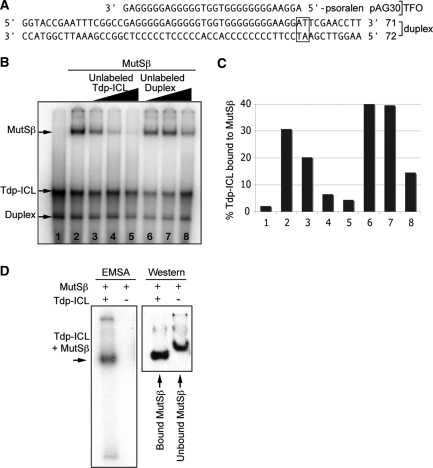
Specific recognition of Tdp-ICLs by MutSβ. (A) Sequences of the 57-bp DNA duplex (complementary oligonucleotides 71 + 72) and the 5′-psoralen-conjugated TFO (pAG30) used to form the Tdp-ICL substrate. The 5′-TpA psoralen crosslinking site is shown in the box. (B) EMSA analysis of MutSβ (43 nM) on Tdp-ICLs (10 nM) with unlabeled Tdp-ICLs or unlabeled DNA duplex used as competitor. The concentrations of the competitors were at 2×, 10× and 50× of the 5′ end γ-32P labeled Tdp-ICL substrate. Purified MutSβ was incubated with labeled Tdp-ICLs at 30°C for 5 min prior to the addition of the competitor DNA. Incubation was then continued for 20 min. The resulting DNA–protein complexes were separated from the free DNA substrate and unbound protein using 6% native PAGE. The gel was then dried and scanned using a PhosphorImager. Lane 1, DNA substrate only; lane 2, MutSβ; lanes 3–5, MutSβ with additional unlabeled competitor Tdp-ICL substrate; lanes 6–8, MutSβ with additional unlabeled competitor duplex DNA. (C) Quantitation of Mutsβ bound to the Tdp-ICL (radioactivity in the band containing MutSβ as a percentage of the total radioactivity loaded in the lanes from Figure 1B). (D) Southwestern blotting analysis of the MutSβ–DNA complex and unbound MutSβ. The MutSβ–DNA complex or MutSβ alone was separated using 4% native PAGE, and then transferred onto a PVDF membrane after exposure to X-ray film. The membrane was probed with an α-MSH2 antibody to detect MutSβ.
A psoralen-crosslinked duplex DNA substrate (ICL only, in the absence of the third strand TFO) was prepared using the same duplex substrate (71 + 72) and same TFO but containing a disulfide link between the psoralen derivative and the TFO (p-s-s-AG30, Midland Certified Reagent Company). After triplex formation and UVA irradiation, the Tdp-ICL was treated with 1/10 volume of 1 M dithiothreitol (DTT) at 65°C for 3 h to release the TFO from the ICL. The duplex ICL was then purified on a 12% denaturing gel, as we have described (19).
Human recombinant proteins
The plasmid expressing subunits MSH2 and MSH3 of MutSβ was constructed by ligation of digested fragments from pFastbacDual-MSH2-MSH6 (22) and pGEM7Zf(+) plasmid harboring a human MSH3 cDNA (23), respectively. The resulting plasmid pFastbacDual-MSH2-MSH3 was used to prepare baculovirus according to the manufacturer's (Invitrogen, Carlsbad, CA, USA) protocol. Recombinant virus was plaque-purified, and high titer stocks were prepared and used to infect Sf9 cells for protein expression. Recombinant MutSβ was purified from cleared lysates of infected Sf9 cells by a chromatographic procedure identical to that described previously for the native human protein (24). XPA–maltose-binding protein fusion protein was expressed and purified from E. coli PR745 (25). The three subunits of RPA (P70, P32 and P14) were expressed by co-infection of Sf9 insect cells and purified by Ni2+-chelate column chromatography, as previously described (26). The XPC–RAD23B–maltose-binding protein fusion complex was expressed and purified in Sf9 or Hi-5 insect cells, as previously described (27).
Electrophoretic mobility-shift assays (EMSAs)
DNA–protein complexes were investigated by EMSAs. The human recombinant purified protein complexes MutSβ (100 ng, 43 nM), XPC–RAD23B (10 ng, 6.5 nM), and XPA (50 ng, 60 nM), and RPA (5 ng, 5 nM) were pre-incubated in binding buffer (37.5 mM Tris–HCl, pH 7.6, 150 mM NaCl, 1.5 mM DTT, 1.5 mM ethylenediaminetetraacetic acid (EDTA), 150 μg/ml bovine serum albumin (BSA), 40 μM ADP, 0.015% Nonidet P-40, 15% glycerol), and then incubated with radiolabeled DNA substrate (10 nM) in a 10 μl reaction volume at 30°C for 20 min. DNA–protein samples were electrophoresed through a 6% (37.5:1 acrylamide:bis-acrylamide) native PAGE with 2.5% glycerol in 1× TGE buffer (25 mM Tris–HCl, pH 8.2, 192 mM glycine, 1 mM EDTA) at 100 V for 1.5 h at 4°C. The gel was dried at 85°C for 2.5 h, and DNA–protein complexes on the gel were visualized using a PhosphorImager Typhoon 9410 (GE Healthcare Life Sciences, Piscataway, NJ, USA). The scanned image was quantified using the ImageQuant software (GE Healthcare Life Sciences) and processed Photoshop CS3 software program (Adobe Systems Incorporated, San Jose, CA, USA).
