Abstract
Targeted gene silencing by RNA interference allows the study of gene function in plants and animals. In cell culture and small animal models, genetic screens can be performed—even tissue-specifically in Drosophila—with genome-wide RNAi libraries. However, a major problem with the use of RNAi approaches is the unavoidable false-positive error caused by off-target effects. Until now, this is minimized by computational RNAi design, comparing RNAi to the mutant phenotype if known, and rescue with a presumed ortholog. The ultimate proof of specificity would be to restore expression of the same gene product in vivo. Here, we present a simple and efficient method to rescue the RNAi-mediated knockdown of two independent genes in Drosophila. By exploiting the degenerate genetic code, we generated Drosophila RNAi Escape Strategy Construct (RESC) rescue proteins containing frequent silent mismatches in the complete RNAi target sequence. RESC products were no longer efficiently silenced by RNAi in cell culture and in vivo. As a proof of principle, we rescue the RNAi-induced loss of function phenotype of the eye color gene white and tracheal defects caused by the knockdown of the heparan sulfate proteoglycan syndecan. Our data suggest that RESC is widely applicable to rescue and validate ubiquitous or tissue-specific RNAi and to perform protein structure–function analysis.
INTRODUCTION
In model organisms, such as yeast, worm, fly and mouse, significant progress has been made to create tools for the manipulation of individual genes, such as random mutagenesis and gene targeting altering the genomic sequence, or RNA interference (RNAi) targeting of mRNA for degradation. Lethality and pleiotropy compromise loss-of-function mutant studies for many genes, rendering identification of late or tissue-specific phenotypes difficult. Gene knockdown by RNAi, when combined with the transgenic Gal4/UAS system in Drosophila (1), is cell-autonomous and inducible in space and time. It is also nontransitive (2,3), allowing isoform-specific knockdown. Drosophila genome-wide RNAi libraries have been generated recently, making large-scale tissue-specific reverse genetic screens possible [(4) and http://www.shigen.nig.ac.jp/fly/nigfly].
A major problem of RNAi, however, is off-target effects (5–7). Even for a fully annotated genome, tolerance for mismatches makes the design of unique RNAi target sequences very difficult. This is particularly compounded in Drosophila, where long double-stranded RNA of several hundred base pairs is usually required for efficient knockdown (8), thus increasing the risk of hitting unrelated genes because of tandem trinucleotide repeats or sequences homologous to the RNAi target (5,6,9,10). Apart from that, activation of the innate immune system (11) and competition with the endogenous RNAi machinery (12) have been reported in mammals.
Clearly, improved RNAi reliability by experimental validation is highly desirable, ideally through rescue of the RNAi phenotype by the targeted gene itself. We set out to test if alteration of the cDNA at the nucleotide level while maintaining the encoding of the native protein would circumvent RNAi-mediated silencing and thus permit rescue. We then used this approach to unravel a novel function for the Drosophila proteoglycan syndecan (sdc) in tracheal development.
MATERIALS AND METHODS
RNAi design and synthesis
The Sdc RNAi target region was selected to be larger than 300 bp, to target all existing splice forms, and not to contain sequences longer than 17 bp that are identical in other loci. UAS::SdcdsRNA was designed as a genomic-cDNA hybrid construct targeting 357 bp of sdc exons 6 and 7. The forward genomic part consisting of sdc exon 6, intron 6 and exon 7 was cloned from gDNA into pUAST, followed by the 3-kb sdc intron 5 to improve genomic stability. Then, exons 6 and 7 cDNA were cloned in reverse orientation and transformed into Sure2 repair-deficient bacteria (Stratagene) to avoid hairpin repair (Figure 4) (13). The UAS::wdsRNA line was obtained from D. Smith (13).
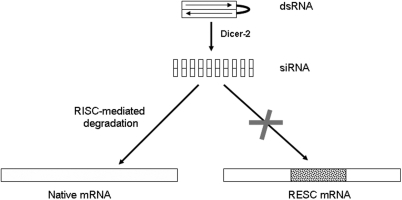
Figure 4.
