Abstract
Array manufacturers originally designed single nucleotide polymorphism (SNP) arrays to genotype human DNA at thousands of SNPs across the genome simultaneously. In the decade since their initial development, the platform's applications have expanded to include the detection and characterization of copy number variation—whether somatic, inherited, or de novo—as well as loss-of-heterozygosity in cancer cells. The technology's impressive contributions to insights in population and molecular genetics have been fueled by advances in computational methodology, and indeed these insights and methodologies have spurred developments in the arrays themselves. This review describes the most commonly used SNP array platforms, surveys the computational methodologies used to convert the raw data into inferences at the DNA level, and details the broad range of applications. Although the long-term future of SNP arrays is unclear, cost considerations ensure their relevance for at least the next several years. Even as emerging technologies seem poised to take over for at least some applications, researchers working with these new sources of data are adopting the computational approaches originally developed for SNP arrays.
INTRODUCTION
Identifying DNA variants that contribute to disease is a central aim in human genetics. Pinpointing these causal loci requires the ability to assess DNA sequence variation on a genome-wide scale. At the vast majority (some 99%) of genomic sites, every human carries the same base residue on both chromosomal homologs. The remainder encodes much of the diversity among humans, including differences in disease susceptibility. Single nucleotide polymorphisms (SNPs)—genome positions at which there are two distinct nucleotide residues (alleles) that each appears in a significant portion of the human population—comprise a major part of these DNA variants. There are some estimated 10 million SNPs in the human genome (1). For simplicity, manufacturers often arbitrarily label the two alleles of a SNP as A and B. Therefore, since each individual usually inherits one copy of each SNP position from each parent, the individual's genotype at a SNP site is typically either AA, AB or BB.
Due to the importance of SNPs, the International HapMap Consortium and others are part of an ongoing effort to identify SNP loci, genotype them in individuals of various ancestries, and uncover their correlation structure in the genome. In the past few years, however, researchers have uncovered copy number variants (CNVs) as important contributors to human genetic variation (2,3). CNVs are defined as chromosomal segments, at least 1000 bases in length, that vary in number of copies from human to human (4). Since their discovery, several high-profile studies have appeared associating CNVs with a variety of common diseases. Recent examples include Alzheimer disease (5), Crohn's disease (6), autism (7,8), psoriasis (9), Parkinson's disease (10) and schizophrenia (11). As the importance of the duplications and deletions that result in these variants is becoming apparent, cataloging them and assessing their frequencies is now an important goal. The Database of Genomic Variants (http://projects.tcag.ca/variation/) is a public effort aiming to comprehensively catalog all human CNVs (and other forms of structural variation) in a manner analogous to that undertaken by the government project dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) for SNPs.
Considerable progress has been made in the technological ability to assay humans for genetic variation. Commercial probe-based SNP array platforms can now genotype, with >99% accuracy, about one million SNPs in an individual in one assay (12,13). Furthermore, the cancer research community has, for some years, been applying these arrays to tumor DNA to find gross copy-number abnormalities in chromosomes. Refinements of the corresponding copy-number detection algorithms subsequently enabled the detection of CNVs in germline DNA from raw SNP array data, and the technology has become the central tool in genome-wide detection of various types of DNA sequence-level human variation.
Over the last decade, the SNP array has been the common thread in an extremely productive synergistic relationship between advances in biological understanding, computational methodology and the technological development in the arrays themselves (Figure 1). Progress in each of these three scientific arenas has spurred progress in the other two, resulting in advances that would have been impossible independently. This review details the history of the development of, applications for, and computational advances associated with SNP arrays. As algorithms used to convert the raw array data into biological inferences have evolved, so have the array platforms developed in response to the biological and computational advances. Furthermore, although ‘next generation’ DNA sequencers may gradually take over some applications (14), computational biologists have already begun borrowing methods initially developed for SNP arrays to analyze sequence data. The review concludes by considering the promise of high-throughput sequencers as tools to assess human genetic variation, contrasting their costs and capabilities with those of SNP arrays.
Figure 1.
