Abstract
Localized clustering of damage is a hallmark of certain DNA-damaging agents, particularly ionizing radiation. The potential for genetic change arising from the effects of clustered damage sites containing combinations of AP sites, 8-oxo-7,8-dihydroguanine (8-oxoG) or 5,6-dihydrothymine is high. To date clusters containing a DNA base lesion that is a strong block to replicative polymerases, have not been explored. Since thymine glycol (Tg) is non-mutagenic but a strong block to replicative polymerases, we have investigated whether clusters containing Tg are highly mutagenic or lead to potentially cytotoxic lesions, when closely opposed to either 8-oxoG or an AP site. Using a bacterial plasmid-based assay and repair assays using cell extracts or purified proteins, we have shown that DNA double-strand breaks (DSBs) arise when Tg is opposite to an AP site, either through attempted base excision repair or at replication. In contrast, 8-oxoG opposite to Tg in a cluster ‘protects’ against DSB formation but does enhance the mutation frequency at the site of 8-oxoG relative to that at a single 8-oxoG, due to the decisive role of endonucleases in the initial stages of processing Tg/8-oxoG clusters, removing Tg to give an intermediate with an abasic site or single-strand break.
INTRODUCTION
In normal aerobic metabolism, reactive oxygen species are produced which may interact with DNA to give a variety of types of lesion. Many of these DNA lesions are chemically indistinguishable from those caused by treatment of cells with ionizing radiations (1,2). The damaging potential of ionizing radiation on biological materials has been proposed to arise largely from the formation of DNA double-strand breaks (DSBs) and clustered DNA damage when two or more lesions occur within one or two helical turns of the DNA by passage of a single radiation track (3,4). Predictions from biophysical models of interactions of radiation tracks with DNA indicate that significant levels of DNA lesions are formed in clusters and that the complexity of the clusters increases with increasing ionization density of the radiation (4–6). Consistent with this prediction is the increased biological effects such as mutagenesis, carcinogenesis, lethality and the reduced reparability of DNA DSBs with increasing ionization density of the radiation (1,2), thereby distinguishing them from readily repairable endogenous damage. Recent studies have verified that clustered DNA damage sites are induced in mammalian cells (7–10) and Escherichia coli (11) by ionizing radiation. The likelihood of clustered damage sites arising endogenously is low, as confirmed recently (12,13).
To prevent biological consequences of single lesions, cells have developed pathways to repair DNA damage (14). However, in the presence of clustered damage, the role of the repair machinery can be complicated depending on the complexity of the cluster. It has been hypothesized that radiation-induced clustered damage sites are less repairable than isolated base lesions caused by aerobic metabolism, and are particularly harmful to cells (3,4). Sites of clustered DNA damage induced in cells by radiation are predicted to consist of closely associated base lesions, such as oxidized guanine [8-oxo-7,8-dihydroguanine (8-oxoG)] and thymine glycol [5,6-dihydroxy-5,6-dihydrothymine or thymine glycol (Tg)] with other types of base lesion or apurinic/apyrimidinic (AP) sites, as verified by both in vitro and in vivo analyses (15–17). We and others have shown using cell extracts or purified proteins that a complex interplay exists between different repair activities in the processing of specific forms of base lesion within a clustered damage site, and this interplay determines the outcome of attempted repair (18–29).
The mutagenic potential of bistranded clustered damage sites containing a mixture of AP sites, 8-oxoG or 5,6-dihydrothymine (DHT) lesions in E. coli is higher in comparison with that of the isolated lesions (30–35). These studies emphasize the importance of the type of lesions, inter-lesion distance and relative orientation of lesions within a cluster for the effective processing of clustered damage sites in the cell. More recently it was shown that the efficiency/abundance of the base glycosylase, such as endonuclease III, in E. coli also plays a decisive role in the initial stages of processing of DHT/8-oxoG clusters, removing DHT initially to give an intermediate with an abasic site or single-strand break (SSB) opposing 8-oxoG (33). However, some types of clustered damage sites may lead to a lethal DSB during attempted repair of the site in E. coli and mammalian cells (11,30,35–37). The generality of the types of clusters which result in DSB or lead to enhanced mutation frequencies detected in E. coli and mammalian cells has now been confirmed using yeast (38).
In this study, we have examined the biological consequences of the cis (5R,6S) isomer of Tg, one of the major oxidative lesions produced by radiation in mammalian cells (16), associated with either 8-oxoG or an AP site within bistranded cluster DNA damage sites. To date, Tg is a known substrate for the glycosylases, endonuclease III (Nth) and endonuclease VIII (Nei) (39–42) and is repaired through the base excision repair (BER) pathways (43). However, when Tg is located directly opposite an AP site within a bistranded cluster damage site, a substantial delay occurs in the rate of repair of the AP site by short-patch BER (44). This reduced efficiency is in part due to a 3-fold reduced rate of incorporation of dAMP opposite Tg by polymerase β (44). Since Tg is a non-mutagenic lesion but is a strong block to both repair and replicative DNA polymerases in vitro (45–50), we report on whether a clustered damage containing Tg when closely opposed to either a 8-oxoG or an AP site is highly mutagenic or potentially cytotoxic, if the cluster is converted into a DSB. The findings significantly extend our understanding on how different types of clusters are processed when containing a lesion which is a block to replicative DNA polymerases. Using wild-type and glycosylase-deficient (fpg, mutY, nth nei and nth nei mutY) strains of E. coli, as well as a combination of these deficiencies, we present the first evidence showing that Tg, a block to replicative DNA polymerases, when present in a cluster with 8-oxoG or an AP site, dictates whether attempted repair of the clustered site leads to either the induction of mutations or a potentially cytotoxic DSB. Therefore, attempted repair of the clustered damaged site containing a block to replicative DNA polymerases may significantly influence the biological outcome of cluster DNA damage sites.
