Abstract
Nucleoside analogs (NAs) represent an important category of prodrugs for the treatment of viral infections and cancer, yet the biological potency of many analogs is compromised by their inefficient activation through cellular 2′-deoxyribonucleoside kinases (dNKs). We herein report the directed evolution and characterization of an orthogonal NA kinase for 3′-deoxythymidine (ddT), using a new FACS-based screening protocol in combination with a fluorescent analog of ddT. Four rounds of random mutagenesis and DNA shuffling of Drosophila melanogaster 2′-deoxynucleoside kinase, followed by FACS analysis, yielded an orthogonal ddT kinase with a 6-fold higher activity for the NA and a 20-fold kcat/KM preference for ddT over thymidine, an overall 10 000-fold change in substrate specificity. The contributions of individual amino acid substitutions in the ddT kinase were evaluated by reverse engineering, enabling a detailed structure–function analysis to rationalize the observed changes in performance. Based on our results, kinase engineering with fluorescent NAs and FACS should prove a highly versatile method for evolving selective kinase:NA pairs and for studying fundamental aspects of the structure–function relationship in dNKs.
INTRODUCTION
In battling existing and newly emerging viral infections and cancer, nucleoside analogs (NAs) represent a prominent and highly potent weapon. Administered as membrane-penetrating nucleosidic prodrugs, the compounds undergo intracellular activation by 2′-deoxynucleoside and 2′-deoxynucleotide kinases into their bioactive triphosphate anabolites. However, inefficient phosphorylation of NAs by endogenous 2′-deoxynucleoside kinases (dNK) limits the potency of existing prodrugs, results in the accumulation of cytotoxic intermediates and is responsible for the failure of a large percentage of new NAs in vivo (1–3). Biochemical and pre-clinical trials have demonstrated that co-administration of an exogenous, broad-specificity dNK via gene therapy can enhance the efficiency of NA prodrug activation (4–8). Nevertheless, even the most promiscuous dNKs from viruses, bacteria and plants show significantly lower activity for NAs compared to their performance with the natural substrates. In addition, the broad substrate specificity of exogenous kinases can create new problems as it interferes with the tightly regulated 2′-deoxynucleoside metabolism. As such, the identification of orthogonal NA kinases, enzymes with changed rather than broader substrate specificity, represents a formidable challenge with great potential benefits for present and future therapeutics.
An alternative to searching for promising dNKs in nature is the application of protein engineering and directed evolution for generating novel enzymes with NA specificity and efficiency. Laboratory evolution experiments to redesign existing kinases have been reported, using site-directed, saturation and random mutagenesis, as well as homologous recombination and chimeragenesis (9–14). However, these efforts have been hampered by the inability to directly screen or select library members for NA phosphorylation. For two decades, researchers in the field have relied on two techniques for dNK library analysis: (i) in vivo selection, using genetic complementation of the auxotrophic Escherichia coli strain KY895 (15) or (ii) in vivo screening on replica plates, testing for cytotoxicity of NAs (9). The auxotrophic selection evaluates library members for thymidine kinase activity, resulting in a bias toward enzymes with, at best, a broader substrate specificity and selecting against truly orthogonal NA kinases. Replica plating, while directly assessing for NA activation, is very laborious and ultimately depends on the cytotoxicity of the phosphorylated NA to the host, a criterion that in our tests in E. coli could not be extended beyond 3′-azido-3′-deoxythymidine (AZT). In light of these biases and functional deficiencies of existing assays, we are arguing that the limited success in identifying active, orthogonal NA kinases can largely be attributed to the inadequate selection or screening techniques.
Inspired by the enzyme library screening methods for novel proteases and glycosyltransferases based on fluorescent substrates (16,17), we envisioned a similar strategy for identifying enzymes that can effectively phosphorylate NAs. Conceptionally, our idea is based on two observations: (i) nucleosides and NAs can be efficiently transported across the cell membrane while their corresponding monophosphates, the product of the kinase-catalyzed phosphoryl transfer, are trapped inside the host cytoplasm and (ii) fluorescent nucleoside analogs (fNAs) with an excitation maxima of >300 nm, which reduces signal interference due to cellular autofluorescence, can serve as substrates for dNKs. While the nucleobases of natural nucleosides do possess intrinsic fluorescence properties under physiological conditions, direct measurements are impractical due to the compounds’ low quantum yields and overlapping absorption maxima with aromatic amino acids in proteins and small-molecule metabolites such as ATP and NADH. Fortunately, relatively small synthetic modifications of a nucleoside's pyrimidine or purine moiety can red-shift its absorption spectrum and increase the quantum yield, enabling detection of these fNAs with high sensitivity in complex mixtures such as the cytoplasm. As summarized in Figure 1, a number of fluorescent 2′-deoxyribonucleosides, used in DNA structure studies, have been reported including the etheno-derivative of adenine (8) (18–20), as well as furano (6) and pyrrolo-pyrimidines (7) and pterines (9,10) (21–23). We predict that the combination of these modified nucleobases with sugar derivatives (11–16), found in NA prodrugs, generates suitable substrates for dNK library screening. As outlined in Figure 2, when bacteria that express individual dNK library members are incubated with a fNA, broad-specificity nucleoside transporters facilitate the efficient translocation of the fluorophor across the membrane (24,25). Production of fNA-monophosphate in the presence of a functional fNA kinase will result in accumulation of the fluorophor in the cytoplasm as its negative charge prevents cellular export. Fluorescence-activated cell sorting (FACS) can subsequently be employed to identify and isolate hosts with the highest fluorescence intensity (17,26).
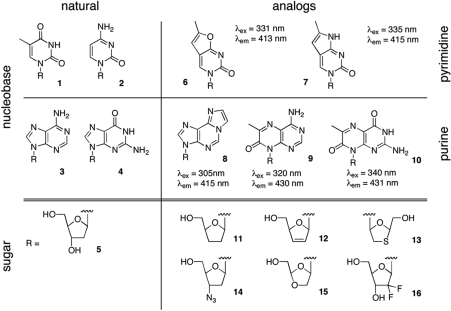
Figure 1.
Overview of modular nucleoside design. In many nucleoside analogs, a natural pyrimidine (1,2) or purine (3,4) moiety is docked to a sugar analog (11–16) instead of the native 2′-deoxyribosyl portion (5). For the creation of fluorescent NAs, an extension of the heterocycles, as exemplified in furano and pyrrolo-pyrimidines (6,7), as well as etheno-adenine (8) and pterines (9,10), red-shifts the excitation/emission spectrum of the nucleobase and increases the quantum yield. When coupled to sugar analogs (11–16), these fluorescent versions of the NA become useful molecular probes for studying their cellular uptake and metabolism (25).