Southwestern analysis
The DNA–protein complexes were separated from the free DNA and protein using native PAGE as described above. The gel was exposed to X-ray film (Eastman Kodak, Rochester, NY, USA) without drying, and then transferred onto a 0.45 μm polyvinylidene fluoride (PVDF) membrane (Milipore, Billerica, MA, USA) using a semi-dry transfer unit (Hoefer, San Francisco, CA, USA) in a transfer buffer [39 mM glycine, 48 mM Tris, 0.1% sodium dodecyl sulfate (SDS), 20% methanol] at 1 mA/cm2 for 1 h. The membrane was washed with Tris–buffered saline containing 0.1% Tween-20 (TBST) and blocked with TBST containing 2% nonfat dry milk powder (Bio-Rad, Hercules, CA, USA). The membrane was then probed with an α-MSH2 antibody (Calbiochem, San Diego, CA, USA) at a dilution of 1:1000 at room temperature for 2 h, washed with TBST, and incubated with a horseradish peroxidase (HRP)-conjugated antibody against mouse IgG (Bio-Rad) at room temperature for 1 h. The membrane was then washed three times with TBST before being developed using an ECL Plus chemiluminescence detection kit (GE Healthcare Life Sciences). Following detection of MSH2, the membrane was stripped using Strip buffer (Pierce, Rockford, IC, USA) and re-probed with an α-RPA/p34 antibody (NeoMarkers, Fremont, CA, USA) at a dilution of 1:500 to detect RPA, or with a maltose-binding protein (MBP)-tagged antibody (α-MBP antibody, New England Biolabs, Beverly, MA, USA) at a dilution of 1:10 000 to detect the XPA-MBP or the XPC–MBP fusion protein.
ChIP assays
ChIP assays were performed using a Simple Chip Enzymatic Chromatin IP Kit (Cell Signaling, Inc., Santa Cruz, CA, USA) according to the manufacturer's recommended protocol with some modification. In brief, 293T human cells (∼1x106 per sample) were transfected with pSupFG1 plasmid only or pSupFG1 plasmid containing a Tdp-ICL. At 24 h after transfection, cells were fixed with 1% formaldehyde for 8 min at room temperature to crosslink DNA and proteins. Crosslinking was quenched with 125 mM glycine, cells were lysed, and chromatin was fragmented by partial digestion with micrococcal nuclease and later sonicated to obtain an average DNA fragment length of 200–500 bp. Following sonication, centrifugation was performed at 12 000 r.p.m. for 10 min at 4°C to remove the soluble supernatant. Next, chromatin was diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl, pH 8.1, 167 mM NaCl) and pre-cleared with protein-G magnetic beads containing salmon sperm DNA and BSA. Approximately 1% of the total chromatin was stored as input DNA and the remaining pre-cleared chromatin (1 ml) was incubated with 5 μg of specific antibody (α-MSH2 antibody, Cell signaling, or α-MSH3 antibody, Abcam, Cambridge, MA, USA) or control α-IgG antibody (mouse IgG; Upstate, Thermeula, CA, USA) and samples were incubated overnight at 4°C on a rotator. Then the immunoprecipitation complexes were isolated by adding protein G magnetic beads, rotating for 2 h at 4°C and beads were collected by applying a magnetic force. Immunoprecipitated complexes were washed sequentially for 5 min three times with low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 150 mM NaCl), one time with high salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 500 mM NaCl), and then twice with TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). The immunocomplexes were then eluted by resuspending the beads in 150 μl of elution buffer (1% SDS, 0.1 M NaHCO3) for 20 min at room temperature. The DNA–protein crosslinks were reversed by heating at 66°C in a water bath for 6 h with the addition of 5 M NaCl to a final concentration of 200 mM. All the samples were then treated with RNase A at 37°C for 30 min, and then with Proteinase K at 45°C for 1.5 h to remove RNA and protein. Next, the DNA fragments were purified using QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA). Fractions of purified ChIP DNA and input DNA were used for PCR analysis. The PCR reaction was performed with AmpliTaq Gold DNA polymerase (Applied Biosystems Inc, Foster City, CA, USA) for 28 cycles of 30 s at 95°C, 30 s at 56°C, and 45 s at 72°C in a Bio-Rad I-cycler. The primers for PCR amplification were: Primer P1, 5′-gcc ccc ctg acg agc atc ac; Primer P2, 5′-tag tta ccg gat aag gcg cag cgg; Primer P3, 5′-aat acc gcg cca cat agc ag; and Primer P4, 5′-agt att caa cat ttc cgt gtc gcc (Figure 2A). Amplified products were separated on 1.5% agarose gels containing ethidium bromide and visualized on an AlphaImager (San Leandro, CA, USA).
Figure 2.