MATERIALS AND METHODS
Substrate oligonucleotides
Oligonucleotides (40-mer) containing a Tg [cis (5R, 6S) thymidine glycol phosphoramidite, Glen Research, Sterling, MA, USA] residue were synthesized at 1 µmol-scale on solid support using the ‘Pac phosphoramidite’ chemistry (Pierce, Milwaukee, WI, USA) with elimination of the 5′-terminal DMTr group (‘trityl-off’ mode). The standard 1 µmol DNA cycle was used on a 392 DNA synthesizer (Applied Biosystems Inc, Palo Alto, CA, USA) with modification: the duration of the condensation was increased 4 times for the modified phosphoramidite (120 s instead of 30 s for a normal nucleoside phosphoramidite). The modified oligonucleotides were then cleaved from the solid support and the alkali-labile protecting groups removed by treatment with 30% ammonia at room temperature for 4 h. The residue dissolved in TEA-3HF was left overnight. Following precipitations with n-butanol followed by ethanol, the oligodeoxynucleotides were purified by denaturing PAGE and desalted on NAP-25 column. Mass measurement by MALDI-TOF in the negative mode confirmed the purity and structure of the modified oligonucleotides. (In negative linear mode, the pseudo-molecular ion found at m/z 12 382.3 is in agreement with the calculated mass of 12 382.1). The cis (5R, 6S) thymidine glycol may undergo slow cis to trans epimerization over 24 h to give an equilibrium concentration containing 87% in the original cis form (51). To minimize the effects of epimerization, freshly prepared oligonucleotides (see later) were used in the assays reported.
The oligonucleotides (40-mer) with either uracil or 8-oxoG modifications were purchased, HPLC purified, from MWG Biotech (Ebersberg, Germany). The sequences of the double-stranded oligonucleotides are presented in Table 1. Strand 2 contains either a uracil or 8-oxoG at a fixed position (position X) opposite to a Tg at variable positions on strand 1 (position Y). The control 1 oligonucleotide contains either uracil or 8-oxoG (position X on strand 2). The control 2 oligonucleotide contains a single Tg (position Y on strand 1). The nomenclature of the relative positions of the two lesions in the clustered DNA damage site was developed by David-Cordonnier et al. (23). A positive or negative number refers to the separation, in base pairs, of one lesion on strand 1 located 5′ (positive number) or 3′ (negative number) to the lesion on strand 2.
Table 1.
Sequence of oligonucleotides used to generate the DNA clustered damage sites
| Position | Sequence | Name |
|---|---|---|
| −5 | 5′-ctcttagtcaggaaYatgtctctatgctgggagcaaaggc | Tg−5 |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | ||
| −1 | 5′-ctcttagtcaggaatatgYctctatgctgggagcaaaggc | Tg −1 |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | ||
| +1 | 5′-ctcttagtcaggaatatgtcYctatgctgggagcaaaggc | Tg + 1 |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | ||
| +5 | 5′-ctcttagtcaggaatatgtctctaYgctgggagcaaaggc | Tg + 5 |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | ||
| control 1 | 5′-ctcttagtcaggaatatgtctctatgctgggagcaaaggc | Control AP, Control 8-oxoG |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | ||
| control 2 | 5′-ctcttagtcaggaatatgYctctatgctgggagcaaaggc | Control Tg |
| 3′-gagaatcagtccttatacagagatacgaccctcgtttccg | ||
| control 3 | 5′-ctcttagtcaggaatatgtctctatgctgggagcaaaggc | No damage |
| 3′-gagaatcagtccttatacagagatacgaccctcgtttccg |
X represents either 8-oxoG, an AP site or a HAP1-SSB (following conversion of uracil to AP site and to a HAP1-SSB as described in Materials and Methods section); Y represents thymine glycol. −5 and −1 indicate the positions on the complementary strand of the X base 3′ from Y base. +1 and +5 are the positions on the complementary strand of the X base 5′ from the Y base. Control 1 is control oligonucleotide containing ether an AP site, a HAP1-SSB or 8-oxoG as a single lesion. Control 2 is the control oligonucleotide containing Tg as a single lesion. Control 3 is the control oligonucleotide containing no damage.
Escherichia coli strains
Isogenic strains CC104 (52), BH980 (mutY::KanR) and BH990 (fpg::KanR mutY::KanR) double mutant strain were a kind gift from Dr S. Boiteux, CEA/Fontenay-aux-Roses, France.
Plasmid preparation
Double-stranded oligonucleotides containing a clustered damage site were ligated into pUC18 plasmid as previously described (32). Briefly, 20 pmol of the complementary oligonucleotides (Table 1) were annealed and 5′-phosphorylated in a forward reaction with T4 polynucleotide kinase. Purified oligonucleotides (5 pmol) were ligated into the Sma1 site of 200 fmol pUC18 plasmid overnight at 16°C with T4 DNA ligase (NEB). After dialysis using 0.025 μm Millipore nitrocellulose filters, 30–50% of plasmid molecules were ligated (open circular form).
Transformation in electro-competent E. coli
An aliquot of 50 ng of the ligation product was added to 60 ml of electro-competent bacteria in a cuvette and electroporated using a Bio-Rad E. coli pulser, set at 1.8 mV. Immediately after electroporation, 500 μl of SOB was added to the transformed bacteria which were then incubated for 1 h at 37°C. Transformants were selected in 5 ml of LB broth containing ampicillin (100 μg/ml) at 37°C for 16 h.
Quantification of mutations of the clustered damage site
Plasmid DNA was retrieved in 50 μl of water from 1.5 ml of the overnight miniculture using a QIAGEN Qiakit-spin miniprep kit. A 15 μl aliquot of the plasmid DNA eluate was added to 5 U of BsmAI at 55°C for 16 h. The samples underwent electrophoresis for 6 h at room temperature on a 1% agarose gel (1 μg/ml ethidium bromide) at 4 V/cm. Following electrophoresis, the gel image was captured under UV light using a CCD camera. Images were analysed using Bio-Rad Quantity One Quantification Software and the mutation frequency calculated as described previously (32,33).
Sequence analysis of plasmid DNA
After the bacteria incubation in SOB (see above), an aliquot was removed and plated onto LB agar containing 100 μg/ml ampicillin. The bacteria were incubated at 37°C overnight and colonies were picked randomly to inoculate 5 ml of LB broth containing 100 μg/ml ampicillin and grown at 37°C overnight. The plasmid DNA was retrieved from the bacteria and prepared for sequencing using an Applied Biosystems Big Dye sequencing kit. The forward and reverse primers, 5′-CTTCGCTATTACGCCAGCTG-3′ and 5′-GGCAGACAGGTTTCCCGACTGGA-3′, allow both plasmid strands to be copied.