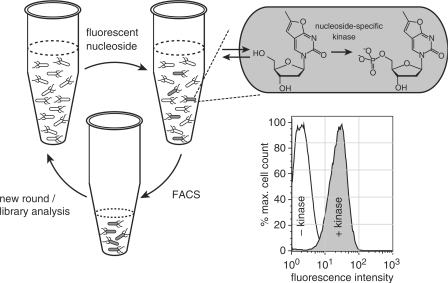
Figure 2.
Schematic overview of the kinase library screening assay with fluorescent NAs. Upon incubation of bacteria with the target fNA, cells that carry a functional kinase variant for the specific substrate will accumulate the fNA monophosphate. Hosts cells with the highest fluorescence intensity can be identified and isolated by fluorescence-activated cell sorting (FACS) for subsequent characterization of the library member or, if desirable, another round of directed evolution. For concept validation, E. coli TOP10 expressing exogenous DmdNK with thymidine kinase activity (+kinase) was compared to host expressing thymidine kinase-deficient human dCK (−kinase). The histogram shows the 20- to 30-fold increase in cellular fluorescence upon incubation with fT in the presence of the exogenous thymidine kinase activity.
We herein report the design and validation of this FACS-based NA kinase screening method, followed by its application for identifying an orthogonal 3′-deoxythymidine (ddT, 1 + 11) kinase from directed evolution libraries of Drosophila melanogaster 2′-deoxynucleoside kinase (DmdNK). We selected ddT for these proof-of-principle experiments as it represents a good example of an NA prodrug whose biological function is compromised due to lack of phosphorylation. As a member of the first generation of NAs tested for antiviral therapy, its chemically synthesized triphosphate proved an effective inhibitor of cellular and viral DNA polymerases in vitro (27,28). Nevertheless, cell culture experiments could not reproduce the high level of antiviral activity, an observation that was largely attributed to inefficient activation by cellular kinases in vivo (28,29).
As a starting point for enzyme evolution, we chose DmdNK, a member of the type-I dNK subfamily that includes the 2′-deoxycytidine kinase, 2′-deoxyguanosine kinase and thymidine kinase 2 from human, as well as several dNKs from insects (30,31). Type-1 dNKs share a common fold but vary widely in their substrate specificities and overall catalytic efficiencies. DmdNK distinguishes itself by showing some of the highest activity among type-1 dNKs for the four natural 2′-deoxyribonucleosides, as well as several NAs (31,32).
MATERIALS AND METHODS
Synthesis of fddT
The fluorescent ddT was prepared in six steps. The details of the experimental procedures and characterization of the intermediates are described in the Supplementary Data.
Directed evolution library construction
Random mutagenesis libraries of wild-type DmdNK were created with the GeneMorph II kit (Stratagene, La Jolla, CA, USA), using an average mutation frequency of two to four nucleotide changes per gene (as determined by DNA sequencing). DNA shuffling libraries were prepared by following the nucleotide exchange and excision technology (NExT) protocol with 33% UTP incorporation (33). DNA fragments from the mutagenesis libraries and parental DmdNK were mixed at a 20:1 ratio for reassembly. Re-engineering of library members for function studies via site-directed mutagenesis was performed by overlap extension PCR, following standard protocols (34). Final PCR products were cloned into pBAD-HisA (Invitrogen, Carlsbad, CA, USA) via NcoI and HindIII restriction sites and electroporated into E. coli TOP10 [F-mcrA D(mrr-hsdRMS-mcrBC) F80lacZDM15 DlacX74 recA1 araD139 D(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG] (Invitrogen, Carlsbad, CA, USA).
Library screening by fluorescence-activated cell sorting
Library screening was performed by inoculating 2 ml LB media supplemented with ampicillin (50 µg/ml) at 37°C until the OD (600 nm) reached ∼0.5. The protein expression was induced with arabinose (0.2%) and incubation was continued for 4 h. The cell culture was mixed with 5–80 µM fNA. For competition experiments, the sample was complemented with an additional 50–800 µM thymidine. After incubation at 37°C for 30–120 min, cells were centrifuged and the pellet was washed three times with the PBS buffer (pH 7.4) before being resuspended in the PBS buffer to ∼1 × 108 cells/ml. Cell sorting was performed on a FACSVantage flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). The event detection was set to forward and side scattering. Sorting was performed at <2000 events/s with excitation by a UV laser (351–364 nm) and emission detection through a band pass filter (424 ± 20 nm). Cells were collected in Eppendorf tubes, plated on LB-agar plates and harvested for the DNA sequence analysis and subsequent rounds of directed evolution.
Kinetic analysis
Selected candidates were subcloned into pMAL-C2x (New England Biolabs, Beverly, MA, USA) for protein overexpression and purification by amylose affinity chromatography according to the manufacturer's protocol. The isolated proteins were >95% pure as determined by SDS–PAGE. Spectrophotometric coupled-enzyme assays to measure substrate phosphorylation were performed at 37°C as described previously (34). The substrate concentration varied from 1 µM to 7 mM. All experiments were performed in triplicates and data were fit to the Michaelis–Menten equation using Origin7 software (OriginLab, Northhampton, MA, USA).
RESULTS
Fluorescent substrates
While the fluorescent analog of thymidine (fT, 5 + 6) is commercially available, a synthetic route for fluorescent ddT (fddT, 6 + 11) was established. Starting with uridine, we used a three-step synthesis to eliminate the 2′ and 3′-hydroxyl groups in the ribose moiety, followed by the assembly of the furano-pyrimidine portion via palladium-catalyzed cross-coupling chemistry (Figure S1 and Supplementary Methods). Consistent with the literature, the excitation and emission wavelengths for both fluorescent nucleosides have red-shifted to 331 nm and 413 nm, respectively. Separately, we evaluated the impact of the larger size of the furano pyrimidine moiety, as well as changes in the protonation of the pyrimidine ring, on the kinetics of DmdNK (Table 1). In comparison to T and ddT, the enzyme's catalytic efficiency with the corresponding fluorescent substrates is reduced by a moderate 3-fold. Based on the steady-state kinetics, the decline in performance is largely attributed to lower turnover rates.
Table 1.
Comparison of normal and fluorescent substrate phosphorylation by 2′-deoxynucleoside kinase
| Substrate | Kcat (s−1) | KM (μM) | Kcat/KM (103 × s−1 M−1) |
|---|---|---|---|
| T | 12.9 ± 0.9 | 2.7 ± 0.5 | 4813 |
| fT | 5.6 ± 0.3 | 3.4 ± 0.7 | 1647 |
| ddT | 0.53 ± 0.03 | 115 ± 22 | 4.6 |
| fddT | 0.18 ± 0.01 | 139 ± 24 | 1.3 |
Kinetic parameters of 2′-deoxyribonucleoside kinase from D. melanogaster for thymidine (T) and 3′-deoxythymidine (ddT), as well as their corresponding fluorescent analogs, fT and fddT.