Synergy between computational methodology, biological inferences and technology. This review aims to showcase SNP arrays at the center of a dynamic synergy across these three fields, each helping to drive advances in the others.
UNDERLYING TECHNOLOGY AND SNP GENOTYPING
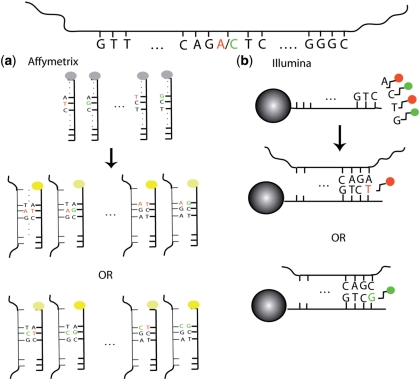
Although the Affymetrix and Illumina SNP arrays work using different chemistries, they have several aspects in common. Both rely on the biochemical principle that nucleotide bases bind to their complementary partners—specifically, A binds to T and C binds to G, in Watson–Crick base pairs. Both array protocols call for the hybridization of fragmented single-stranded DNA to arrays containing hundreds of thousands of unique nucleotide probe sequences. Each probe is designed to bind to a target DNA subsequence. A specific hypothetical example for one SNP is shown in Figure 2. In both cases, specialized equipment can produce a measure of the signal intensity associated with each probe and its target after hybridization. The underlying principle is that the signal intensity depends upon the amount of target DNA in the sample, as well as the affinity between target and probe. Extensive processing and analysis of these raw intensity measures yield SNP genotype inferences. Both manufacturers report genotyping accuracy well over 99.5%. This section details some of the computational algorithms that have been developed to convert the set of probe intensities into genotypes.
Figure 2.
Overview of SNP array technology. At the top is the fragment of DNA harboring an A/C SNP to be interrogated by the probes shown. (a) In the Affymetrix assay, there are 25-mer probes for both alleles, and the location of the SNP locus varies from probe to probe. The DNA binds to both probes regardless of the allele it carries, but it does so more efficiently when it is complementary to all 25 bases (bright yellow) rather than mismatching the SNP site (dimmer yellow). This impeded binding manifests itself in a dimmer signal. (b) Attached to each Illumina bead is a 50-mer sequence complementary to the sequence adjacent to the SNP site. The single-base extension (T or G) that is complementary to the allele carried by the DNA (A or C, respectively) then binds and results in the appropriately-colored signal (red or green, respectively). For both platforms, the computational algorithms convert the raw signals into inferences regarding the presence or absence of each of the two alleles.
Affymetrix platform
Affymetrix was the first to commercially produce SNP arrays, nearly a decade ago. The HuSNP assay, initially prototyped in Wang et al. (15), was designed to genotype 1494 SNPs on one chip. Subsequent versions increased stepwise from 10 000 to 100 000 to 500 000, and finally to nearly one million SNPs in the current release. Every SNP site is interrogated by a set of probes that are each 25-nt long. A probe is designed to be complementary, or very nearly complementary, to a portion of the DNA sequence harboring the SNP site (Figure 2a). In the first few versions of the array, each SNP was interrogated by between 24 and 40 distinct probe sequences, forming a probe set. Within a set, each probe is associated with either allele A or allele B. Additionally, each probe is either a perfect match (PM; perfectly complementary to one of the target alleles), or a mismatch (MM; identical to a perfect match probe except that the center base is altered so as to be perfectly complementary to neither allele). The idea of the mismatch probe comes from mRNA expression arrays (16), and their purpose is to measure background noise. The scheme yields quartets comprised of four types of probes: PMA, MMA, PMB and MMB. The computational goal is to convert these 8–10 probe quartet intensity measures from raw array data into a genotype inference—AA, AB or BB.