Illustration of the RESC rescue principle. Small interfering nucleotide duplexes (siRNA) derived from cleavage of long, double-stranded RNAi by the RNase Dcr-2 guides the RNA-induced silencing complex guides (RISC) complex to complementary mRNA that is subsequently cleaved and degraded. RESC constructs are mismatched in the RNAi target region and therefore escape RISC-mediated degradation.
RESC design
The boundaries of the RESC sequence were defined by the RNAi target boundaries. In total, 235 silent mutations were introduced into the 687-bp w RNAi target region and 72, 33 and 17 mutations to obtain 6-mer, 12-mer or 24-mer changes into the 357-bp Sdc RNAi target region (Supplementary Figure 1). GC content, restriction sites, splice sites, repetitive nucleotide sequences and unavoidable gaps (Methionine, Tryptophan) were taken into account, leaving stretches of maximally five nucleotides unchanged. Preferably, amino acids with different codon usage were swapped. Finally, the RESC sequence was verified at amino acid level to be wild type (align p).
wRESC synthesis
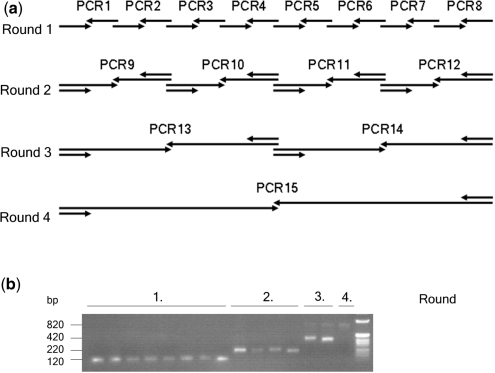
wRESC was synthesized with 16 desalted oligonucleotide primers (Invitrogen) in alternating directions, each 70 nucleotides in length (+/–1 or 2 bp, G or C at 3′ end) and with 20-bp overlap each. To reach unique restriction sites for further cloning, the two flanking primers were extended over the target region to contain wild-type sequence at their 5′ end. Primer pairs (first round) or purified PCR fragments together with the two outside primers (from round 2 on) were subjected to standard PCR elongation (Figure 1b). Finally, a 795-bp BspEI–SnaBI fragment was obtained from four rounds standard PCR and cloned to exchange the wild type w BspEI–SnaBI fragment in pUAST.
Figure 1.
Generation of RESC DNA from oligonucleotides. (a) Illustration of primer positions and PCR reactions required to generate a RESC DNA fragment in four rounds of PCR starting from 16 oligonucleotides. In each round, the two corresponding PCR products and outside primers from the previous round are used for amplification. Arrows indicate direction of elongation. (b) Agarose gel showing wRESC PCR products and their size obtained in four PCR rounds as illustrated in (a).
sdcRESC synthesis
For sdcRESC every 6th, 12th or 24th nucleotide within the exon 6/7 RNAi target region was mutated (Supplementary Figure 1). sdcRESC was synthesized with eight primers in alternating directions, 72–76 bp in length (+/–1 or 2 bp, G or C at the 3′ end) and with 22–25-bp overlap. To reach the unique KspI restriction site in exon 5 the RESC sequence was extended by wild-type sequence. The RESC fragment was obtained from three rounds standard PCR (Figure 1b) and cloned to exchange the sdc wild-type sequence in pUAST-sdc. All inserts were verified by sequencing.
Cell culture
Two Mio Drosophila S2 cells per 1 ml Drosophila serum-free medium (Invitrogen) were transiently transfected with 5 μl Cellfectin (Invitrogen) and 3 μg total DNA (pUAST-sdc with or without RESC modification, pUAST-SdcdsRNA or empty vector, and pMT-Gal4, 1:1:1 ratio), induced with 700 μM CuSO4 5 h after transfection, and harvested 48 h later.