FACS-based screening
Implementing the proposed kinase screen by cellular entrapment of fluorescent NAs is critically dependent on three aspects regarding the host organism. Firstly, the host must be able to efficiently take up fluorescent and non-fluorescent substrates. Secondly, it should be devoid of endogenous kinases with activity for the fluorescent nucleosides to improve the signal-to-noise ratio and avoid false positives. Finally, the host must facilitate the expression of functional 2′-deoxynucleoside kinases.
Escherichia coli is a host organism that meets all three criteria. The bacterium possesses broad-specificity nucleoside transporters that facilitate efficient transmembrane equilibration of regular NAs and their fluorescent counterparts including furano-pyrimidine derivatives (24). Furthermore, no interference of host enzymes with the fddT was detected. Although E. coli carries an endogenous thymidine kinase, the enzyme belongs to a separate subfamily [type-II (35,36)] with very strict substrate specificity and no detectable in vitro activity for the fluorescent T or ddT analogs (data not shown). These findings were confirmed by in vivo validation experiments described below that did not observe intracellular accumulation of fluorescent nucleosides in the absence of an exogenous kinase. Finally, E. coli is a well-established host organism for the overexpression of numerous exogenous kinases including DmdNK and human 2′-deoxycytidine kinase (dCK) (34,37). These two enzymes were used in our validation experiments with fT; DmdNK is the positive control as it possesses high thymidine kinase activity and dCK is the negative control with no detectable activity for thymidine.
The validation of our fNA screen also included the evaluation of multiple expression vectors with various promoters to identify suitable conditions for tunable and homogeneous kinase production. Initial experiments were performed with DmdNK under control of lac and T7 promoters (Figure S2). Following induction of the protein expression, bacterial cultures were incubated with fT and analyzed by flow cytometry. The latter data indicate highly variable protein expression levels in individual cells in the bacterial population. In contrast, uniformly high kinase expression was observed in the same experiments, using E. coli TOP10 in combination with DNA vectors carrying the tightly regulated arabinose PBAD promoter (38). Consequently, all experiments for validation of our screening method and for analysis of the directed evolution libraries were performed in this expression system.
A typical flow cytometry histogram for the host carrying either dCK or DmdNK is shown in Figure 2. Under identical assay conditions, the presence of an exogenous dNK with thymidine kinase activity results in a 20- to 30-fold increase in cellular fluorescence upon incubation with fT. More importantly, the signal difference is sufficient to allow for ∼1000-fold enrichment of E. coli cells carrying a kinase with the desired substrate specificity through cell sorting in the flow cytometer. The enrichment factor was calculated based on results from sorting experiments with defined mixtures of DmdNK and dCK (Figure S3). Separately, we verified the absence of cytotoxic effects of the fNAs. The viability of bacteria expressing DmdNK in the presence of up to 200 µM of the fluorescent analog was unaffected (data not shown).
Directed evolution
Next, our FACS-based screening technique was applied toward the laboratory evolution of an orthogonal ddT kinase, starting with the fruitfly enzyme DmdNK. Following three rounds of random mutagenesis and one round of DNA shuffling, libraries were screened for kinases with increased activity for fddT. Evaluating between 300 000 and 500 000 cells per round of FACS, the sorting gate was set to collect the top 0.5% of highly fluorescent cells. The selection pressure for NA activity was gradually raised over subsequent rounds of screening. To favor kinases that tightly bind fddT, its concentration was lowered from initially 80 µM to 5 µM in the final round. Furthermore, the exposure time of cells to fluorescent substrate was shortened from 120 to 30 min to favor enzymes with higher fNA turnover. Finally, the addition of up to 800 µM thymidine to compete with fNA binding gave preference to library members with reduced affinity for the native substrates.
After the final round of directed evolution, 13 selected candidates were characterized by DNA sequencing. The analysis indicated that the genes carry four to eight expressed mutations (Figure S4). Besides several one-time substitutions, eight positions (T85M, S123L, A147V, E172V, Y179F, H193Y, Q198H and S224C) were found in two or more sequences. Among these variants, the substitutions in positions 147, 198 and 224 were considered neutral, involving conservative changes in regions distant to the active site, and were therefore excluded from further analysis. A preliminary screening of the remaining enzymes for substrate specificity quickly identified the three variants with a mutation in position E172 as the most promising candidates. Variant R4.V1 (round 4; variant 1) carries a total of eight amino acid substitutions (T85M, S123L, A147V, E172V, D189E, H193Y, N210D, S223G), R4.V3 contains four changes (T85M, E172V, Y179F and H193Y) and R4.V9 has six mutations (T85M, S123L, N130D, E172V, H193Y and L203I). The three variants were overexpressed and purified to homogeneity for subsequent kinetic characterization.
Kinetic analysis
For the overexpression of the selected kinases, library members were fused to maltose-binding protein (MBP) as a purification tag that has the additional benefit of increasing enzyme stability. We established in separate experiments that the N-terminal MBP does not have a significant effect on the kinetic properties of the wild-type enzymes (data not shown). Following purification, we measured the steady-state kinetics of R4.V1, R4.V3 and R4.V9 to evaluate their catalytic performance with the four natural 2′-deoxynucleosides, as well as the two NAs ddT and fddT (Table 2). All three candidates showed a similar overall shift in the catalytic performance. For T and dC, the kcat/KM values declined by three to four orders of magnitude. The weaker performance resulted from a drop in catalytic activities and higher Michaelis–Menten constants. In the case of dA and dG, the catalytic efficiency dropped by two to three orders of magnitude, largely due to declines in turnover. In contrast, the catalytic performance of the three variants with ddT and fddT remained unchanged relative to DmdNK (<2-fold). While the kinetic parameters for ddT stayed the same, the KM values for fddT dropped 5- to 10-fold but were accompanied by a decline in activity of similar magnitude, leaving the specificity constant unchanged.
Table 2.