With each version of the array developed by the manufacturer, the computational community has responded with corresponding algorithmic development. The algorithms have, in turn, influenced array design. For example, in deciding which SNPs to include on each new version of the array, Affymetrix has chosen those for which the current computational algorithm performs best. For the 10K version of their array (17,18), they adopted a partitioning around medoids (PAM)-based algorithm (19). Interestingly, PAM is a statistical methodology developed with social science applications in mind (20), but computational biologists adapted the approach to meet the needs of the technology. Briefly, at each SNP the algorithm uses the maximum difference between PMA and MM probes as a proxy for allele A abundance, and the analogous measure for allele B abundance. Unsupervised clustering, across many samples, of the ratio of A abundance to the sum of A and B abundances yields the three genotype classes. Any point not sufficiently close to its closest cluster center is given an indeterminate ‘No Call’ genotype. To attain a high level of genotypic heterogeneity, the manufacturers trained their algorithm on an ethnically diverse panel of 133 individuals.
The algorithm used for the 10K version relies on examining probe intensities across multiple arrays. In order to compare these values fairly, it is crucial to first normalize the intensities to take into account non-biological differences such as overall array brightness. Normalization aims to correct these technological biases in probe intensity by homogenizing, to some degree, the intensity distributions of the arrays. Normalization methods initially proposed were adopted from the mRNA expression microarray literature, and include cyclic lowess (21), invariant-set normalization (22) and others. These normalization methods developed by the computational research community are relatively sophisticated. Surprisingly, however, current consensus seems to have instead settled on a very simple approach. Quantile normalization (23) is a non-parametric method that ensures that all arrays in the study have precisely the same probe intensity distribution. The basic algorithm can be programmed in one line of computer code in most languages. One simply replaces the nth highest probe intensity value of each array with the mean of the nth highest probe intensity values across all arrays. The effect is to ensure that an array's highest-intensity probe has the same value across arrays, as does the second highest, and so on. However, the intensity ranks of the probes within each array remain unchanged.
With the advent of the 100K array, the manufacturer switched to a dynamic model algorithm (24). Cutler et al. originally developed the algorithm for a different Affymetrix product, the sequencing array. Unlike the PAM-based algorithm, the dynamic model approach operates without a need for training data. The idea is to represent the three genotypes by three different models relating genotype to the signal intensity values of each probe quartet. The AA model stipulates that the PMA intensity predominates, while the intensities of the other three probes have smaller (and approximately equal) means. Similarly, the BB model stipulates a PMB foreground and approximately equal background for the other three. The AB model assumes equal PMA and PMB means in the foreground, and equal MMA and MMB means in the background. The algorithm also adds a null model of equal means across all probe types, corresponding to a genotype No Call. The score for each model is the difference between the model likelihood and the highest likelihood among the other models. This yields four scores for each probe quartet. Finally, a Wilcoxon signed rank test is performed against the null hypothesis of median score (across quartets) equal to zero for each model. A significant P-value gives a corresponding genotype call.
When Affymetrix introduced the 500K array, they initially used the dynamic model algorithm. However, the computational community again responded to the evolving platform with yet another approach (25), which the manufacturer subsequently modified and called Bayesian Robust Linear Model with Mahalanobis distance classifier (BRLMM). This algorithm ignores the MM probes, based on the assertion that they add no improvement over using the PM probes alone. The method explicitly models log-transformed probe intensity as a stochastic function DNA quantity, including probe-specific effects as a term. For a fixed SNP, the model is
where Iij is the (normalized) probe intensity in array j for probe i of the probe set interrogating the SNP, fi represents the probe-specific effect, tj represents the genotype-specific effect (the quantity of interest), and eij is an error term. BRLMM fits the model using median polish (26), separately for each of the A and B alleles. The result is a pair of signal values (one for each allele) at the SNP for each array. Following a ‘cluster center stretch’ transformation, the algorithm clusters the pairs. The posterior distribution of a Bayesian procedure determines the cluster centers and variance/covariances. Finally, the method assigns genotypes based upon the transformed pairs’ Mahalanobis distance from the cluster centers. ‘No Calls’ are made when the distance from the closest cluster center is more than half the distance to the second closest. For the 500K array, as well as its successor the 6.0 array (see below), model training was performed using the 270 HapMap samples (27).