Preparation of 5′-end labelled oligonucleotides
Oligonucleotide (0.2 µg) was 5′-end-labelled using 10 U of T4 polynucleotide kinase (Invitrogen, Paisley, UK) with 25 μCi [γ-32P]ATP (6000 Ci/mmol, 10 mCi/ml, Perkin Elmer, Waltham, MA, USA) in 20 μl buffer (70 mM Tris–HCl pH 7.6, 10 mM MgCl2, 100 mM KCl, 1mM β-2-mercaptoethanol) for 30 min at 37°C. Following purification on a 12% denaturing polyacrylamide gel, the labelled oligonucleotide was hybridized with a 2-fold excess of the non-radiolabelled complementary strand in Tris–EDTA buffer at pH 8 through heating 5 min at 80°C followed by cooling at room temperature over 1 h. Efficient annealing of the oligonucleotides was verified on a 12% native polyacrylamide gel.
Preparation of an AP site
The purified double-stranded oligonucleotides which contained a uracil residue were treated with 1 U of uracil-DNA-glycosylase (UDG) (Invitrogen, Paisley, UK) in 100 μl buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EDTA) for 30 min at 37°C to produce an AP site. After ethanol precipitation, the AP site containing oligonucleotides were used in the repair assays.
Preparation of a HAP1-SSB
The purified double-stranded oligonucleotides which contained an AP site were treated with 25 ng HAP1 in 50 μl buffer (20 mM HEPES pH 7.9, 100 mM KCl, 1 mM MgCl2, 0.2 mM EDTA, 20% glycerol) for 30 min at 37°C to produce a SSB with 3′-OH and 5′-deoxyribose 5′-phosphate (dRP) termini. After ethanol precipitation, the HAP1-SSB containing oligonucleotides were used in the repair assays.
Preparation of nuclear extracts
The nuclear extracts were prepared as previously described (29) from Ku80-deficient xrs5 cells. Briefly, the cells were harvested in exponential phase in α-complemented minimum Eagle's medium (Sigma-Aldrich, Poole, UK) supplemented with 10% foetal bovine serum (Mycoplex, PAA Laboratories, Teddington, UK), 100 U/ml penicillin (GIBCO-Invitrogen, Paisley, UK), 100 mg/ml streptomycin (GIBCO-Invitrogen, Paisley, UK) and 0.1% l-glutamine. The pelleted cells were resuspended in an equal volume of buffer (10 mM HEPES pH 7.9, 100 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) and incubated on ice for 15 min. The cells were lysed by drawing the cell suspension into a 0.5 μm diameter needle 10 times and the nuclei were collected by a brief centrifugation at 12 000g at 4°C. The nuclear proteins were extracted by incubation in 2/3 volume high salt buffer (20 mM HEPES pH 7.9, 420 mM NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) for 30 min with agitation on ice. Following centrifugation for 10 min at 12 000 g, 4°C, the supernatant was dialysed twice over a total period of 16 h against 1 l of buffer (20 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, 0.5 mM PMSF). The protein concentration, determined using the Bradford colorimetric technique, was found to be 5.6 mg/ml. Aliquots of nuclear extracts were stored at −80°C.
Repair assays
For analysis of the AP site or the HAP1-SSB BER process, the double-stranded oligonucleotides (10 000 c.p.m., 0.75 fmol) were incubated with 1 μg of nuclear extract in 5 μl repair buffer (70 mM Tris–HCl pH 7.5, 10 mM MgCl2, 10 mM DTT, 4 mM ATP, 40 mM phosphocreatine, 1.6 μg/ml phosphocreatine kinase, 0.1 mM each of dATP, dCTP, dGTP, dTTP) at 37°C for a set time between 0 and 60 min. The concentrations of extract had been optimized from titration studies (data not shown). To stop the reactions, 5 μl denaturing stop solution (98% formamide, 2 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol) was added. The samples were then subjected to electrophoresis on a 12% denaturing polyacrylamide gel containing 8 M urea in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3) for 60 min at a constant power of 90 W. The dried gel was exposed to a Bio-Rad PhosphorImager screen for visualization of repair products using phosphorimaging technology (Bio-Rad, Molecular Imager FX) and quantified with Quantity One software (Bio-Rad, Hercules, CA, USA). When following the time dependence of repair of the AP site or of HAP1-SSB, the intensity of the bands representing either single-stranded DNA, single-stranded DNA with one or five bases added (before ligation; see Results section) or rejoined DNA [ligation of the SSB following addition of the missing base(s)] is expressed as a percentage of the total intensities for all bands. The efficiencies of repair of an AP site or a HAP1-SSB contained within a clustered damage site with Tg were compared with those for the repair of the respective single lesions in the control oligonucleotide. The errors represent standard deviations of the mean from at least three experiments.
A second repair assay, using dideoxynucleoside triphosphate, was performed under the same conditions as the repair assay using deoxynucleoside triphosphate with substitution of 10 μM ddATP for 0.1 mM dATP.
Cleavage assays for SSB analysis
The double-stranded oligonucleotides (10 000 c.p.m., 0.75 fmol) were incubated with increasing amounts of purified glycosylases, endo- or exonuclease or xrs5 nuclear extracts in 5 μl buffer. Fpg, Nth and xrs5 buffer contained 20 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, 0.5 mM PMSF. Reactions were carried out at 37°C for 30 min. Exo III reaction buffer is composed of 25 mM CaCl2, 625 mM NaCl, 330 mM Tris–HCl, 50 mM DTT and Endo VIII buffer, 10 mM Tris–HCl, 75 mM NaCl, 1 mM EDTA, pH 8. Reactions were carried out at 37°C for 15 min. To stop the reactions, 5 μl denaturing stop solution (98% formamide, 2 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol) was added. The samples were then subjected to electrophoresis on a 12% denaturing polyacrylamide gel containing 8 M urea in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3) or 12% native polyacrylamide gel for 45 min at a constant power of 90 W. The dried gel was exposed to a Bio-Rad PhosphorImager screen for visualization of repair products using phosphorimaging technology (Bio-Rad, Molecular Imager FX) and quantified with Quantity One software (Bio-Rad). The incision/excision efficiencies were determined for each protein or nuclear extract, comparing the single lesion in the control oligonucleotides with the same lesion within the clustered damaged site, for the same amount of protein.