Western blot
S2 cells or whole anesthetized flies were homogenized in lysis buffer (1% Triton X-100, 140 mM NaCl, 10 mM Tris) with complete protease inhibitor cocktail (Roche), boiled in reducing SDS sample buffer and loaded on 4–15% Biorad Ready Tris–HCl gels, and transferred to positively charged Zeta-probe blotting membrane (Biorad). The membrane was blocked with 0.5% Casein (Merck) in 0.6 M NaCl, incubated with α-Sdc (14) or α-tubulin antibody, and detected with alkaline phosphatase-conjugated secondary antibody (Promega) and CSPG (Tropix) on Amersham Hyperfilm.
Fly lines
Fly stocks were kept on standard fly food. Transgenic flies (sdcRESC, wRESC) were generated by standard P-element-mediated germline transformation. Transgenic lines were generated from sdcRESC with 6-mer changes. UAS::wdsRNA flies were obtained from D. Smith, btl-Gal4 from the Japan National Institute of Genetics, all other lines from the Bloomington Stock Center. sdc cDNA was obtained from J. Lincecum, sdc23 stock from G. Vorbrüggen.
Phenotype analysis
The tracheal morphology of third instar larvae was analyzed in a filet preparation by stereomicroscopy. Larvae were cold anesthetized in 4°C PBS and dissected with iris spring scissors (Fine Science Tools) from the ventral side. Although the lumen of the tracheae is easily visible with bright field microscopy, this does not reliably reflect morphology because air filling is not always complete. Hence, reporter lines were generated that express membrane-bound CD8-GFP fusion protein in the trachea with btl::Gal4. Eye colors of 5–7-day-old adult flies were analyzed under white light with a stereomicroscope (Leica).
Primers
sdc RESC, 6-mer changes
For 1
Atcgatggtccgcggatcggtggcaacgatggagatattacagagcgcggaccgggtgctggtggtagcaatgtg
For 2
Tcaatagtcagccgtccgatacaaaaggcattgatcataggccgaacggaaacgaggtggtgataatgagtgag
For 3
Tcccaaccgggaatactggcagctgtgattggaggtgcggtcgtcggcctcctgtgcgcgattctcgtcgtgatg
For 4
Gacgaaggatcgtatgccctggatgagcctaagcgttcgccagccaataattcctatgccaagaacgcgaacaac
Rev 1
TTTGTATCGGACGGCTGACTATTG ACGTTGGTGTTCGGGTCTAGTTCATGCACATTGCT ACCACCAGCACCCGG
Rev 2
CAGCTGCCAGTATTCCCGGTTGGGAGAAGAA ACTGGACGTCCGATCATCCTCACTCATTATCACCACCTCGTTTC
Rev 3
CATCCAGGGCATACGATCCTTCGTC TTTCTTTCGCATTCGGTAGACGATAAACATCACGACG AGAATCGCGCACAG
Rev 4
GAGGTACCCTCGAGCCGGAGC TCGCATATTCTCACGCATAGAATTCTCTGTTGTT CGCGTTCTTGGCATAGG
sdc RESC, 12-mer changes
For 1
Atcgatggtccgcggatcggtggcaacgatggagatattacagagcgcggaccgggtgctggtggcagcaatgtg
For 2