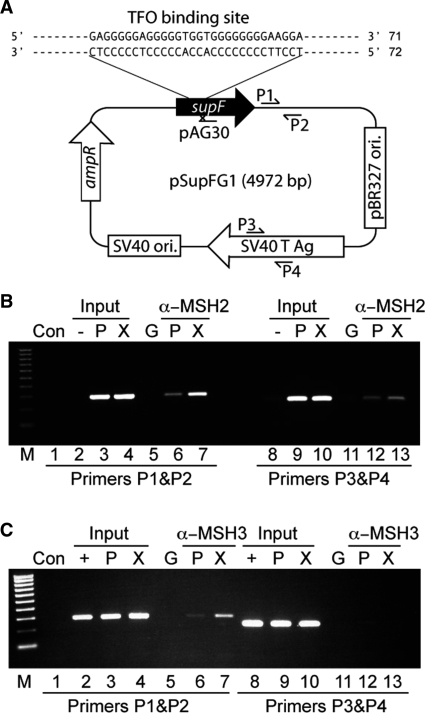
MSH2 binds to a Tdp-ICL in human cells. (A) Schematic representation of plasmid, pSupFG1, containing the same TFO binding site as in the synthetic duplex 71 + 72, adjacent to a psoralen crosslinking site, and primer sites (P1, P2, P3 and P4) on the plasmid used for ChIP analysis. (B and C) Representative agarose gels of PCR products from ChIP assays demonstrating the binding of the α-MSH2 or α-MSH3 antibody to DNA near the site of the Tdp-ICL. Purified DNA was analyzed by standard PCR methods using primers P1 and P2 (near the TFO binding and psoralen crosslinking site) and control primers P3 and P4 (∼2 kb from the TFO binding and psoralen crosslinking site). Lane 1, PCR control without input template; lanes 2–7, PCR products amplified with primers P1 & P2; lanes 8–13, PCR products amplified with primers P3 & P4; C, PCR control without template; ‘–’ template from untransfected cells; ‘+’ template from the cells transfected with plasmid; G, pulldowns with α-IgG antibody; P, template from the cells transfected with the control plasmid; X, template from the cells transfected with the plasmid containing the Tdp-ICL. M, 100 bp DNA ladder (Bio-Rad).
Antibody supershift assays
DNA–protein complexes were formed under the same reaction conditions as described above. After incubation at 30°C for 5 min, 0.2 μg of an antibody against RPA (α-RPA/p34 antibody), XPA–MBP (α-MBP antibody), or MSH3 (α-MSH3 antibody) was added to the reaction, and incubation was continued at 30°C for 20 min. Samples were electrophoresed through a 4% native PAGE (37.5:1 acrylamide/bis-acrylamide) in 1× TGE buffer at 4°C (25 mA) for 2–3 h. The gels were then dried and visualized using autoradiography.
RESULTS
Recognition of Tdp-ICLs by MutSβ
Tdp-ICLs and ICLs alone (in the absence of the TFO) represent complex DNA lesions that may be recognized by proteins from more than one DNA repair mechanism. Similar to NER mutant cell lines, cells deficient in the MMR protein MSH2 exhibited impaired repair of Tdp-ICLs or ICLs only in vivo (20). Gel-shift assays have demonstrated that MutSβ recognizes duplex ICLs induced by psoralen (15). To determine if MutSβ could recognize psoralen ICLs in the presence of a triplex structure (i.e. Tdp-ICLs), we performed EMSAs with purified human recombinant MutSβ protein complex and radiolabeled Tdp-ICL substrate. The Tdp-ICL substrate contains a 57-bp DNA duplex of two 5′-end-32P-labeled complementary oligonucleotides (71 + 72; Figure 1A). The duplex was crosslinked to a 5′-psoralen-coupled 30-nt TFO (pAG30; Figure 1A), which has been used previously to study the function of NER proteins in the recognition of Tdp-ICLs (17,18,21). By EMSA analysis, we found that purified MutSβ [at 43 nM, the estimated apparent Kd (Kapp); data not shown] bound to the Tdp-ICL substrate (at 10 nM), forming a complex that migrated more slowly than the unbound Tdp-ICL (Figure 1B, indicated by arrows). We observed a similar mobility shift when we incubated MutSβ with a PAGE purified Tdp-ICL substrate (Figure S1), while MutSβ (at the same concentration) showed no detectable binding to the non-damaged DNA duplex control substrate. To determine the specificity of MutSβ binding to the Tdp-ICL, increasing amounts of unlabeled Tdp-ICL were added to the reaction as a competitor. Unlabeled Tdp-ICL effectively competed with labeled Tdp-ICL substrate, leaving few detectable shifted complexes (of MutSβ bound to the radiolabeled Tdp-ICL) on the gel (Figure 1B and C, compare lanes 2 and 5). In contrast, addition of the same amount of unlabeled duplex DNA competitor in the reaction served as a much less efficient competitor (Figure 1C). Higher concentrations of the unlabeled duplex substrate (50×) were able to compete for MutSβ binding to some extent, demonstrating that MutSβ can bind with low affinity to duplex DNA non-specifically under the conditions of our assay (Figure 1B and C, compare lanes 5 with 8). The presence of MutSβ in the slower migrating complex was confirmed by Southwestern blotting analysis (Figure 1D), which showed that Tdp-ICL-bound MutSβ migrated slightly faster than the unbound MutSβ protein in the absence of DNA substrate. Taken together, these results suggest that MutSβ binds to Tdp-ICLs with high affinity and specificity.
To investigate the extent to which MutSβ recognized the ICL versus the triple-helical structure formed by the TFO (in addition to the ICL induced by psoralen), we analyzed the binding of MutSβ to a Tdp-ICL (triplex ICL) compared to a duplex ICL after removal of the TFO from the Tdp-ICL substrate. MutSβ bound to the psoralen ICL in the absence of triplex formation, consistent with previous findings (15). The binding affinity of MutSβ for the duplex ICL was slightly less than that of the Tdp-ICL (Figure S2), indicating that the triple helical structure enhances the recognition of Tdp-ICLs by MutSβ. We were unable to examine binding of MutSβ to a DNA triplex in the absence of the ICL (triplex only), because the triplex structure was not stable under the conditions of our assays.