Kinetic properties of wild-type and engineered kinases
|
DmdNK |
R4.V1 (T85M, S123L, A147V, E172V, D189E, H193Y, N210D, S223G) |
R4.V3 (T85M, E172V, Y179, H193Y) |
R4.V9 (T85M, S123L, N130D, E172V, H193Y, L2031) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat/KM | kcat/KM | kcat/KM | kcat/KM | |||||||||
| kcat (s−1) | KM (μM) | (103 × s−1 M−1) | kcat (s−1) | KM (μM) | (103 × s−1 M−1) | kcat (s−1) | KM (μM) | (103 × s−1 M−1) | kcat (s−1) | KM (μM) | (103 × s−1 M−1) | |
| T | 12.9 ± 0.9 | 2.7 ± 0.5 | 4813 | 0.09 ± 0.01 | 286 ± 27 | 0.3 | 0.03 ± 0.003 | 319 ± 88 | 0.1 | 0.08 ± 0.01 | 111 ± 14 | 0.7 |
| (−143) | (−106) | (−16 000) | (−430) | (−118) | (−48 000) | (−160) | (−41) | (−6875) | ||||
| dC | 11.7 ± 0.7 | 2.0 ± 0.4 | 5850 | 0.09 ± 0.01 | 156 ± 7 | 0.6 | 0.03 ± 0.002 | 95 ± 19 | 0.36 | 0.11 ± 0.01 | 46 ± 13 | 2.3 |
| (−130) | (−78) | (−9750) | (−390) | (−48) | (−16 250) | (−106) | (−23) | (−2543) | ||||
| dA | 15.8 ± 0.4 | 98 ± 3 | 161 | 0.08 ± 0.01 | 1560 ± 346 | 0.05 | <0.01 | >3000 | – | 0.16 ± 0.03 | 1734 ± 619 | 0.09 |
| (−197) | (−16) | (−3220) | – | – | – | (−99) | (−18) | (−1789) | ||||
| dG | 12.3 ± 0.4 | 450 ± 54 | 28 | 0.04 ± 0.01 | 1180 ± 300 | 0.03 | <0.04 | >3000 | – | 0.19 ± 0.01 | 1545 ± 154 | 0.12 |
| (−307) | (−2.6) | (−933) | – | – | – | (−65) | (−3.4) | (−233) | ||||
| ddT | 0.53 ± 0.03 | 115 ± 22 | 4.6 | 0.27 ± 0.02 | 102 ± 33 | 2.6 | 0.49 ± 0.01 | 167 ± 9 | 3 | 0.24 ± 0.01 | 101 ± 21 | 2.4 |
| (−2) | (+1.1) | (−1.8) | (−1.1) | (−1.5) | (−1.5) | (−2.2) | (+1.1) | (−1.9) | ||||
| fddT | 0.18 ± 0.01 | 139 ± 24 | 1.3 | 0.020 ± 0.001 | 25 ± 3 | 0.8 | 0.021 ± 0.001 | 10 ± 1 | 1.9 | 0.017 ± 0.001 | 13 ± 2 | 1.3 |
| (−9) | (+5.6) | (−1.6) | (−9) | (+14) | (+1.5) | (−11) | (+11) | (1) | ||||
Kinetic parameters for natural 2′-deoxyribonucleosides (T, dC, dA, dG) and nucleoside analogs (ddT, fddT) of wild-type enzyme (DmdNK) and selected candidates from the directed evolution experiments after four rounds. Numbers in parentheses are fold change in catalytic efficiency for the particular substrate (kcat/KM [variant]/kcat/KM [DmdNK]).
Among the three kinase variants, R4.V3 showed the most favorable change in performance. The substrate specificity of R4.V3, defined by [kcat/KM(NA)]/[kcat/KM(T)], indicates a 20- and 30-fold preference for ddT and fddT over T. These results suggest a change in substrate specificity by over four orders of magnitude, effectively reversing the enzyme's substrate preference compared to DmdNK whose kinetic data show a 1000- to 3700-fold greater performance for T over fddT and ddT, respectively. Intrigued by the observation that the fewest number of mutations cause the most significant changes in function, we further investigated the individual contributions of the four mutations in R4.V3 by reverse engineering.
Reverse engineering
The role of the four mutations in R4.V3 (Figure 3) was evaluated by reversing individual amino acid substitutions, followed by overexpression, purification and kinetic characterization of the new variant (Table 3). The most dramatic change was observed in R4.V3-[172]. Replacing V172 with the original glutamate raises the level of activity for the native substrates by two to three orders of magnitude. R4.V3-[172] shows parent-like turnover numbers but 2- to 16-fold higher KM values compared to DmdNK, compromising its wild-type-like overall performance. The F179Y substitution in R4.V3-[179] also contributes toward the recovery of the wild-type activity, yielding a 10-fold higher catalytic efficiency than R.V3 for the native substrates as a result of improvements in kcat and KM values. Together, the two mutations are responsible for R4.V3 discriminating against natural substrates, yet not even one of the two substitutions seems to have a major effect on the kinetic parameters for ddT (<2-fold).
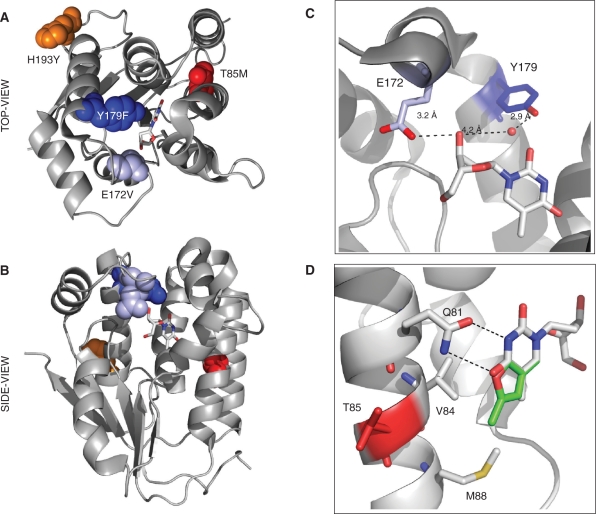
Figure 3.
Structure model of R4.V3. The locations of the four mutations are highlighted in DmdNK with thymidine, bound in the phosphoryl acceptor-binding site (PDB access no.: 1OT3 (40)). The top (A) and side view (B) of DmdNK show the close proximity of E172V (light blue) and Y179F (dark blue) to the substrate while H193Y (orange) and T85M (red) are located in more distant locations to the active site. (C) The close-up view of the active site shows the direct hydrogen-bonding interactions of E172 with the 3′-OH group of the substrate's ribose moiety. The OH group of Y179 contacts the same position on the substrate via bridging water. (D) The modeled overlay of thymidine (gray) and fT (green) in the active site of DmdNK indicates a potential steric clash of the fluorescent substrate's furano moiety with V84 and M88. The T85M mutation is believed to cause a slight conformational shift of the helical region, creating a more favorable binding pocket for substrates with modifications in the C4/C5 position of the nucleobase. Figures were created with PyMOL software (DeLano Sci., Palo Alto, CA).