Tgaatagtcagccctccgatacaaagggcattgatcacaggccgaacggcaacgaggtggtcataatgagcgag
For 3
Tcccagcccggaatactggctgctgtgattggcggtgcggtcgttggcctcctctgcgccattctcgtggtcatg
For 4
Gacgagggatcgtatgcgctggatgagccaaagcgttcgccggccaataattcgtatgccaaaaatgcgaacaac
Rev 1
TTTGTATCGGAGGGCTGACTATTCA CGTTGGTGTTGGGGTCTAATTCGTGCACATTGCTG CCACCAGCACCCGG
Rev 2
CAGCAGCCAGTATTCCGGGCT GGGAGAAGAAGCTGGACGTGCGATCATCCTCGCTCATTATGAC CACCTCGTTGC
Rev 3
CATCCAGCGCA TACGATCCCTCGTCTTTCTTCCTCATTCGGTACACGATAA ACATGACCACGAGAATGGCGCAGAG
Rev 4
AGAGGTACCCTCGAGC GCGAGTCGCATTATCTCACGCGTAGAATTCGCGG TTGTTCGCATTTTTGGCATACG
sdc RESC, 24-mer changes
For 1
Atcgatggtccgcggatcggtggcaacgatggagatattacagagcgcggaccgggtgctggtggcagcaacgtg
For 2
Tgaatagtcagccctccgacacaaagggcattgatcacaggcccaacggcaacgaggtggtcatcatgagcgag
For 3
Tcccagcccggaattctggctgctgtgattggcggtgccgtcgttggcctcctctgcgccatactcgtggtcatg
For 4
Gacgagggatcctatgcgctggatgagccaaagagatcgccggccaataattcgtatgcgaaaaatgcgaacaac
Rev 1
TTTGTGTCGGAGGGCTGACTAT TCACGTTCGTGTTGGGGTCTAATTCGTGCACGTTGC TGCCACCAGCACCCGG
Rev 2
CAGCAGCCAGAATTCCGGGCTG GGAGAAGAAGCTCGACGTGCGATCATCCTCGCTCATGATG ACCACCTCGTTGC
Rev 3
CATCCAGCGCATAGGATCCCTCGTC TTTCTTCCTCATGCGGTACACGATAAACATGACCACGAG TATGGCGCAGAG
Rev 4
AGAGGTACCCTCGAGC GCGAGTGCCATTATCTCACGCGTAGAACTCGCGGTT GTTCGCATTTTTCGCATACG
w RESC primers
For 1
Ggctccggatggcggcagctggtcaaccggacacgcggactattctgcaacgagcgacacatcccagctc
For 2
Gagtcgcgtaccctggagagttgctggcagtcatgggtagctcaggcgcgggcaagaccacgctcctgaac
For 3
Ccaggtatccccgtcgggcatgaggctcctgaacgggcagccagtcgatgcaaaagaaatgcaagcgcgg
For 4
Attgggtcgctcaccgcgcgagagcatctcatctttcaagcgatggtccgaatgccgcggcacctcacttac
For 5
Tcatacaagaactcagcttgtcgaagtgccaacataccattataggcgtcccgggacgggtcaagggactaag
For 6
Tgcatcggaagcgctgacggaccctccactgcttatatgtgacgaacctacgagtggcctagatagtttc
For 7
Aaactcagccaaaagggaaagacggtgattctcactatccaccaacctagctctgaactattcgaactgttc
For 8
Gagtcgcctttctaggaaccccttcggaggcagtagatttcttctcatacgtgggtgcccagtgtcctac
Rev 1
CTCTCCAGGGTACGCGAC TCCACAAACATTCTTCAGTAAGTGCTTTCTAGG AGCTGGGATGTGTCGCTCG
Rev 2
TGCCCGACGGGGATACCTGG ATCCCTTGTGGGGACCGGAAAGCCAGCGCGTTCAG GAGCGTGGTCTTGCC
Rev 3
CGCGCGGTGAGCGACC CAATGAATAGATCGTCTTGTTGCACGTACGCACACCGCG CTTGCATTTCTTTTG
Rev 4
CAAGCTGAGTTCTTGTATGA CTTGGTCGACTCGTGCGACGCGTTGGCGGTAAGTGAG GTGCCGCGGCATTC
Rev 5
CCGTCAGCGCTTCCGA TGCAAAGGCTAGCCTTTTCCGCTCGCCTCCGCTTAGTCCCTTG ACCCGTCCCGG
Rev 6
TTTCCCTTTTGGCTGAGT TTCTTAAGGACTTGCACCACGGAATGAGCAGTGAAACT ATCTAGGCCACTCG
Rev 7
GGTTCCTAGAAAGGCG ACTCGCCCTTCAGCCATAAGCAGAATCTTATCGAACAGT TCGAATAGTTCAGAG
Rev 8
CCTACGTAAAAGTCCG CCGGATTGTAGTTGGTAGGACACTGGGCACCCAC.