MutSβ interacts with Tdp-ICLs in human cells
We have shown that MutSβ binds Tdp-ICLs using an in vitro purified system. To confirm that this is a physiologically relevant interaction, we performed ChIP experiments to assess the binding of MutSβ in human cells. The pSupFG1 plasmid (Figure 2A) containing a Tdp-ICL (at the same location as that of the 71 + 72 synthetic duplex substrate; see Figure 1A), or pSupFG1 plasmid alone (18) were transfected into 293T human cells. Twenty-four hours after transfection, cells were harvested and chromatin was immunoprecipited (as described in the ‘Materials and Methods’ section) using an α-MSH2, α-MSH3, or a control α-IgG antibody. Input and immunoprecipited DNA samples were amplified by semi-quantative PCR (Figure 2B and C) with specific primers (P1 and P2) near the ICL and another set of primers (P3 and P4) ∼2 kb from the ICL to serve as an internal control for the efficiency of immunoprecipitation. As shown in Figure 2B and C, we observed that the α-MSH2 and α-MSH3 antibodies were able to pull down more pSupFG1 DNA containing the Tdp-ICL as compared to the pSupFG1 control plasmid in the absence of the Tdp-ICL (Figure 2B and C, compare lanes 6 and 7). As expected, the amount of total DNA from cell lysates was similar in each sample as assessed by the amount of DNA amplified using either primer set (Figure 2B and C, ‘Input’ lanes). An IgG antibody was used as an antibody specificity control, and as expected, we did not observe a signal in the IgG antibody samples (Figure 2B and C, lanes 5 and 11). This result indicates that the human mismatch repair protein complex, MutSβ recognizes and binds Tdp-ICL-damaged DNA to a greater extent than it does to non-damaged DNA in human cells, consistent with our EMSA data (see Figure 1).
Formation of MutSβ and XPA–RPA complexes on Tdp-ICLs
Previously, we observed that proteins in the NER pathway (XPA, RPA, and XPC–RAD23B) function in the recognition of Tdp-ICLs (17,18,21). To determine whether MutSβ interacts with these NER proteins in the recognition of Tdp-ICLs, we conducted EMSA analysis with purified recombinant human MutSβ and XPA–RPA at their estimated Kapp concentrations [XPA 60 nM; RPA 4 nM; (18)]. Both the XPA–RPA complex and the MutSβ heterodimer recognized the Tdp-ICL individually, as evidenced by shifted substrates detected by EMSA (Figure 3A, indicated by arrows). However, when both XPA–RPA and MutSβ were incubated simultaneously with the Tdp-ICL substrate, a slowly migrating band was detected on the gel, suggesting that both protein complexes bound the Tdp-ICL and together formed a multimeric complex with the Tdp-ICL (Figure 3A, lane 4).
Figure 3.
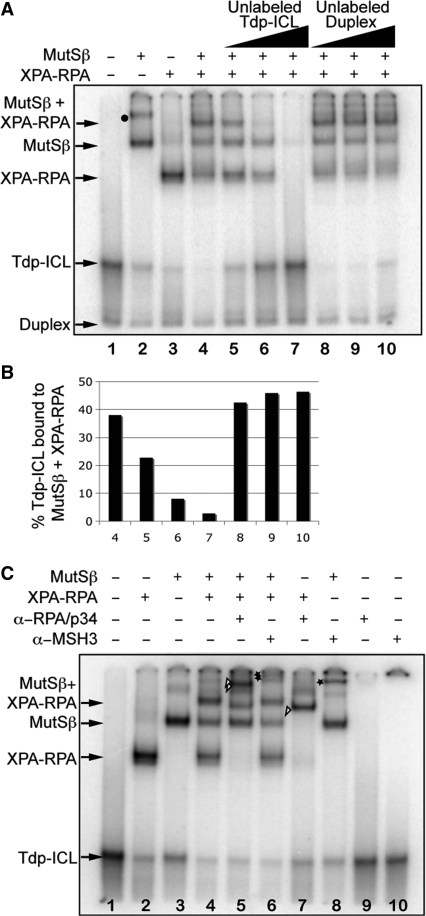
Formation of complexes of MutSβ and XPA–RPA on Tdp-ICLs. (A) The purified human recombinant protein complexes MutSβ (43 nM) and XPA–RPA (XPA at 60 nM; RPA at 4 nM, pre-incubated) were added alone or together to Tdp-ICLs (10 nM). The protein–DNA complexes are indicated by arrows. An unlabeled Tdp-ICL or DNA duplex competitor was added to the reaction at 2×, 10× and 50× the [γ-32P]dATP-labeled Tdp-ICL substrate concentration (10 nM). Lane 1, DNA substrate only; lanes 2–4, DNA substrate with indicated proteins; lanes 5–7, MutSβ and XPA–RPA with increased unlabeled Tdp-ICL competitor; lanes 8–10, MutSβ and XPA–RPA with increased unlabeled DNA duplex competitor. Binding of multiple MutSβ molecules to Tdp-ICLs is indicated with a black dot. (B) Quantitation of MutSβ and XPA–RPA bound to the Tdp-ICL (radioactivity in the band containing both MutSβ and XPA–RPA as a percentage of the total radioactivity loaded in the lanes from Figure 3A). (C) Antibody supershift assay of MutSβ and XPA–RPA on the Tdp-ICL. The purified protein complexes were incubated with the Tdp-ICLs at 30°C for 5 min and then incubated with an antibody (α-RPA/p34 or α-MSH3, 0.2 μg each) for 20 min. The DNA–protein complexes were separated from the free DNA substrate using 6% native PAGE. The protein–DNA complexes are indicated by arrows. The triangle marks the XPA–RPA–Tdp-ICL complex shifted by an α-RPA/p34 antibody; the double triangle marks the MutSβ–XPA–RPA–Tdp-ICL complex shifted by an α-RPA/p34 antibody; the star marks the MutSβ–Tdp-ICL complex shifted by an α-MSH3 antibody; the double star marks the MutSβ–XPA–RPA–Tdp-ICL complex shifted by an α-MSH3 antibody.