Reconstitution of short-patch BER with purified proteins
The [32P]-5′-end labelled double-stranded oligonucleotides (10 000 c.p.m., 0.75 fmol) were incubated for 20 min at 37°C in the absence or presence of purified BER enzymes, e.g. 10 ng HAP1 or 10 ng HAP1 + 2 ng pol β in a 10 µl reaction solution containing 80 mM HEPES pH 7.9, 10 mM MgCl2, 2 mM dithiothreitol (DTT), 200 µM EDTA, 4 mM ATP, 800 µg/ml bovine serum albumin, 40 µM each of dATP, dTTP, dGTP and dCTP. The reactions were stopped by the addition of 10 µl denaturating stop solution (98% formamide, 2 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol). The samples were subsequently incubated at 90°C for 3 min and then subjected to electrophoresis on a 20% denaturating polyacrylamide gel containing 8 M urea in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3).
RESULTS
The mutability of clustered DNA damage in bacteria strains
A plasmid-based assay previously developed in our laboratory (32) has been used to investigate in E. coli the mutability of Tg-containing clustered damage sites also containing either uracil, a precursor to an AP site, or 8-oxoG on the opposite strand. The oligonucleotide constructs containing either uracil or 8-oxoG are at a fixed position in a BsmAI restriction site and Tg at various positions from the fixed lesion are shown in Table 1. It was confirmed that when either 8-oxoG or Tg as single lesions are present within the restriction site, BsmAI, incision is inhibited (data not shown). Mutation frequencies were assessed from the efficiency of BsmAI incision as described previously (32).
Clustered damage sites containing uracil and Tg lead to loss of colonies after transformation
Following transformation of the plasmid constructs containing uracil and Tg in the bistranded clusters into wild-type E. coli, the number of surviving colonies is <10–20% of that for the corresponding plasmid constructs containing no damage or either uracil or Tg as a single lesion (data not shown). We have previously shown that uracil is rapidly converted into an AP site in E. coli (32). Sequence analysis of plasmids obtained from the few colonies surviving showed only a low frequency of mutation with the majority being either loss of the adenine opposite to the site of Tg (+1 and −5 oligonucleotides) or deletion of Tg (+5, 0 or −1 oligonucleotides).
Clustered damage sites containing 8-oxoG and Tg lead to enhancement of the mutagenic potential of 8-oxoG
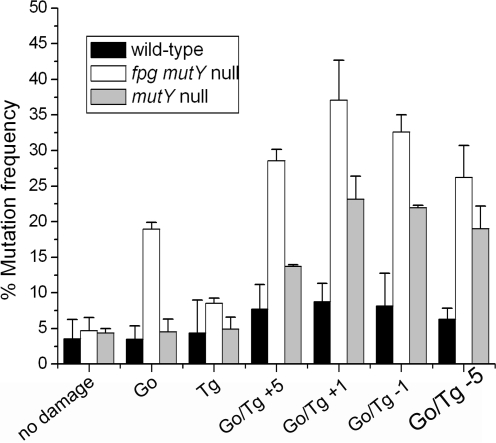
In contrast to clusters containing U/Tg, the transformation frequencies, i.e. the number of colonies lost is similar for plasmid constructs containing clusters with 8-oxoG/Tg to those with single or no damage sites in both wild-type and mutant strains. Bistranded clusters containing 8-oxoG and Tg do, however, lead to enhanced mutation frequencies compared with those for either 8-oxoG or Tg as single lesions in wild-type and particularly in mutY and fpg mutY mutant E. coli strains as shown in Figure 1, with the gel data shown in Supplementary Figure 1. The highest mutation frequencies for these clusters (37%) were seen in the fpg mutY mutant strain. The mutation frequency decreases with increasing separation between 8-oxoG and Tg particularly in the mutY and fpg mutY null strains. Since Tg is a substrate for Nth or Nei, the mutation frequencies with the −1 cluster containing 8-oxoG/Tg and either Tg or 8-oxoG as a single lesion were also determined in nth nei and nth nei mutY mutants (Supplementary Figure 2). The main difference seen is a small increase in the mutation frequency with the cluster in the triple mutant relative to that with the mutY null strain. At present we do not have an explanation for the mutation frequency seen with the nth nei mutY mutant relative to that seen with the nth nei mutant for the 8-oxoG/Tg-1 cluster, as the similarity of the mutation frequencies with the nth nei mutY mutant and the mutY null strain infers that the mutations occur at 8-oxoG.
Figure 1.
Mutation frequency of the Go/Tg clustered damage site transformed into wild type, mutY strain and fpg mutY strains of E. coli. The types of clusters are shown along the x-axis. Error bars display the standard errors of the mean from three experiments.
To identify the types of mutations, plasmids were obtained from individual colonies following fpg mutY transformation with the +1 and −1 clustered 8-oxoG/Tg. Those plasmids which showed a mutant band following restriction with BsmAI were sequenced. The main mutation seen in >95% of the individual colonies containing mutant plasmid is a G:C to T:A transversion at the site of 8-oxoG with no mutations detected at the Tg site. A deletion of the base pair at the site of 8-oxoG was also detected. Following restriction of the plasmids obtained from individual colonies with BsmA1 as described previously (33), the percentage of mutant band compared with non-mutant band ranges from 10% to 100% due to the presence of both incomplete and complete mutations as shown in Supplementary Figure 3.
Repair of clustered damaged sites in vitro
Having seen reduced transformation frequencies (colony numbers) and/or mutagenic effects of clustered damaged sites containing Tg opposite either U or 8-oxoG in E. coli, we wanted to investigate further the efficiency of repair of these cluster damage sites.
Efficiency of repair of an AP site or a SSB is retarded in the presence of Tg
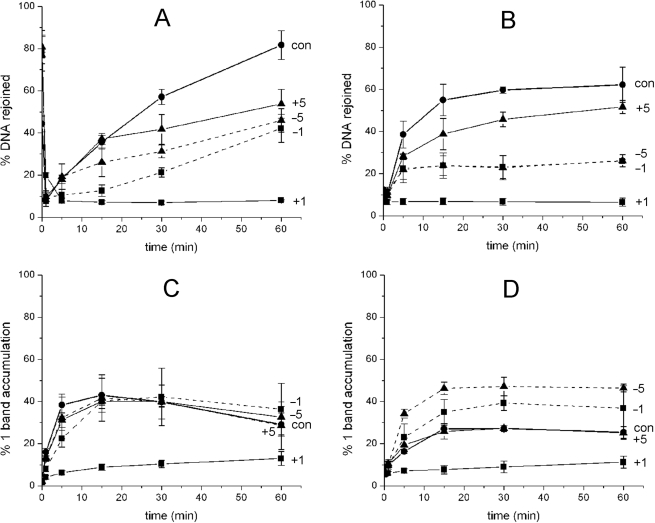
To investigate whether the efficiency of repair of either an AP site or a HAP1-SSB is affected when present in a bistranded clustered site containing Tg, the cluster-containing oligonucleotides were treated with xrs5 nuclear extracts for 0–60 min. Supplementary Figure 4A shows a representative profile from a phosphorimaging scan allowing each stage of BER to be monitored over 60 min i.e. incision of an AP site (control) within 1 min to a SSB, followed by addition of bases by polymerases and ultimately restoration of a repaired intact 40-mer. With the exception of the cluster containing an AP site at position +1 to Tg, the time course of repair of the AP site, following its incision to a SSB, is reduced in the presence of Tg relative to that of the control for each of the clusters (Figure 2A). Repair of the pre-formed HAP1-SSB when in the cluster is also reduced (Figure 2B, representative gel is shown Supplementary Figure 4B) but to a greater extent than that seen with an AP site, especially for the −1 and −5 clusters where the efficiency is reduced by ∼2.5-fold. With these latter clusters, the initial rate of rejoining of the AP site appears to be independent of the presence of Tg when at positions −1 or −5.