RESULTS
Rescue of RNAi-mediated w loss-of-function
RNAi specificity is ideally demonstrated by re-expression of the knocked-down gene. However, a transgene containing the original RNAi target sequence is subject to RNAi as well, and deletion of the target sequence while maintaining gene function is possible only if noncoding sequence is targeted. A possible way to circumvent this problem may be to introduce silent mismatches into the rescue construct such that the mRNA sequence is altered while maintaining the protein coding sequence. The degenerate nature of the genetic code permits sequence alterations for 18 out of the 20 amino acids and the stop codon (exceptions are Methionine and Tryptophan), allowing approximately one change every third nucleotide of a cDNA. We reasoned that even when considering the tolerance of the RNAi for some mismatches, this degree of alteration would be sufficient to escape the RNAi machinery.
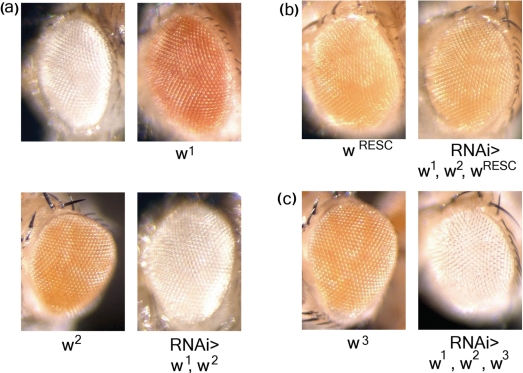
To test this idea in vivo and in vitro, we chose to rescue two genes in Drosophila, one with a known and one with a novel phenotype. The white (w) gene is required for red eye color in Drosophila and was previously shown to be effectively knocked down by RNAi in vivo (13,15) (Figure 2a). We decided to rescue w with a rescue construct containing 235 silent mutations in the 687-bp target sequence and named it wRESC (RNAi Escape Strategy Construct). Since RNAi in Drosophila is not transitive, only the w RNAi target sequence but not the flanking sequences were mismatched. To introduce this high number of mutations, site-directed mutagenesis by which only a few new mutations can be generated per round of PCR is not an efficient approach. In addition, a natural RESC template that could be used for amplification does not exist. Hence, we decided to create the new sequence completely from custom-synthesized oligonucleotides. We divided the target fragment including overlapping wild-type sequence on both ends for further cloning into 16 oligonucleotides, 70–75-nt long and with an overlap of 20–25 nt. By successive elongation of the oligonucleotides and purification of intermediate fragments the final wRESC fragment was obtained after four rounds of PCR (Figure 1a and b). The wRESC fragment was cloned to replace the corresponding part of the wild-type sequence of w in a standard vector used for P-element-mediated germline transformation, where w routinely serves as a marker gene expressed under its own promoter. The original vector in which the w sequence was unaltered served as a control. Several lines of both w and wRESC transgenic flies were obtained which restore eye color to w null mutant flies, showing that the wRESC transgene is functional (Figure 2b). However, whereas w RNAi efficiently targeted combinations of up to three w transgenes, resulting in complete loss of eye color (Figure 2a and c), wRESC was not targeted, as judged by the fact that wRESC and w RNAi + wRESC flies show identical eye color (Figure 2b).
Figure 2.
Photographs (100×) from eyes of age-matched adult flies demonstrating eye color rescue with wRESC. w RNAi causes total loss of eye color, which is restored with wRESC but not w3. Genotypes are (a) w1118 (upper left); w1118;tub::Gal4,w+(1)/+ (upper right); w1118;;UAS::wdsRNA,w+(2)/+ (lower left); w1118;;tub::Gal4,w+(1)/UAS::wdsRNA,w+(2) (lower right). (b) w1118,w+(RESC)/w1118 (left); w1118,w+(RESC)/w1118;; tub::Gal4,w+(1)/UAS::wdsRNA,w+(2) (right) (c) w1118,w+(3)/w1118 (left); w1118,w+(3)/w1118;; tub::Gal4,w+(1)/UAS::wdsRNA,w+(2) (right).