In this slower migrating complex, binding of XPA–RPA and MutSβ to the DNA substrate appeared to be specific to the Tdp-ICL, as the radiolabeled complex was diminished in the presence of unlabeled Tdp-ICL but not by unlabeled duplex DNA (Figure 3A and B, compare lane 7 with lane 10). We observed similar binding of XPA–RPA and MutSβ to purified Tdp-ICLs (Figure S1), but not to the non-damaged duplex DNA, which further suggested that these proteins bind specifically to the Tdp-ICL. The binding affinity of XPA–RPA to duplex ICLs (in the absence of the triplex structure) was approximately an order of magnitude lower than for Tdp-ICLs. XPA–RPA and MutSβ bound together to a duplex ICL substrate (in the absence of the TFO), but again the binding affinity was approximately an order of magnitude lower than for Tdp-ICLs (data not shown), suggesting that TFO-directed triplex formation enhanced the recognition of the ICL by repair proteins.
The presence of both XPA–RPA and MutSβ in the higher-order complex was demonstrated by antibody supershift analysis (Figure 3C) and further confirmed by Western blotting analysis (Figure S3). We observed a supershift of the slower migrating band on the gel when an α-RPA/p34 antibody was added to the reaction (Figure 3C, indicated by double triangles), and a supershifted band on the gel (Figure 3C, indicated by double stars) with the addition of an α-MSH3 antibody. We further confirmed the presence of this α-MSH3-shifted complex on a 4% native PAGE (Figure S4). Together, the presence of XPA, RPA, and MutSβ with the Tdp-ICLs in this complex was confirmed using Western blotting (Figure S3) and antibody supershift analysis (Figures 3C and S4), indicating that MutSβ and XPA–RPA bound together on Tdp-ICLs.
Interaction between MutSβ and XPA–RPA on Tdp-ICLs
When we incubated both MutSβ and XPA–RPA with a Tdp-ICL, a higher-order complex formed in addition to two faster migrating complexes representing discrete binding of either MutSβ or XPA–RPA to the Tdp-ICL (see Figure 3). Decreasing the amount of the DNA substrate in the reactions resulted in an increase in the ratio of the slower migrating complex (containing both MutSβ and XPA–RPA bound to the Tdp-ICL) to the faster migrating complexes (containing Tdp-ICL with either MutSβ or XPA–RPA; Figure 4A and B). This interaction appeared to be specific to the Tdp-ICL, because the total amount of non-damaged duplex DNA substrate remained similar in all samples (Figure 4B).
Figure 4.
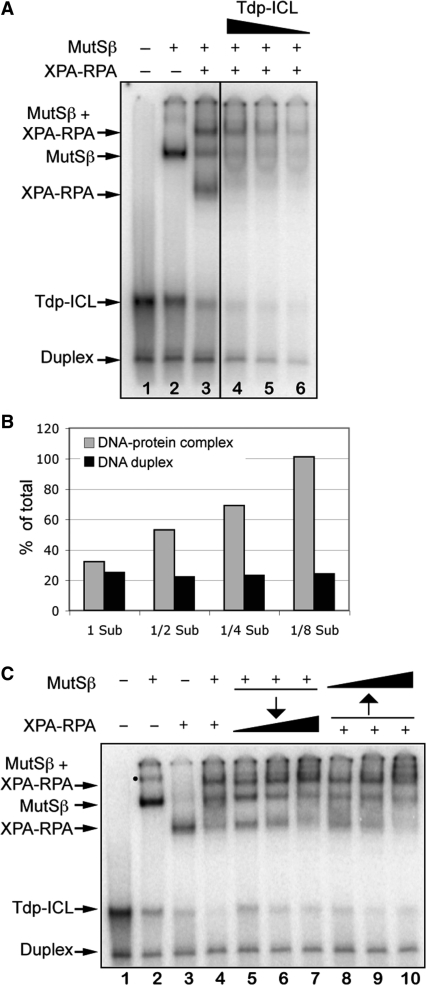
MutSβ and XPA–RPA interact on Tdp-ICLs. (A) EMSA analysis of purified MutSβ and XPA–RPA complexes with decreasing concentrations of the Tdp-ICL substrates. Lane 1, DNA substrate; lane 2, MutSβ; lane 3, XPA–RPA and MutSβ; lanes 4–6, MutSβ and XPA–RPA with decreasing amounts of DNA substrate (1/2×, 1/4× and 1/8×, respectively, of 10 nM). The protein–DNA complexes are indicated by arrows. (B) Quantitation of MutSβ–XPA–RPA–Tdp-ICL complex bands and duplex DNA bands as the percentage of total DNA substrate (Sub) loaded in lanes 3–6 in Figure 4A. (C) EMSA analysis of the Tdp-ICLs with sequential addition of purified MutSβ or XPA–RPA. Lane 1, DNA substrate; lane 2, MutSβ; lane 3; XPA and RPA; lane 4, MutSβ and XPA–RPA; lanes 5–7, samples were incubated with MutSβ at 30°C for 10 min, then XPA–RPA (at 1×, 2× and 5× of 60 nM/4 nM) was added, and the samples were incubated for another 10 min. Lanes 8–10, samples were incubated with XPA–RPA at 30°C for 10 min, then MutSβ (at 1×, 2× and 5× of 43 nM) was added, and samples were incubated for another 10 min. The DNA–protein complexes were separated from free DNA substrate using 6% native PAGE. The black dot marks the multiple binding of MutSβ to the Tdp-ICL.