Table 3.
Impart of individual mutations on kinetic properties of engineered kinase
| R4.V3-[85] |
R4.V3-[172] |
R4.V3-[179] |
R4.V3-[193] |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat/KM | kcat/KM | kcat/KM | kcat/KM | |||||||||
| kcat (s−1) | KM (μM) | (103 × s−1 M−1) | kcat (s−1) | KM (μM) | (103 × s−1 M−1) | kcat (s−1) | KM (μM) | (103 × s−1 M−1) | kcat (s−1) | KM (μM) | (103 × s−1 M−1) | |
| T | 0.13 ± 0.01 | 92 ± 14 | 1.4 | 13.9 ± 0.9 | 43 ± 9 | 326 | 0.22 ± 0.01 | 188 ± 29 | 1.2 | 0.08 ± 0.01 | 251 ± 36 | 0.3 |
| (−100) | (−34) | (−3438) | (−1.1) | (−22) | (−14) | (−58) | (−70) | (−4000) | (−161) | (−93) | (−16 000) | |
| dC | 0.13 ± 0.01 | 334 ± 65 | 0.39 | 11.7 ± 0.5 | 4 ± 1 | 2925 | 0.15 ± 0.01 | 35 ± 3 | 4.3 | 0.05 ± 0.01 | 105 ± 15 | 0.46 |
| (−90) | (−167) | (−15 000) | (1) | (−2) | (−2) | (−78) | (−18) | (−1360) | (−234) | (−53) | (−12 700) | |
| dA | <0.01 | >3000 | – | 12.4 ± 0.7 | 218 ± 36 | 57 | 0.25 ± 0.05 | 3000 ± 500 | 0.08 | <0.01 | >3000 | – |
| – | – | – | (−1.2) | (−2) | (−2.5) | (−63) | (−31) | (−2000) | – | – | – | |
| dG | <0.01 | >3000 | – | 4.3 ± 0.1 | 721 ± 22 | 6 | 0.06 ± 0.02 | 1500 ± 200 | 0.04 | 0.05 ± 0.01 | 1100 ± 243 | 0.05 |
| – | – | – | (−3) | (−1.6) | (−4.5) | (−200) | (−3) | (−700) | (−246) | (−2.5) | (−560) | |
| ddT | 1.36 ± 0.01 | 49 ± 3 | 28 | 0.49 ± 0.03 | 186 ± 28 | 2.6 | 0.48 ± 0.01 | 124 ± 15 | 3.9 | 0.84 ± 0.02 | 154 ± 16 | 5.5 |
| (+2.6) | (+2.3) | (+6) | (−1.1) | (−1.6) | (−1.8) | (−1.1) | (−1.1) | (−1.2) | (+1.7) | (−1.3) | (+1.2) | |
| fddT | 0.03 ± 0.002 | 81 ± 17 | 0.4 | 0.10 ± 0.01 | 14 ± 3 | 7.5 | 0.04 ± 0.01 | 14 ± 1 | 2.5 | 0.02 ± 0.01 | 10 ± 1 | 2.5 |
| (−6) | (+1.7) | (−3) | (−1.8) | (+10) | (+5.7) | (−4.5) | (+10) | (−2) | (−9) | (+14) | (−2) | |
Kinetic parameters of R4.V3 with natural 2′-deoxyribonucleosides (T, dC, dA, dG) and nucleoside analogs (ddT, fddT) after reverse engineering the four mutations. Numbers in parenthesis are fold change in catalytic efficiency for the particular substrate (kcat/KM [variant]/kcat/KM [DmdNK]).
As for the two mutations distant to the active site, swapping Y193 back to the original histidine in R4.V3-[193] leads to only minor changes in the kinetic performance, indicating that the mutation is not relevant for the observed changes in substrate specificity. In contrast, the reversion of T85M shows an unexpected effect on the enzyme's substrate specificity. While leaving the catalytic efficiency for 2′-deoxycytidine and the purine substrates unaffected, R4.V3-[85] exhibits 10- and 14-fold higher kcat/KM values for ddT and T, respectively. For both substrates, the improvements result from approximately 3-fold higher turnover numbers and 3-fold lower apparent binding constants, raising questions about the selection for T85M during the directed evolution experiments. An indicator for the functional role of position 85 is provided by the change in the kinetic properties of R4.V3-[85] for fddT. The enzyme's 8-fold rise in KM(fddT) compared to R4.V3, which translates into a 5-fold lower catalytic efficiency, hints that the substitution is involved in remodeling the substrate nucleobase binding pocket.
DISCUSSION
Deoxynucleoside kinases are important targets for protein engineering, due to their role in NA prodrug activation and potential application in suicide gene therapy. Ideally, an engineered kinase should exhibit high activity for the NA while effectively discriminating against phosphorylation of native substrates to avoid unbalancing the cellular dNTP pool (39). In the absence of known native kinases for NAs, the tailoring of enzymes by various directed evolution strategies have been attempted, yet has had limited success due to a lack of selection or screening methods to directly identify library members with the desired NA activity (9–14). Here, we have established a novel screening method, using fluorescent analogs of natural 2′-deoxynucleosides and NAs in combination with FACS, to identify and isolate enzymes with enhanced specificity and activity for these substrates.
For the proof of principle, we used fddT as the target substrate to search for an orthogonal ddT kinase. After four rounds of directed evolution, kinetic analysis of selected candidates revealed a dramatic shift from the parental enzyme's preference for the 2′-deoxyribosyl moiety to the 2′,3′-dideoxyribose derivative. Our most successful variant R4.V3 shows a change in substrate specificity by more than four orders of magnitude, now favoring ddT over dC and T by an 8- and 30-fold higher catalytic efficiency, respectively. As such, our screening protocol has been effective in identifying ‘loss-of-function’ variants that gain NA specificity by suppressing efficient phosphorylation of natural 2′-deoxynucleosides. Nevertheless, a desired simultaneous ‘gain-in-function’ for fddT (and ddT) was not observed in R4.V3. While the 10-fold increase in kcat/KM for ddT in R4.V3-[T85] indicates the possibility for improvement, we hypothesize that more substantial increases in activity for NAs with a 2′,3′-dideoxyribose moiety might be difficult to achieve due to the sugar's deficiency in functional groups that compromises productive substrate binding.