Sdc RESC in vitro
Available mutants of the single fly homolog of the vertebrate transmembrane heparan sulfate proteoglycan syndecan (sdc) are all derived from imprecise excisions causing deletions that potentially affect a nearby gene on the complementary strand called Smad anchor for receptor activation (sara). sara shares the enhancer region with sdc, making sdc an ideal candidate to confirm the validity of the RESC strategy.
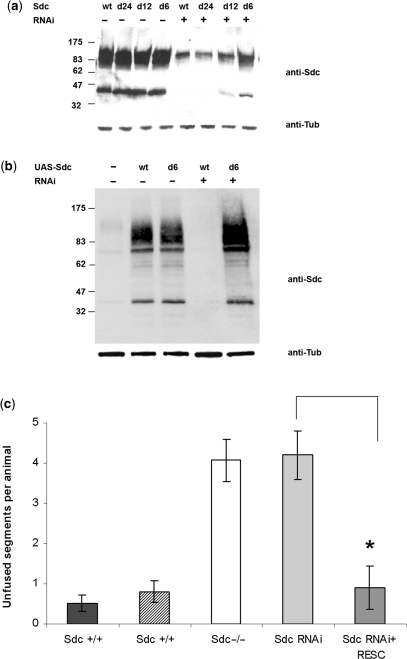
While a maximum number of nucleotides were mismatched in wRESC, we now first tested to what extent the nucleotides in sdcRESC had to be exchanged in order to escape RNAi. We generated three UAS::sdcRESC vectors for rescue experiments, with mismatches every 6th, 12th or 24th bp in exons 6 and 7, giving rise to 72, 33 and 17 mutations in the 357-bp target region, respectively (Supplementary Figure 1). In Drosophila S2 cells in culture, all sdc constructs were expressed at similar levels in the absence of RNAi, but under sdc RNAi conditions the expression levels decreased with increasing similarity between hairpin sequence and target region in the rescue construct. Expression of sdcRESC with 24-mer-mismatch spacing was not different from wild-type sdc, suggesting that a mutation every 24th base pair is insufficient to escape silencing by RNAi. The 12-mer changes resulted in some level of escape; however, the strongest effect was observed with the 6-mer construct (Figure 3a).
Figure 3.
sdcRESC rescue in vitro and in vivo. (a) αSdc western blot from Drosophila S2 cells transiently transfected with pMT-Gal4, with (+) or without (–) UAS::SdcdsRNA, and UAS::sdc with wild-type (wt), or RESC sequence (Δ24, 24-mer changes; Δ12, 12-mer changes; Δ6, 6-mer changes). (b) αSdc western blot from adult flies ubiquitously expressing Sdc without (wt) or with (Δ6) RESC sequence, in the presence (+) or absence (–) of ubiquitous Sdc RNAi. (c) Quantification of unfused segments of tracheal dorsal branches per animal, in wild-type (sdc+/+), sdc23 homozygous (sdc−/−) mutants, tracheal Sdc RNAi alone, or with tracheal rescue sdcRESC. n ≥ 10; error bars = CI; *P < 0.01.
Sdc RESC in vivo
To confirm the RESC effect in vivo, transgenic flies were generated that inducibly express sdcRESC with 6-mer mismatches. In the absence of RNAi, both sdc and sdcRESC were efficiently translated, while under RNAi conditions most wild-type sdc but very little, if any, sdcRESC was degraded (Figure 3b).
Both, sdc mutants and RNAi animals, show an almost identical nonfusion phenotype in the dorsal branches of the tracheal system (Schulz et al., submitted for publication). When sdcRESC was expressed in the tracheae of sdc RNAi animals, the dorsal branch phenotype was completely reversed (Figure 3c), confirming that Sdc is necessary for tracheal development and that the RESC technique permits tissue-specific rescue of RNAi-mediated phenotypes.