In order to determine if the binding of one of the protein complexes to the Tdp-ICL substrate influenced the binding of the other protein complex, we performed order-of-addition experiments. We observed that either protein complex alone could recognize Tdp-ICLs within 1 min after incubation (data not shown). As shown in Figure 4C, the higher-order complex containing XPA–RPA, MutSβ, and the Tdp-ICL was formed on a pre-formed complex of XPA–RPA–Tdp-ICL or on a pre-formed complex of MutSβ–Tdp-ICL. Thus, interaction between MutSβ and XPA–RPA occurs on Tdp-ICLs regardless of their order of addition to the reaction.
Independent binding of Tdp-ICLs to MutSβ and XPC–RAD23B at Kapp protein concentrations
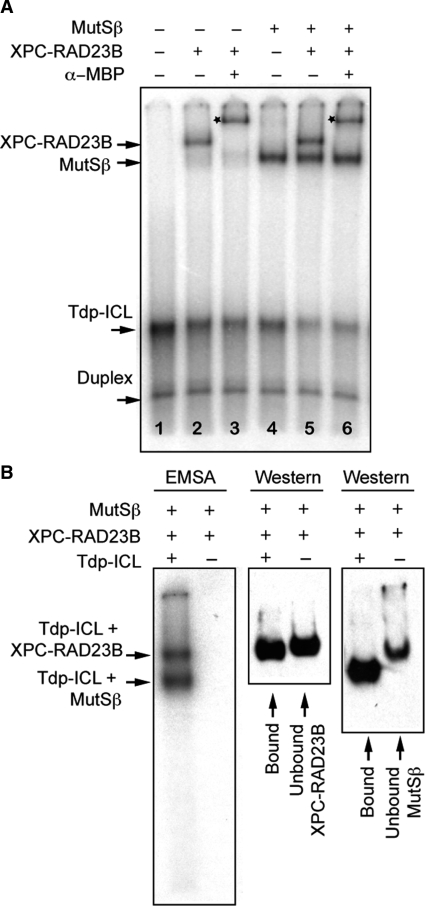
XPC–RAD23B is thought to be the first damage recognition factor in the NER pathway to bind DNA damage (13,28). Because we observed that XPA–RPA interacted with MutSβ on Tdp-ICLs, we were compelled to determine whether MutSβ interacted with XPC–RAD23B on TFO-directed psoralen ICLs. To further delineate the proteins involved in the recognition of Tdp-ICLs, we conducted EMSA analysis with MutSβ and XPC–RAD23B as described above for MutSβ and XPA–RPA. At its Kapp concentration, XPC–RAD23B (6.5 nM) formed a complex with the Tdp-ICL (Figure 5A, lane 2), consistent with our previously published results (17). In contrast to the interaction identified between MutSβ and XPA–RPA, incubation of MutSβ and XPC–RAD23B with Tdp-ICLs resulted in the formation of two discrete complexes with Tdp-ICLs (Figure 5A, lane 5). Southwestern blot analysis confirmed that the two individual complexes were composed of either MutSβ with Tdp-ICLs or XPC–RAD23B with Tdp-ICLs (Figure 5B). In the presence of an α-MBP antibody, which was able to recognize the MBP-fused XPC protein, only the band containing XPC–RAD23B was supershifted, while the complex containing MutSβ bound to Tdp-ICLs maintained its mobility on the gel (Figure 5A), suggesting that XPC–RAD23B and MutSβ bound to Tdp-ICLs in an independent fashion. We also observed independent binding of these two complexes with purified Tdp-ICLs (Figure S5) or duplex ICLs (data not shown) at Kapp concentrations. Similar to that of XPA–RPA, the binding of XPC–RAD23B to duplex ICLs (in the absence of the triplex structure) was decreased, consistent with our previous report (17). Taken together, these data suggest that binding of either XPC–RAD23B or MutSβ to psoralen ICLs may exclude the binding of the other protein complex to the same molecule. This differential binding of the lesion by the NER and MMR protein complexes may result in differential processing of the lesion. However, either of these complexes can interact with XPA–RPA on Tdp-ICL (as shown above and ref. 17), such that the binding of MutSβ to psoralen ICLs might also facilitate repair of the lesion by NER proteins. Similar to our results with XPA–RPA, XPC–RAD23B and MutSβ also formed a complex together with Tdp-ICLs at increased protein concentrations (data not shown).