Nevertheless, our evolved ddT kinase performs very favorably under physiological conditions. Despite the largely unchanged kinetic parameters of R4.V3 for ddT and fddT in vitro, the true functional benefit of R4.V3 in regard to selective ddT phosphorylation is apparent under competitive substrate conditions in vivo (Figure 4). The incubation of bacteria, expressing either R4.V3 or DmdNK, with the same fddT/T substrate mixture shows substantially higher cellular fluorescence in the hosts carrying the engineered kinase (Figure 4B). Rather than selecting a candidate with a greatly enhanced catalytic activity for fddT, the ability of R4.V3 to eliminate the competition of natural 2′-deoxynucleosides for occupancy of the active site seems an equally successful strategy for increasing NA phosphorylation in vivo. As the intracellular concentration of natural deoxynucleosides normally does not exceed 10 µM (39), the KM values of R4.V3 above 90 µM for the native substrates should largely eliminate competition for the active site.
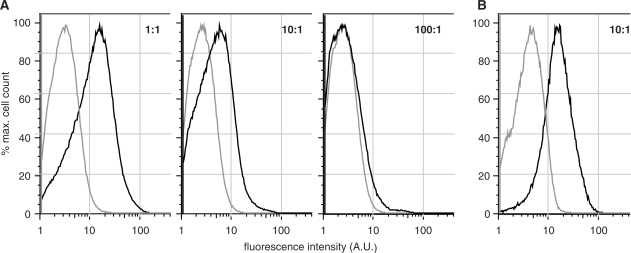
Figure 4.
In vivo competition experiments in E. coli, studying the cellular accumulation of fluorescent ddT in the presence of increasing amounts of thymidine (1–100-fold molar excess of thymidine). (A) Bacteria expressing members of the fourth-round kinase library (black line) consistently outperform cells expressing DmdNK (gray line). At 80 µM fddT + 80 µM thymidine (1:1), the library shows approximately 10-fold higher fluorescence intensity over the wild-type enzyme. Although the favorable ratio decreases with increasing thymidine concentration, a direct consequence of the two substrates’ competition for the active site, the evolved kinases slightly surpass DmdNK even at 8 mM thymidine (100:1). (B) Cellular accumulation of fddT (10 µM) in bacteria expressing R4.V3 (black) or DmdNK (gray) in the presence of a 10-fold molar excess of thymidine (100 µM). The 5-fold higher fluorescence intensity for R4.V3 confirms kinetic data of improved substrate specificity for the evolved kinase and demonstrates its relevance to performance in the complex cellular environment.
We next evaluated the contributions of the four mutations in R4.V3 toward the observed functional changes by reverse engineering. The substitutions in positions 172 and 179 show the most dramatic impact on substrate specificity and are likely a direct consequence of the ribose modifications in fddT (Figure 3C). In DmdNK, the two residues are part of the active site and, according to the crystal structure, form hydrogen-bonding interactions directly (E172) or via a water molecule (Y179) to the 3′-hydroxyl group of the phosphoryl acceptor (40). Mutations in these two otherwise highly conserved positions have not previously been observed in engineered kinases, a fact that we attribute to biases resulting from the library analysis for thymidine kinase activity in the E. coli auxotroph KY895. The two active site mutations E172V and Y179F not only eliminate the favorable hydrogen-bonding interactions but also actively discriminate against the native substrates by optimizing the binding pocket for the more hydrophobic 2′,3′-dideoxyribose moiety of ddT. Consistent with this interpretation, our kinetic data suggest that simultaneous reversion of these two mutations restores the catalytic activity for the native substrates to wild-type levels (Table 3).
In contrast, the changes in residues 85 and 193 are not obvious as the two positions lack proximity to the substrate. While our study confirms the mutation in position 193 as neutral, the reversion of T85M in R4.V3-[85] results in a 10-fold improvement in the catalytic performance for ddT and T while diminishing kcat/KM for fddT by raising KM by a similar magnitude. Based on the observation that the effect is thymine specific, we argue that T85M is a response to substitutions in C-4/5 of the nucleobase of fddT. The introduction of the furano moiety in our fNAs adds an extended, sterically rigid substituent at C-4/5 of the pyrimidine. This extra bulk leads to unfavorable interactions and potential steric clashes with side chains of V84 and M88 in the active site binding pocket, an effect that the enzyme could compensate for by mutating the neighboring T85 (Figure 3D). As the side chain of T85 points away from the active site, packing tightly against a neighboring helix in the protein structure, we speculate that the mutation at position 85 results in a slight repositioning of the helix that carries residues 84–88, thereby enlarging the binding pocket and allowing for more favorable binding interactions of the furano–pyrimidine moiety. Mutations in position 85 have previously been observed in the engineering of DmdNK and were rationalized by the residue's importance in defining the topology of the phosphoryl acceptor-binding site (11). While efforts to further investigate this hypothesis are in progress, the results from our reverse engineering indicate that the effect of T85M is strictly additive, enabling us to generate an improved kinase for the non-fluorescent ddT by simply reversing this mutation. Furthermore, the kinetic data for R4.V3-[85] show that the two active site mutations in positions 172 and 179 are not neutral in regard to ddT phosphorylation but are responsible for a 6-fold increase in the catalytic efficiency over the parental DmdNK.
The comparison of our engineered kinases with variants from previous directed evolution experiments further emphasizes the benefits of direct screening for NA activation. The best variants of HSV–thymidine kinase (9,13) and DmdNK (11,12,41) typically show <2-fold gains in kcat/KM for the NA. Furthermore, mutants selected by genetic complementation or cytotoxicity screening retain high activity for the native substrate. The most selective mutants of HSV–thymidine kinase show a preference for NA over thymidine (kcat/KM(NA) or kcat/KM(thymidine)) of 0.63 for AZT (9) and 0.17 for ganciclovir (13). For DmdNK, the highest NA preference reported for AZT is 0.14 (11) while ddC reaches 0.39 (41). In contrast, the NA preference of R4.V3[T85] for ddT is 20. The ability of an evolved NA kinase to discriminate against native 2′-deoxynucleosides is advantageous as it improves its performance in vivo by reducing competition for substrate binding to the enzyme active site. As shown in Figure 4, a series of flow cytometry experiments demonstrates the preferred phosphorylation of fddT by R4.V3, even in the presence of up to 100-fold molar excess of the competing native thymidine. In addition, the elevated specificity also reduces the risk of potential side effects due to fluctuations in the cellular dNTP concentration. Although the long-term effects of auxiliary dNKs on a host organism were not addressed in our study, preventing the unregulated phosphorylation of natural 2′-deoxynucleosides should minimize their potential to interfere with other metabolic processes and DNA replication fidelity.