DISCUSSION
The discovery of RNAi and its exploitation for the versatile, rapid and efficient study of gene function has the potential to revolutionize genetics and genetic screens in small model organisms such as Drosophila and Caenorhabditis elegans. However, a major drawback of RNAi-mediated gene targeting is off-target effects, especially in Drosophila where long dsRNA give rise to many short dsRNA species. Thus far, off-target effects were addressed by in silico analysis of the RNAi sequence, use of more than one nonoverlapping RNAi per targeted gene, rescue by a putative ortholog or paralog, or comparison to known mutant phenotypes (16,17). The latter is of particular importance for conclusions about biological function but limited by the fact that null alleles are only available for a minority of genes, and that even among those not all show easily discernable phenotypes. The most powerful proof of RNAi specificity would be rescue by the endogenous target gene itself. Here, we present a simple method that exploits the degenerate genetic code to introduce frequent silent mismatches in the entire target region in an otherwise wild-type rescue construct. On average, exchange of every third base in a coding sequence is possible. Our data indicate that exchanging every sixth nucleotide—equivalent to three to four mismatches in a ∼21-bp dsRNA—only in the segment targeted by the RNAi is sufficient to rescue RNAi mediated knockdown. In contrast to the in vivo experiments, the rescue appears to be incomplete in vitro. Yet, as opposed to the stable expression under in vivo conditions, the expression of the transgenes in cell culture was achieved by triple transient transfections, and it is unlikely that all cells with knockdown also express the rescue construct. Constructs with mismatch of every 12th nucleotide or less, yielding ∼21-bp fragments with only two peripheral or a single central mismatch, were still largely degraded. This confirms that RNAi target recognition is extremely sequence specific and several mismatches per short dsRNA still allows for some translation (15,18,19).
When RNAi is performed with long hairpins of up to 1000 bp such as in Drosophila (8), site-directed mutagenesis would be too time consuming to introduce the high number of mutations required to modify the complete target sequence of the rescue construct. Instead, we suggest introducing mutations by using long, overlapping oligonucleotides as templates for amplification and synthesis of the entire RESC fragment. Assuming oligonucleotides that are 80-nt long and overlap by 20 bp, synthesis of a target region of 500 bp or less requires three PCR rounds or seven reactions with 8 primers, for target regions between 500 and 1000 bp four PCR rounds with 16 primers suffice. Due to the transitive nature of Drosophila RNAi, it is sufficient to mismatch only the direct RNAi target sequence, thus the size of the RESC fragment is independent from the total gene product size.
To simplify future RESC design, we created a simple and user-friendly web-based tool to generate RESC and the necessary oligonucleotides sequences for any gene at: https://med.kuleuven.be/cme-mg/lng/RESC/.
To demonstrate the feasibility and functionality of RESC, we rescued phenotypes associated with two independent genes in vitro and in vivo. The eye color phenotype of the w gene was restored by wRESC containing mismatches in 212 of 229 codons of the target region, but not in the nontargeted sequence. The targeting of sdc represents a common case where despite the availability of putative null alleles, their specificity cannot be completely established. In this case the combination of RNAi and RESC reveals novel tissue-autonomous and gene-specific functions. Furthermore, once a RESC construct is generated it can be exploited further in rescue-based structure–function analysis.
The combination of the transgenic RNAi and RESC rescue with the Gal4/UAS system and its variations offers several advantages. First, wild-type protein can be knocked down and exchanged for by wild-type or mutant protein using the same promoter at any time during development, and in any chosen tissue. Second, tissue-specific expression avoids ‘anatomical off-target effects’, in contrast to systemic RNA injection. Third, without Gal4 both UAS::dsRNA and UAS::RESC can be kept in a combined stock in off-modus, avoiding modifier accumulation over several generations of stock keeping. Fourth, the effects are dominant, making screens more feasible.