Figure 5.
Independent binding of MutSβ and XPC–RAD23B to Tdp-ICLs at low (Kapp) protein concentrations. (A) EMSA analysis of purified human recombinant MutSβ (43 nM) and XPC–RAD23B (6.5 nM) with Tdp-ICLs (10 nM). An α-MBP antibody was used to supershift the XPC–RAD23B–Tdp-ICL complex (by identification of the MBP-tagged XPC protein). Lane 1, DNA substrate; lane 2, XPC–RAD23B; lane 3, XPC–RAD23B and α-MBP antibody; lane 4, MutSβ; lane 5, XPC–RAD23B and MutSβ; lane 6, XPC–RAD23B, α-MBP antibody and MutSβ. The star marks the protein–DNA complex shifted by an α-MBP antibody. (B) Southwestern blot analysis of the DNA-protein complexes and unbound proteins using 4% native PAGE. The gels were exposed to X-ray film and then transferred to a PVDF membrane. The membrane was probed with the α-MSH2 antibody and then stripped and blotted again with the α-MBP antibody.
DISCUSSION
Psoralen-modified TFOs have been used to introduce site-specific ICLs in genomic DNA in cells, which may provide a means to improve upon ICL-based chemotherapeutic strategies. In this study, we found that the MMR complex MutSβ bound to TFO-directed psoralen ICLs with high specificity and affinity, and interacted with the NER complex XPA–RPA, in recognizing these lesions in our purified in vitro system.
The recognition and processing of Tdp-ICLs may differ from that of duplex ICLs, as TFO binding to the DNA duplex may prevent binding and/or cleavage of damaged DNA strands by repair proteins (29). However, the presence of the third strand TFO adjacent to the duplex ICLs may increase the structural distortion at the site of damage, and as a consequence, recruit repair proteins more efficiently. In support of this idea, Christensen et al. (19) demonstrated that the bacterial UvrABC nuclease incised TFO-directed psoralen ICLs with a similar pattern, but greater efficiency than psoralen ICLs alone. Consistent with this, the work described here demonstrated that the NER and MMR damage recognition proteins bound the TFO-directed psoralen ICLs with greater affinity than they did the psoralen ICLs in the absence of the TFO, and thus the TFO may promote more efficient repair of ICLs. Despite their structural differences, both the TFO-directed ICLs and the ICLs alone were recognized by the same protein complexes, suggesting that the triplex structure itself may be repaired in a fashion similar to that of the ICL alone. Therefore, psoralen-modified TFOs can be used to facilitate mechanistic studies of ICL processing and repair by creating ICLs at specific sites in the genome. In addition, direct substrate comparisons can be made between Tdp-ICLs and ICLs (in the absence of the TFO) by facile removal of a disulfide linked TFO-psoralen conjugate following crosslink formation, as we have demonstrated in this study and in a previous report (19).
Although the binding of both NER and MMR complexes to Tdp-ICLs was detected in our in vitro system, we cannot rule out the possibility that the binding of these two complexes on the same molecule may lead to the differential processing of the lesions. For example, some lesions may be repaired in an error-free pathway with MutSβ, while others are repaired via an error-prone pathway with NER, as suggested previously (16,18,20). However, it maybe too simple to classify these protein–protein, and DNA–protein interactions into two independent pathways. In fact, the data presented here suggest that the binding of MutSβ to Tdp-ICLs may facilitate the recruitment of the XPA–RPA complex (required for both global and transcription-coupled NER), but not the XPC–RAD23B complex (required only for global NER) to sites of DNA damage. These results provide a plausible explanation for the requirement for XPA, but not XPC in TFO-induced mutagenesis in mammalian cells (30,31). In normal cells, the concentration of MutSβ is lower than NER proteins [MutSβ: ∼1 × 103 (24); XPC: ∼5 × 104; RPA: ∼3 × 105; XPA: ∼5 × 104 molecules per cell (32–34)]. However, we cannot rule out that during active replication, the participation of MutSβ in ICL processing is likely to take place where the local concentration of MutSβ may be higher, such that an interaction with NER factors on ICLs is favored. In fact, at higher protein concentrations (i.e. 5-fold above the Kapp concentration of either XPC–RAD23B or MutSβ), we observed the formation of a higher-order complex containing both XPC–RAD23B and MutSβ bound to Tdp-ICLs (data not shown), suggesting that MutSβ and XPC–RAD23B may act together to recognize the Tdp-ICL when the relative concentrations of the proteins are higher than that of the damaged DNA substrate.
The canonical function of MMR is to recognize mismatched basepairs during replication to ensure genomic integrity (35). In addition to this function MutSβ has been found to promote trinucleotide repeat (TNR) expansion and to prevent the repair of TNR hairpins, perhaps due to its decreased ATPase activity after binding to imperfect hairpins (36). In addition, MSH2 can bind to repetitive slipped-strand DNA structures and prevent homologous recombination at those sites (37). The psoralen ICL-induced sensitivity of cells deficient in MutSβ, but not in MLH1 demonstrates that MutSβ functions in ICL processing independent of its role in MMR (20,38).