Finally, we anticipate directed evolution of kinases in combination with fNA screening to be a general strategy for identifying orthogonal NA kinases. Similar to the synthesis of fddT, other sugar moieties (for example 12–16) can be combined with one of the fluorescent nucleobases (6–10), giving access to a large number of potential substrates for FACS-based screening of kinase libraries. Similarly, the screening protocol can be adapted to other members of the type-1 dNK family. Human dNKs are of particular interest for creating orthogonal NA kinases as they reduce the risk of undesirable immunogenic response in clinical applications. Separately, the screening of kinase libraries with fluorescent substrates could be adapted to the eukaryotic expression system such as Saccharomyces cerevisiae or mammalian cell lines. Both are well suited for FACS and could further optimize the search process for novel NA kinases. Such novel kinases could not only improve the potency of existing prodrugs but also offer a second chance for NAs that failed in previous clinical studies due to a lack of phosphorylation by endogenous dNKs. As such, the development of kinases tailored to a NA of choice sets the stage for clinical studies testing the co-administration of enzyme plus small-molecule prodrug and potentially introduces a large number of new antiviral and cancer prodrugs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (GM69958 to S.L.); a grant to the Emory Center for AIDS Research (AI050409) from the National Institutes of Health and institutional funding from the Emory University Health Science Center. Funding for open access charge: National Institutes of Health (GM69958).
Conflict of interest statement. None declared
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Lutz laboratory for helpful discussions and comments on the manuscript.
REFERENCES
- 1.Arner ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol. Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 2.Gentry GA. Viral thymidine kinases and their relatives. Pharmacol. Ther. 1992;54:319–355. doi: 10.1016/0163-7258(92)90006-l. [DOI] [PubMed] [Google Scholar]
- 3.Shi J, McAtee JJ, Schlueter Wirtz S, Tharnish P, Juodawlkis A, Liotta DC, Schinazi RF. Synthesis and biological evaluation of 2′,3′-didehydro-2′,3′-dideoxy-5-fluorocytidine (D4FC) analogues: discovery of carbocyclic nucleoside triphosphates with potent inhibitory activity against HIV-1 reverse transcriptase. J. Med. Chem. 1999;42:859–867. doi: 10.1021/jm980510s. [DOI] [PubMed] [Google Scholar]
- 4.Culver KW, Ram Z, Wallbridge S, Ishii H, Oldfield EH, Blaese RM. In vivo gene transfer with retroviral vector-producer cells for treatment of experimental brain tumors. Science. 1992;256:1550–1552. doi: 10.1126/science.1317968. [DOI] [PubMed] [Google Scholar]
- 5.Jordheim LP, Galmarini CM, Dumontet C. Gemcitabine resistance due to deoxycytidine kinase deficiency can be reverted by fruitfly deoxynucleoside kinase, DmdNK, in human uterine sarcoma cells. Cancer Chemother. Pharmacol. 2006;58:547–554. doi: 10.1007/s00280-006-0195-8. [DOI] [PubMed] [Google Scholar]
- 6.Moolten FL. Tumor chemosensitivity conferred by inserted herpes thymidine kinase genes: paradigm for a prospective cancer control strategy. Cancer Res. 1986;46:5276–5281. [PubMed] [Google Scholar]
- 7.Solaroli N, Johansson M, Balzarini J, Karlsson A. Enhanced toxicity of purine nucleoside analogs in cells expressing Drosophila melanogaster nucleoside kinase mutants. Gene Ther. 2007;14:86–92. doi: 10.1038/sj.gt.3302835. [DOI] [PubMed] [Google Scholar]
- 8.Zheng X, Johansson M, Karlsson A. Retroviral transduction of cancer cell lines with the gene encoding Drosophila melanogaster multisubstrate deoxyribonucleoside kinase. J. Biol. Chem. 2000;275:39125–39129. doi: 10.1074/jbc.M006212200. [DOI] [PubMed] [Google Scholar]
- 9.Christians FC, Scapozza L, Crameri A, Folkers G, Stemmer WP. Directed evolution of thymidine kinase for AZT phosphorylation using DNA family shuffling. Nat. Biotechnol. 1999;17:259–264. doi: 10.1038/7003. [DOI] [PubMed] [Google Scholar]
- 10.Gerth ML, Lutz S. Non-homologous recombination of deoxyribonucleoside kinases from human and Drosophila melanogaster yields human-like enzymes with novel activities. J. Mol. Biol. 2007;370:742–751. doi: 10.1016/j.jmb.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knecht W, Munch-Petersen B, Piskur J. Identification of residues involved in the specificity and regulation of the highly efficient multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster. J. Mol. Biol. 2000;301:827–837. doi: 10.1006/jmbi.2000.3990. [DOI] [PubMed] [Google Scholar]
- 12.Knecht W, Sandrini MP, Johansson K, Eklund H, Munch-Petersen B, Piskur J. A few amino acid substitutions can convert deoxyribonucleoside kinase specificity from pyrimidines to purines. EMBO J. 2002;21:1873–1880. doi: 10.1093/emboj/21.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokoris MS, Black ME. Characterization of Herpes Simplex Virus type 1 thymidine kinase mutants engineered for improved ganciclovir or acyclovir activity. Protein Sci. 2002;11:2267–2272. doi: 10.1110/ps.2460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munir KM, French DC, Loeb LA. Thymidine kinase mutants obtained by random sequence selection. Proc. Natl Acad. Sci. USA. 1993;90:4012–4016. doi: 10.1073/pnas.90.9.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igarashi K, Hiraga S, Yura T. A deoxythymidine kinase deficient mutant of Escherichia coli: II. Mapping and transduction studies with phage phi 80. Genetics. 1967;57:643–654. doi: 10.1093/genetics/57.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lairson LL, Watts AG, Wakarchuk WW, Withers SG. Using substrate engineering to harness enzymatic promiscuity and expand biological catalysis. Nat. Chem. Biol. 2006;2:724–728. doi: 10.1038/nchembio828. [DOI] [PubMed] [Google Scholar]
- 17.Olsen MJ, Stephens D, Griffiths D, Daugherty P, Georgiou G, Iverson BL. Function-based isolation of novel enzymes from a large library. Nat. Biotechnol. 2000;18:1071–1074. doi: 10.1038/80267. [DOI] [PubMed] [Google Scholar]