Figure 2.
The effect of Tg on the time for rejoining of an AP site (A) or a HAP1-SSB (B) when in different clustered sites following incubation with xrs5 nuclear extracts. The extent of accumulation of SSB+1 base during repair of an AP site (C) or a HAP1-SSB (D) when ddATP replaces dATP in the repair buffer. AP site control (con, filled circle: solid line) and AP/Tg at positions +1 (filled square: solid line), +5 (filled triangle: solid line), −1 (filled square: dashed line), −5 (filled triangle: dashed line). To assist the reader, lines have been drawn between the points to guide the eye only and do not represent fitted curves.
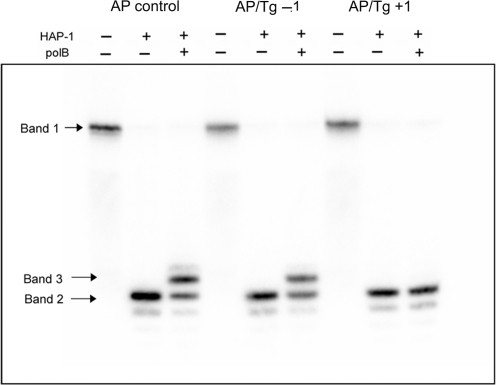
When either an AP site or HAP1-SSB is at position +1 to Tg in the cluster, their repair is inhibited (Figure 2A and B). Additionally as seen in Supplementary Figure 4, the repair intermediate (SSB+1 base) which should have been formed through addition of a single base by polymerase(s) present in the xrs5 extracts is not detected. Using purified polβ and HAP1, it was confirmed that HAP1 incises the AP site when bistrandedly clustered with Tg at +1 or −1 whereas polβ only inserts a base into the resulting SSB when Tg is at −1 but not when at +1 as shown in Figure 3. This observation is consistent with the known polymerase block of Tg (50).
Figure 3.
Effect of Tg on the incision of an AP site and addition of the first base when at positions +1 and −1 in a cluster by the proteins HAP-1 and HAP-1 + pol β, respectively.
Repair of either an AP site or HAP1-SSB in a bistranded cluster with Tg occurs by both long and short-patch BER
Oligonucleotides containing either an AP site or HAP1-SSB as a single lesion, as previously shown using xrs5 nuclear extract (24), or in a cluster with Tg at +5 (Figure 2A and B) repair mainly occurs via short-patch BER as seen in Supplementary Figure 4 from the addition of one base prior to ligation. With the cluster containing Tg at +1, repair does not occur or is very inefficient. In contrast, during repair of either an HAP1-SSB or an AP site within a cluster containing Tg at −1 or −5, the sequential addition of more than one base occurs, representing a mix of short- and long-patch BER (Supplementary Figure 4). To verify that addition of >1 base occurs, dATP was substituted by ddATP, which is incorporated as the second base downstream from the repair gap and as a consequence would lead to reduced level of repair product by long-patch BER pathway. Under these conditions and particularly during the repair of the HAP1-SSB with Tg at −1 or −5, the SSB+1 intermediate accumulates to higher levels within 15 min than seen with the control HAP1-SSB (Figure 2C and D) and the level of repair product at 60 min is low (data not shown). Using HAP1-SSB or when clustered with Tg at +5, we verified that the majority of the HAP1-SSB are rejoined in cell extracts by short-patch repair, by substituting ddATP or ddTTP for the corresponding dNTPs, the second and fifth dNTP to be incorporated, respectively. ddATP prevents long-patch repair and ddTTP prevents chain elongation. The differences in the levels of rejoining for HAP1-SSB or when clustered with Tg at +5 are consistent with short-patch repair and not chain elongation, with a contribution from long-patch repair processes (Supplementary Figure 5). Few, if any, HAP1-SSB are rejoined if dGTP is substituted with ddGTP, the first base to be incorporated (Supplementary Figure 5). All the findings with cell extracts confirm that repair of HAP1-SSB and, to a lesser extent, an AP site also proceeds by long-patch BER, particularly when Tg is present at −1 and −5.
The efficiency of incision of an AP site in the presence of Tg
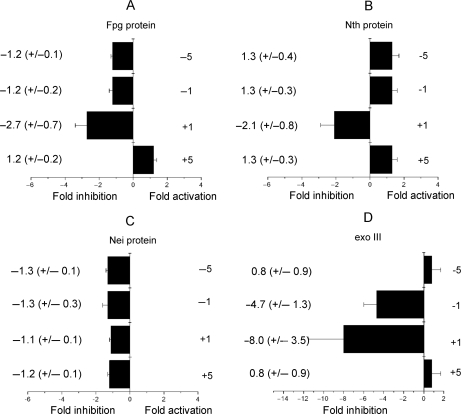
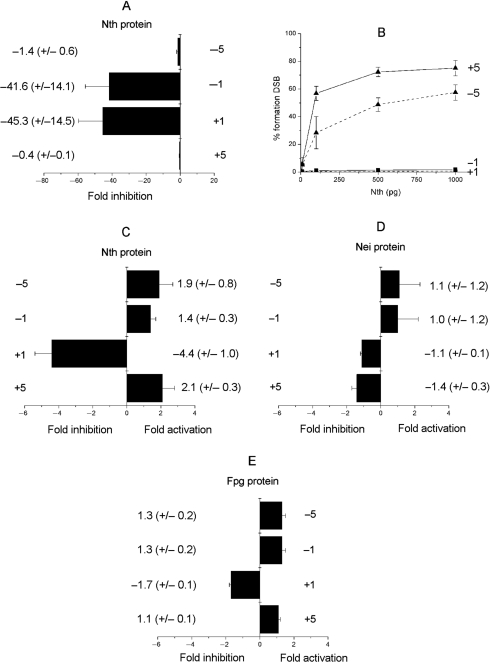
As an AP site is rapidly incised by xrs5 cell extracts when clustered with Tg (Figure 2A), the dependence of the efficiency of incision of an AP site by Fpg, Nth and Nei glycosylases on the proximity of Tg in the various bistranded clusters was determined and is shown in Figure 4A–C. Only minor changes in the efficiency of incision of an AP site, compared with that for the control AP site, occur when Tg is situated in any of the tested positions. In contrast, the incision of an AP site by E. coli exonuclease III is impaired in the presence of an opposing Tg when at –1 or +1 (Figure 4D), consistent with previous findings (26).