Our technique has implications beyond Drosophila. In mammals, RNAi is already successfully used to knock down dominant mutant human disease genes such as Huntingtin (20,21), but in human gene therapy a problem may arise from targeting of the endogenous wild-type copy (20). A RESC-based approach rescue might be a valuable strategy to prevent this.
The main limitations of the Gal4-driven RNAi/RESC combination are intrinsic to the Gal4/UAS system. First, RESC expression levels are not endogenous and, due to position effects, subject to variation between the transgenic lines. The latter may be overcome by using a P-element replacement strategy or φC31 integrase to target a common locus. Overexpression is more difficult to avoid when using the Gal4/UAS system alone, since lowering expression by decreasing the temperature will also affect RNAi performance. To uncouple RNAi and rescue, two independent systems with different induction modes, such as the Gal4 and lexA systems may be used simultaneously (22). Second, the use of tissue-specific driver lines can only cause knockdown in cells where the target mRNA is present, but it induces expression in all cells expressing the driver, which is not necessarily identical. Here, the endogenous promoter of the gene of interest would be ideal. Another limitation is that synonymous codons are not necessarily expressed with the same accuracy (23) and at the same level (24), but this is probably of minor importance given the efficiency of the rescue we observe. In our web-based tool we suggest the most frequently used alternative codon.
An alternative to the RESC approach might be the use of cDNA from other Drosophila species. However, finding a related species with the right balance between sufficient change at the nucleotide level to avoid knockdown and no critical change at the amino acid level in order to fully restore function is not an easy task.
In summary, the combination of RESC rescue with large-scale RNAi libraries are a powerful approach to study gene function in vitro and tissue-specifically in vivo with high reliability because off-target effects can be excluded experimentally.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
The National Fund for Scientific Research - Flanders (FWO) (grant G.0498.05 to G.D.) and grants G.0542.08N and G.0543.08N (to B.A.H); Flanders Institute for Biotechnology (VIB) (to G.D. and B.A.H.); Interuniversity Attraction Poles (IUAP VI-20) Program of the Belgian Federal Government (to G.D.); and K.U.Leuven (to B.A.H.; Impuls, CREA, GOA; and to G.D., GOA). Funding for open access charge: VIB.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Helga Ceulemans for excellent technical assistance, Stein Aerts (Leuven) for establishing the online tool to generate RESC constructs, the Bloomington Drosophila Stock Center, the National Institute of Genetics (Shizuoka), Dean Smith (Dallas), and Gerd Vorbrüggen (Göttingen) for providing fly stocks, and John Lincecum (Boston) for sdc cDNA and antibody.
REFERENCES
- 1.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 2.Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celotto AM, Graveley BR. Exon-specific RNAi: a tool for dissecting the functional relevance of alternative splicing. RNA. 2002;8:718–724. doi: 10.1017/s1355838202021064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- 7.Moffat J, Reiling JH, Sabatini DM. Off-target effects associated with long dsRNAs in Drosophila RNAi screens. Trends Pharmacol. Sci. 2007;28:149–151. doi: 10.1016/j.tips.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aleman LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 11.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell. Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 12.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 13.Kalidas S, Smith DP. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–184. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- 14.Spring J, Paine-Saunders SE, Hynes RO, Bernfield M. Drosophila syndecan: conservation of a cell-surface heparan sulfate proteoglycan. Proc. Natl Acad. Sci. USA. 1994;91:3334–3338. doi: 10.1073/pnas.91.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 16.Arziman Z, Horn T, Boutros M. E-RNAi: a web application to design optimized RNAi constructs. Nucleic Acids Res. 2005;33:W582–W588. doi: 10.1093/nar/gki468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature. 2007;445:785–788. doi: 10.1038/nature05528. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 22.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 23.Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlini DB, Stephan W. In vivo introduction of unpreferred synonymous codons into the Drosophila Adh gene results in reduced levels of ADH protein. Genetics. 2003;163:239–243. doi: 10.1093/genetics/163.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.