- 18.Barrio JR, Secrist JA, 3rd, Leonard NJ. Fluorescent adenosine and cytidine derivatives. Biochem. Biophys. Res. Commun. 1972;46:597–604. doi: 10.1016/s0006-291x(72)80181-5. [DOI] [PubMed] [Google Scholar]
- 19.Boryski J, Golankiewicz B, De Clercq E. Synthesis and antiviral activity of novel N-substituted derivatives of acyclovir. J. Med. Chem. 1988;31:1351–1355. doi: 10.1021/jm00402a017. [DOI] [PubMed] [Google Scholar]
- 20.Secrist JA, 3rd, Barrio JR, Leonard NJ. A fluorescent modification of adenosine triphosphate with activity in enzyme systems: 1,N6-ethenoadenosine triphosphate. Science. 1972;175:646–647. doi: 10.1126/science.175.4022.646. [DOI] [PubMed] [Google Scholar]
- 21.Bergstrom DE, Inoue H, Reddy PA. Pyrido[2,3-D]pyrimidine nucleosides—synthesis via cyclization of C-5-substituted cytidines. J. Org. Chem. 1982;47:2174–2178. [Google Scholar]
- 22.Berry DA, Jung KY, Wise DS, Sercel AD, Pearson WH, Mackie H, Randolph JB, Somers RL. Pyrrolo-dC and pyrrolo-C: fluorescent analogs of cytidine and 2′-deoxycytidine for the study of oligonucleotides. Tetrahedron Lett. 2004;45:2457–2461. [Google Scholar]
- 23.Hawkins ME, Pfleiderer W, Balis FM, Porter D, Knutson JR. Fluorescence properties of pteridine nucleoside analogs as monomers and incorporated into oligonucleotides. Anal. Biochem. 1997;244:86–95. doi: 10.1006/abio.1996.9879. [DOI] [PubMed] [Google Scholar]
- 24.Patching SG, Baldwin SA, Baldwin AD, Young JD, Gallagher MP, Henderson PJ, Herbert RB. The nucleoside transport proteins, NupC and NupG, from Escherichia coli: specific structural motifs necessary for the binding of ligands. Org. Biomol. Chem. 2005;3:462–470. doi: 10.1039/b414739a. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Sun XJ, Smith KM, Visser F, Carpenter P, Barron G, Peng YS, Robins MJ, Baldwin SA, Young JD, et al. Studies of nucleoside transporters using novel autofluorescent nucleoside probes. Biochemistry. 2006;45:1087–1098. doi: 10.1021/bi0520535. [DOI] [PubMed] [Google Scholar]
- 26.Georgiou G. Analysis of large libraries of protein mutants using flow cytometry. Adv. Protein Chem. 2000;55:293–315. doi: 10.1016/s0065-3233(01)55007-x. [DOI] [PubMed] [Google Scholar]
- 27.Fisher MA, Yadav PN, Yadav J, Kristol D, Arnold E, Modak MJ. Identification of a pharmacophore for nucleoside analog inhibitors directed at HIV-1 reverse transcriptase. J. Mol. Recognit. 1994;7:211–214. doi: 10.1002/jmr.300070309. [DOI] [PubMed] [Google Scholar]
- 28.Waqar MA, Evans MJ, Manly KF, Hughes RG, Huberman JA. Effects of 2′,3′-dideoxynucleosides on mammalian cells and viruses. J. Cell Physiol. 1984;121:402–408. doi: 10.1002/jcp.1041210218. [DOI] [PubMed] [Google Scholar]
- 29.Balzarini J, Kang GJ, Dalal M, Herdewijn P, De Clercq E, Broder S, Johns DG. The anti-HTLV-III (anti-HIV) and cytotoxic activity of 2′,3′-didehydro-2′,3′-dideoxyribonucleosides: a comparison with their parental 2′,3′-dideoxyribonucleosides. Mol. Pharmacol. 1987;32:162–167. [PubMed] [Google Scholar]
- 30.Eriksson S, Munch-Petersen B, Johansson K, Eklund H. Structure and function of cellular deoxyribonucleoside kinases. Cell Mol. Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munch-Petersen B, Knecht W, Lenz C, Sondergaard L, Piskur J. Functional expression of a multisubstrate deoxyribonucleoside kinase from Drosophila melanogaster and its C-terminal deletion mutants. J. Biol. Chem. 2000;275:6673–6679. doi: 10.1074/jbc.275.9.6673. [DOI] [PubMed] [Google Scholar]
- 32.Munch-Petersen B, Piskur J, Sondergaard L. Four deoxynucleoside kinase activities from Drosophila melanogaster are contained within a single monomeric enzyme, a new multifunctional deoxynucleoside kinase. J. Biol. Chem. 1998;273:3926–3931. doi: 10.1074/jbc.273.7.3926. [DOI] [PubMed] [Google Scholar]
- 33.Muller KM, Stebel SC, Knall S, Zipf G, Bernauer HS, Arndt KM. Nucleotide exchange and excision technology (NExT) DNA shuffling: a robust method for DNA fragmentation and directed evolution. Nucleic Acids Res. 2005;33:e117. doi: 10.1093/nar/gni116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerth ML, Lutz S. Mutagenesis of non-conserved active site residues improves the activity and narrows the specificity of human thymidine kinase 2. Biochem. Biophys. Res. Commun. 2007;354:802–807. doi: 10.1016/j.bbrc.2007.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welin M, Kosinska U, Mikkelsen NE, Carnrot C, Zhu C, Wang L, Eriksson S, Munch-Petersen B, Eklund H. Structures of thymidine kinase 1 of human and mycoplasmic origin. Proc. Natl Acad. Sci. USA. 2004;101:17970–17975. doi: 10.1073/pnas.0406332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segura-Pena D, Lutz S, Monnerjahn C, Konrad M, Lavie A. Binding of ATP to TK1-like enzymes is associated with a conformational change in the quaternary structure. J. Mol. Biol. 2007;369:129–141. doi: 10.1016/j.jmb.2007.02.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyidogan P, Lutz S. Systematic exploration of active site mutations on human deoxycytidine kinase substrate specificity. Biochemistry. 2008;47:4711–4720. doi: 10.1021/bi800157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song S, Pursell ZF, Copeland WC, Longley MJ, Kunkel TA, Mathews CK. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc. Natl Acad. Sci. USA. 2005;102:4990–4995. doi: 10.1073/pnas.0500253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikkelsen NE, Johansson K, Karlsson A, Knecht W, Andersen G, Piskur J, Munch-Petersen B, Eklund H. Structural basis for feedback inhibition of the deoxyribonucleoside salvage pathway: studies of the Drosophila deoxyribonucleoside kinase. Biochemistry. 2003;42:5706–5712. doi: 10.1021/bi0340043. [DOI] [PubMed] [Google Scholar]
- 41.Knecht W, Rozpedowska E, Le Breton C, Willer M, Gojkovic Z, Sandrini MP, Joergensen T, Hasholt L, Munch-Petersen B, Piskur J. Drosophila deoxyribonucleoside kinase mutants with enhanced ability to phosphorylate purine analogs. Gene Ther. 2007;14:1278–1286. doi: 10.1038/sj.gt.3302982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.