Figure 4.
Effect of Tg on the efficiency of incision of an AP site in the different clustered sites by (A) Fpg (1 pg to 10 ng), (B) Nth (1 pg to 10 ng), (C) Nei (0.05–1 U) and (D) exo III (0.01–0.1 U) in double-stranded oligonucleotides. The fold inhibition/activation was obtained from comparison of the concentration dependence for incision of the AP site with that for the control AP site. The error bars represent the standard deviation from three different experiments.
Efficiency of excision of a Tg in the presence of an AP site or 8-oxoG
We next asked if the rate of excision of Tg is influenced by an AP site (or resulting SSB) when in a bistranded cluster. Tg as a single lesion is not excised from the oligonucleotide at the highest amount of Fpg (10 ng) or nuclear extracts (0.01–0.5 μg) used. With Nth, the efficiency of excision of Tg when positioned at +1 or −1 to an AP site is drastically reduced by ∼40-fold relative to that of the control which contains a single Tg (Figure 5A). In contrast, the efficiency of excision of Tg by Nth is not affected when an AP site is at position +5 or −5 to Tg. This observation together with the observation that Tg does not influence the rate of incision of an AP site by Nth is consistent with the formation of a DSB when the AP site/Tg clusters +5 or −5 were treated with Nth (Figure 5B). However, DSBs are not formed when Tg and an AP site are separated by only one base in a bistranded cluster (Figure 5B), reflecting the reduced efficiency of Nth to excise Tg in these clusters.
Figure 5.
Effect of an AP site on (A) the efficiency of excision of Tg. by Nth (1 pg to 1 ng) and (B) the dependence for formation of a DSB on the concentration of Nth determined following native PAGE. Effect of 8-oxoG on the efficiency of excision of Tg. by Nth (1 pg to 10 ng) (C) or Nei (0.05–1 U) (D). Effect of Tg on the efficiency of excision of 8-oxoG by Fpg (1 pg to 10 ng) (E). The fold inhibition was obtained from comparison with the control containing only Tg (A–D) or 8-oxoG (E). The error bars represent the standard deviation from three different experiments.
With Nei, an AP site and Tg, but not 8-oxoG, when present as single lesions, are removed with high efficiency (Supplementary Figure 5). Since the formation of a DSB was not observed when the Tg/AP site clusters were treated with Nei, it is inferred that Tg is not excised by Nei when clustered with an AP site. The inefficient excision of Tg by Nei was confirmed when clustered with an AP site at any of the positions tested (data not shown).
The efficiency of excision of Tg by Nth and Nei proteins in the presence of 8-oxoG is shown in Figure 5C and D. The greatest decrease in efficiency of excision of Tg was seen with Nth when 8-oxoG is present in the +1 position to Tg (a 4.4-fold reduction) relative to that seen for Tg when present as a single lesion. The efficiency of excision of Tg by Nth when 8-oxoG is present at any of the other positions tested is only slightly modified. Similarly the efficiency of excision of Tg by Nei was not affected when 8-oxoG is present at all the positions tested. Since Nei does not excise 8-oxoG from double-strand oligonucleotide (Supplementary Figure 6), it was confirmed that DSBs are not formed after treatment of Tg/8-oxoG bistranded clusters with Nei (data not shown).
Efficiency of 8-oxoG excision in the presence of Tg
The influence of Tg on the efficiency of excision of 8-oxoG by Fpg was determined using the oligonucleotides described in Table 1 containing either a single 8-oxoG or 8-oxoG in a bistranded cluster with Tg. The presence of Tg at any of the tested positions does not significantly affect the efficiency of excision of 8-oxoG by Fpg (Figure 5E).
DISCUSSION
We have shown that Tg, a lesion which is a block to replicative polymerases but non-mutagenic in E. coli (45–50) enhances the mutagenic potential of 8-oxoG when both are present within a bistranded clustered damage site. This result suggests that the presence of Tg has interfered with the repair of 8-oxoG, despite the in vitro experimental data indicating that Tg does not significantly influence the excision efficiency of 8-oxoG by Fpg (Figure 5E). Consistent with previous observations with DHT in a bistranded cluster with 8-oxoG in E. coli (33), it is inferred that sequential removal of the lesions occurs, with Tg being removed initially by Nth to give an AP site opposing the 8-oxoG lesion. This suggestion is consistent with our observation that the efficiency of excision of Tg by Nth or Nei is essentially unaffected by the presence of 8-oxoG. The AP site resulting from excision of Tg is then converted rapidly to an SSB by the associated AP lyase activity of Nth (25,53,54). The resulting AP/8-oxoG clustered damage substrate can be highly mutagenic (32), since the presence of the AP site (or SSB) retards the excision of 8-oxoG so that it persists into plasmid replication. Any consequences arising from the polymerase blocking action of Tg appear to be minimized when in a clustered site with 8-oxoG, a finding supported by the similar transformation efficiencies for plasmid constructs containing Tg only or when clustered with 8-oxoG. Additional evidence pointing to the initial removal of Tg from the clustered Tg/8-oxoG site comes from the observation that if 8-oxoG had been removed initially then an AP/Tg cluster would have been formed as an intermediate. Plasmid constructs containing Tg/AP sites bistranded clusters lead to a drastic reduction in the number of viable E. coli colonies due to conversion of the cluster into a DSB or for the +1 cluster due to the replicative polymerase block by Tg, leading to a replication-induced DSB (see later).