The role of MutSβ in processing ICLs in cells is still not clear. Based on our previous work, it is not unreasonable to assume that the binding of MutSβ to the ICL eventually results in its efficient processing and removal from the DNA (20). In addition to NER and MMR factors, proteins functioning in base excision repair may excise monoadducts induced by psoralen (39,40). ICLs may also be repaired by proteins from different repair pathways in a cell-cycle-dependent fashion (41). ICL processing and repair in human cells is more complicated than in bacterial cells, and likely involves a combination of or competition between proteins functioning in different classified repair pathways. Further investigations on the interaction between MutSβ, NER proteins, and downstream repair factors on ICLs are warranted.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health/NCI grants [CA097175 and CA093729 to K.M.V]. P.M. is an Investigator of the Howard Hughes Medical Institute. Funding for open access charge: CA097175.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mr Juan Culajay and Ms Sarah Henninger for technical assistance. We acknowledge Dr Rick Finch for useful discussions.
REFERENCES
- 1.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol. Cell. Biol. 2000;20:8283–8289. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 3.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 4.Momtaz K, Fitzpatrick TB. The benefits and risks of long-term PUVA photochemotherapy. Dermatol. Clin. 1998;16:227–234. doi: 10.1016/s0733-8635(05)70005-x. [DOI] [PubMed] [Google Scholar]
- 5.Jain A, Wang G, Vasquez KM. DNA triple helices: Biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen LA, Finch RA, Booker AJ, Vasquez KM. Targeting oncogenes to improve breast cancer chemotherapy. Cancer Res. 2006;66:4089–4094. doi: 10.1158/0008-5472.CAN-05-4288. [DOI] [PubMed] [Google Scholar]
- 7.Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl Acad. Sci. USA. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sladek FM, Munn MM, Rupp WD, Howard-Flanders P. In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 1989;264:6755–6765. [PubMed] [Google Scholar]
- 9.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 10.Cheng S, Sancar A, Hearst JE. RecA-dependent incision of psoralen-crosslinked DNA by (A)BC excinuclease. Nucleic Acids Res. 1991;19:657–663. doi: 10.1093/nar/19.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 13.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 14.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutSβ is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair. 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc. Natl Acad. Sci. USA. 2002;99:5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen LA, Wang H, Van Houten B, Vasquez KM. Efficient processing of TFO-directed psoralen DNA interstrand crosslinks by the UvrABC nuclease. Nucleic Acids Res. 2008;36:7136–7145. doi: 10.1093/nar/gkn880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 22.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSalpha.hMutLalpha.heteroduplex complexes. J. Biol. Chem. 2001;276:33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 23.Fujii H, Shimada T. Isolation and characterization of cDNA clones derived from the divergently transcribed gene in the region upstream from the human dihydrofolate reductase gene. J. Biol. Chem. 1989;264:10057–10064. [PubMed] [Google Scholar]
- 24.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Peterson CA, Lu X, Legerski RJ. Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision repair. Mol. Cell Biol. 1995;15:1993–1998. doi: 10.1128/mcb.15.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen J, Cotmore SF, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakasugi M, Sancar A. Order of assembly of human DNA repair excision nuclease. J. Biol. Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 28.Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- 29.Duval-Valentin G, Takasugi M, Helene C, Sage E. Triple helix-directed psoralen crosslinks are recognized by Uvr(A)BC excinuclease. J. Mol. Biol. 1998;278:815–825. doi: 10.1006/jmbi.1998.1728. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, Xu XS, Yang J, Wang G. Defining the function of XPC protein in psoralen and cisplatin-mediated DNA repair and mutagenesis. Carcinogenesis. 2003;24:1111–1121. doi: 10.1093/carcin/bgg051. [DOI] [PubMed] [Google Scholar]
- 32.van der Spek PJ, Eker A, Rademakers S, Visser C, Sugasawa K, Masutani C, Hanaoka F, Bootsma D, Hoeijmakers JH. XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 1996;24:2551–2559. doi: 10.1093/nar/24.13.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seroussi E, Lavi S. Replication protein A is the major single-stranded DNA binding protein detected in mammalian cell extracts by gel retardation assays and UV cross-linking of long and short single-stranded DNA molecules. J. Biol. Chem. 1993;268:7147–7154. [PubMed] [Google Scholar]
- 34.Cleaver JE, Charles WC, McDowell ML, Sadinski WJ, Mitchell DL. Overexpression of the XPA repair gene increases resistance to ultraviolet radiation in human cells by selective repair of DNA damage. Cancer Res. 1995;55:6152–6160. [PubMed] [Google Scholar]
- 35.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen BA, Yang Z, Lai M, Gajec M, Badger JD, 2nd, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, McMurray CT. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 37.Pearson CE, Ewel A, Acharya S, Fishel RA, Sinden RR. Human MSH2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]
- 38.Wu Q, Vasquez KM. Human MLH1 protein participates in genomic damage checkpoint signaling in response to DNA interstrand crosslinks, while MSH2 functions in DNA repair. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000189. e1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couve-Privat S, Mace G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiencke JK, Wiemels J. Genotoxicity of 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) Mutat. Res. 1995;339:91–119. doi: 10.1016/0165-1110(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 41.McHugh PJ, Sarkar S. DNA interstrand cross-link repair in the cell cycle: a critical role for polymerase zeta in G1 phase. Cell Cycle. 2006;5:1044–1047. doi: 10.4161/cc.5.10.2763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.