Most strikingly in the present study, we see that the frequencies of mutation for the Tg/8-oxoG clusters are very similar to those from our recent findings with DHT/8-oxoG, 8-oxoG/8-oxoG (33) and AP/8-oxoG (32) clustered lesions in the various E. coli strains, clearly supporting the sequential repair model where the same key intermediate (AP/8-oxoG or SSB/8-oxoG) is involved. Further, the majority of the mutations occur at the site of 8-oxoG, consistent with the initial excision of Tg from the Tg/8-oxoG cluster. That MutY has the most important anti-mutagenic role with Tg/8-oxoG clusters is consistent with the findings with DHT/8-oxoG, 8-oxoG/8-oxoG (33) and AP/8-oxoG (32) clusters. We have also shown that bistranded clustered damage initially consisting of Tg opposing 8-oxoG gives rise to incomplete mutations after bacterial processing; as previously described for DHT/8-oxoG clusters (33). In principle, potentially mutagenic damage on one strand of plasmid DNA should give rise to both mutant and non-mutant progeny, providing that damage survives until DNA replication, as discussed previously with DHT/8-oxoG clusters (33). The proportion of each plasmid DNA strand represented in the final progeny will depend on the efficiency with which the two strands are replicated, and on the probability of conversion of the damage to a mutation.
In contrast to the observations with 8-oxoG/Tg clusters, the effect of the replicative polymerase blocking action of Tg is suggested to be reflected in the transformation frequency of E. coli when Tg is in a bistranded cluster with an AP site when compared with that for Tg as a single lesion. We have confirmed using purified proteins that Tg acts as a polymerase block when clustered with an AP site at +1 but not at −1 (Figure 3), consistent with the known polymerase block of Tg (50) and that the repair of the AP site when at −1 occurs primarily through the short-patch BER involving addition of the base into the gap.
Since Tg does not modify the efficiency of incision of an AP site by several glycosylases (Figure 4), the AP site is converted rapidly into a SSB. However, the subsequent repair of the SSB will be impaired at all tested positions (Figure 2B). For instance, base addition was not seen during attempted repair of the resulting SSB when Tg is located in +1 position due to the polymerase block of Tg (see above). For a SSB resulting from incision of the AP site at −1 to Tg, the rejoining of the SSB is reduced when compared with that of a single SSB (Figure 2B), leading to extension of the lifetime of the SSB. The AP site positioned at either +1 or −1 strongly retards the excision of Tg by Nth (Figure 5A) and Nei, so that Tg is predicted to persist to give a SSB/Tg intermediate. If this intermediate is not repaired before undergoing replication due to lifetime extension of the SSB, the SSB-carrying strand may be lost as previously discussed (32,33) and replication of the Tg-carrying strand occurs until the replicative polymerase(s) reaches the blocking Tg, leading to plasmid loss. As a consequence, the processing of a cluster containing Tg and an AP site at +1 or −1 is suggested to lead to the loss of plasmid and therefore colony loss in E. coli. When Tg is at position −1 or +1, a proportion of the SSB may be repaired prior to replication. However, the repair of the SSB is only a minor pathway as only 10–20% of E. coli colonies survive to give low levels of mutations.
Repair of the SSB is also reduced when Tg is present at either +5 or −5 (Figure 2B). However, the presence of an AP site, or the resulting SSB, at these positions does not now retard the excision of Tg by Nth, so that a DSB may be formed. Therefore, in addition to plasmid loss at replication (discussed above) colony loss may also arise through the formation of DSB by incision of both lesions in the bistranded cluster. The loss of colonies with the Tg/AP site cluster +5 or −5 is consistent with the in vitro experimental data showing that DSBs are formed when these clusters are treated with Nth. Low transformation frequencies have previously been seen within bistranded clusters containing two AP sites (35).
From comparison of the repair of bistranded clusters containing Tg with bistranded clusters containing DHT (Byrne,S. et al., submitted for publication) opposite a SSB or AP site, the efficiency of repair of the SSB is reduced similarly by Tg and DHT when present on the opposing strand in both the 3′ and 5′ orientation by nuclear extracts. However, the efficiency of the repair of the AP site is only reduced when DHT is present 3′ to the AP site in contrast to a reduction seen when Tg is in both the 3′ and 5′ orientation except for the +1 cluster when Tg acts as polymerase block. Consistent with our previous findings with clusters containing 8-oxoG with an AP site or HAP1-SSB (24) or DHT/AP site (Byrne et al., submitted for publication), repair of a HAP1-SSB cluster containing Tg occurs by both short- and long-patch repair and not by strand elongation involving polymerases.
The processing of radiation-induced clustered DNA damage sites containing Tg can give rise to different outcomes, influenced by the lesion on the opposite strand to that containing Tg. Whereas Tg opposite to an AP site gives rise to potentially cytotoxic lesions, 8-oxoG opposite to Tg in a cluster ‘protects’ against potentially cytotoxic lesions but enhances mutagenicity, particularly at the site of 8-oxoG.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
European Commission [contract MCRTN-CT-2003-505086 and RISC-RAD (FI6R-CT-2003-508842)]; Medical Research Council UK. Funding for open access charge: Medical Research Council, UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Murat Saparbev (Institute Gustave Roussy, France) for providing us with purified HAP1 and Nth, Dr Serge Boiteux (CEA, France) for providing Fpg and Dr Didier Gasparutto (CEA/Grenoble-UJF, France) for help in preparation of the thymine glycol containing oligonucleotides.
REFERENCES
- 1.O’Neill P, Wardman P. Radiation chemistry comes before radiation biology. Int. J. Radiat. Biol. 2009;85:9–25. doi: 10.1080/09553000802640401. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Carell T, Cellia L, Chatgilialoglu C, Gimisis T, Miranda M, O’Neill P, Ravanat J-L, Robert M. DNA damage and radical reactions: mechanistic aspects, formation in cells and repair studies. Chimia. 2008;62:742–749. [Google Scholar]
- 3.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 4.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 5.Nikjoo H, O’Neill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat. Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Nikjoo H, O’Neill P, Goodhead DT, Terrissol M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int. J. Radiat. Biol. 1997;71:467–483. doi: 10.1080/095530097143798. [DOI] [PubMed] [Google Scholar]
- 7.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl Acad. Sci. USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland BM, Bennett PV, Sutherland JC, Laval J. Clustered DNA damages induced by X-rays in human cells. Radiat. Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgakilas AG. Processing of DNA damage clusters in human cells: current status of knowledge. Mol. BioSys. 2008;4:30–35. doi: 10.1039/b713178j. [DOI] [PubMed] [Google Scholar]
- 11.Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl Acad. Sci. USA. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutherland BM, Bennett PV, Cintron NS, Guida P, Laval J. Low levels of endogenous oxidative damage cluster levels in unirradiated viral and human DNAs. Free Radic. Biol. Med. 2003;35:495–503. doi: 10.1016/s0891-5849(03)00327-7. [DOI] [PubMed] [Google Scholar]
- 13.Bennett PV, Cintron NS, Gros L, Laval J, Sutherland BM. Are endogenous clustered DNA damages induced in human cells? Free Radic. Biol. Med. 2004;37:488–499. doi: 10.1016/j.freeradbiomed.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg EC, Walker GC, Siede W, Wood RD, Schulz RA, Ellenberg T. DNA Repair and Mutagenesis. 2nd. Washington D.C: ASM Press; 2005. [Google Scholar]
- 15.Pouget J-P, Frelon S, Ravanat J-L, Testard I, Odin F, Cadet J. Formation of modified DNA bases in cells exposed either to gamma radiation or to high-LET particles. Radiat. Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Cadet J, Douki T, Ravanat J-L. Oxidatively generated damage to the guanine moiety of DNA: mechanic aspects and formation in cells. Acc. Chem. Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 17.Frelon S, Douki T, Ravanat J-L, Pouget JP, Tornabene C, Cadet J. High-performance liquid chromatography-tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem. Res. Toxicol. 13:1002–1010. doi: 10.1021/tx000085h. [DOI] [PubMed] [Google Scholar]
- 18.Weinfeld M, Rasouli-Nia A, Chaudhry MA, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat. Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Lomax ME, Gulston MK, O’Neill P. Chemical aspects of clustered DNA damage induction by ionising radiation. Radiat. Prot. Dosimetry. 2002;99:63–68. doi: 10.1093/oxfordjournals.rpd.a006840. [DOI] [PubMed] [Google Scholar]
- 20.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhry MA, Weinfeld M. The action of Escherichia coli endonuclease III on multiply damaged sites in DNA. J. Mol. Biol. 1995;249:914–922. doi: 10.1006/jmbi.1995.0348. [DOI] [PubMed] [Google Scholar]
- 22.David-Cordonnier MH, Boiteux S, O’Neill P. Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 23.David-Cordonnier MH, Cunniffe SM, Hickson ID, O'Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 24.Lomax ME, Cunniffe S, O’Neill P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhry MA, Weinfeld M. Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 26.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 27.Mourgues S, Lomax ME, O’Neill P. Base excision repair processing of abasic site/single strand break lesions within clustered damage site associated with XRCC1 deficiency. Nucleic Acids Res. 2007;35:7676–7687. doi: 10.1093/nar/gkm947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eot-Houllier G, Gonera M, Gasparutto D, Giustranti C, Sage E. Interplay between DNA N-glycosylases/AP lyases at multiply damaged sites and biological consequences. Nucleic Acids Res. 2007;35:3355–3366. doi: 10.1093/nar/gkm190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomax ME, Cunniffe S, O’Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 30.D'Souza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 32.Pearson CG, Shikazono N, Thacker J, O’Neill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shikazono N, Pearson C, O’Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malyarchuk S, Brame KL, Youngblood R, Shi R, Harrison L. Two clustered 8-oxo-7,8-dihydroguanine (8-oxodG) lesions increase the point mutation frequency of 8-oxodG, but do not result in double strand breaks or deletions in Escherichia coli. Nucleic Acids Res. 2004;32:5721–5731. doi: 10.1093/nar/gkh911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gulston M, de Lara C, Jenner T, Davis E, O’Neill P. Processing of clustered DNA damage generates additional DSB in mammalian cells post-irradiation. Nucleic Acids. Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;36:4872–4882. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozmin SG, Sedletska Y, Reynaud-Angelin A, Gasparutto D, Sage E. The formation of double-strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucleic Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harrison L, Hatahet Z, Purmal AA, Wallace SS. Multiply damaged sites in DNA: interactions with Escherichia coli endonucleases III and VIII. Nucleic Acids Res. 1998;26:932–941. doi: 10.1093/nar/26.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 41.Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 42.Miller H, Fernandes AS, Zaika E, McTigue MM, Torres MC, Wente M, Iden CR, Grollman AP. Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 2004;32:338–345. doi: 10.1093/nar/gkh190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dianov GL, Thybo T, Dianova I, Lipinski LJ, Bohr VA. Single nucleotide patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J. Biol. Chem. 2000;275:11809–11813. doi: 10.1074/jbc.275.16.11809. [DOI] [PubMed] [Google Scholar]
- 44.H. Budworth H, Dianov GL. Mode of inhibition of short-patch base excision repair by thymine glycol within clustered DNA lesions. J. Biol. Chem. 2003;278:9378–9381. doi: 10.1074/jbc.M212068200. [DOI] [PubMed] [Google Scholar]
- 45.Clark JM, Beardsley GP. Functional-effects of cis-thymine glycol lesions on DNA-synthesis in vitro. Biochemistry. 1987;26:5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 46.Clark JM, Beardsley GP. Thymine glycol lesions terminate chain elongation by DNA-polymerase-I in vitro. Nucleic Acids Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chem. Res. Toxicol. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 48.Ide H, Kow YW, S.S. Wallace SS. Thymine glycols and urea residues in m13-DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans J, Maccabee M, Hatahet Z, Courcelle J, Bockrath R, Ide H, Wallace SS. Thymine ring saturation and fragmentation products - lesion bypass, mis insertion and implications for mutagenesis. Mutat. Res. 1993;299:147–156. doi: 10.1016/0165-1218(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 50.Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. USA. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lustig ML, Cadet J, Boorstein RJ, Teebor GW. Synthesis of the diastereomers of thymine glycol, determination of concentrations and rates of interconversion of their cis-trans epimers at equilibrium and demonstration of differential alkali lability within DNA. Nucleic Acids Res. 1992;20:4839–4845. doi: 10.1093/nar/20.18.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailly V, Verly WG. Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem. J. 1987;242:565–572. doi: 10.1042/bj2420565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J, Linn S. The mechanisms of action of E. coli endonuclease III and T4 UV endonuclease (endonuclease V) at AP sites. Nucleic Acids Res. 1988;16:1135–1141. doi: 10.1093/nar/16.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.