The most recent version of the Affymetrix array has seen several changes, largely driven by the academic sector. The industry collaborated with computational researchers, whose observations (28–30) led the fundamental changes in probe composition to optimize the limited space available on the array. Each SNP on the Human SNP Array 6.0 is interrogated only by six or eight perfect match probes—three or four replicates of the same probe for each of the two alleles (31). Therefore, intensity data for each SNP consists of two sets of repeated measurements. Furthermore, the SNP probe sets are augmented with nearly 1 million copy number probes, which are meant to interrogate regions of the genome that do not harbor SNPs, but rather may be polymorphic with regard to copy number. Each such copy number site is interrogated by only one probe (see below). Academic researchers developed a new genotyping algorithm, termed Birdseed (32), which Affymetrix adopted in the software that is marketed with the 6.0 array. From the raw A and B (normalized) probe intensities, Birdseed obtains a pair of summarized A and B signals using a median polish procedure similar to that used in BRLMM. Internally-run array data from 270 individuals (27) determines expected locations of the clusters formed by plotting A signals versus B signals a priori. Birdseed fits the signals from the test samples to a two-dimensional Gaussian mixture model using an expectation–maximization (EM) procedure (33), with initialization provided by the a priori expected locations. The EM procedure results in genotypes for each SNP, giving a confidence score for each genotype based on the call's proximity to its cluster. Thresholds may be set for these confidence scores to generate No Calls.
Illumina platform
Like the Affymetrix platform, the Illumina BeadArray has gradually increased in capacity over the years—from 100 000 SNPs (Human-1) to the current (HumanHap1M) one million, with intermediary steps 240 000, 317 000, 550 000 and 650 000. However, from a data analysis perspective, the array data output format has remained relatively consistent—one raw measurement for the A allele and one for the B allele at each SNP (Figure 2b). As a result of the stability in data output (and fewer years on the market), there has been considerably less algorithmic evolution.
The raw file from a single HumanHap1M array consists of some two million data points, conceptually some one million pairs,
The computational workhorse in the Illumina protocol is its normalization procedure. Instead of normalizing across arrays, the manufacturer's software performs internal normalization on each sample individually, without relying on multiple arrays, using the same six transformation parameters for all allele pairs. The parameters capture appropriate factors for shifting, scaling, and rotating the X- and Y-coordinates, and are inferred using the pairs themselves, following outlier removal. The goal is to produce a pair of raw allele-specific copy measurements at each SNP. The method uses these pairs for genotype calls. Specifically, for each SNP, define a transformed ratio θ of the normalized allele intensities
Each of the three genotypes—AA, AB or BB—represents a cluster in one-dimensional θ space (34,35). Proximity to a cluster determines a test sample's genotype, and cluster separation determines SNP quality score. It should be noted that, like the current version of the Affymetrix array, the HumanHap1M also includes copy number probes meant to interrogate non-SNP human genetic variation.
Application: determining ancestry from multiple genotypes
One benefit of the ability to generate many genotypes easily and quickly is convenient assessment of individual ancestry. This is particularly important in disease association studies involving individuals with diverse ancestries. In such studies, population stratification can produce a false apparent association between disease and SNP genotype when ancestry differs between affected and unaffected groups, as any SNP with allele frequencies differing by ancestry may appear to be associated with disease. The necessity of accounting for ancestry in population-based studies immediately underscores the advantage of SNP array-based studies. Rather than relying on self-reported ancestry, which is notoriously problematic (36), the researcher can automatically infer detailed information in this regard without the need for additional data collection.
The most widely applied method to assess ancestry based on multiple genotypes is the structure program (37). Structure assumes that there are K subpopulations in the sample, with the underlying idea that each genotyped locus will have different allele frequencies in each of the K subpopulations, enabling distinction between them. After inference using a Markov chain Monte Carlo method, structure outputs probabilities for each individual having ancestry from each subpopulation. When structure assigns to an individual substantial (non-zero) probability values for more than one subpopulation, it can signal a mixed ancestral background. With fewer than 100 SNP genotypes, structure can easily classify the HapMap samples into African, Asian and European ancestries. Furthermore, varying degrees of European and African ancestries can be discerned in African American individuals.
The structure methodology was developed before SNP arrays became commercially available, and hundreds of thousands of genotypes is clearly overkill with regard to the algorithm's necessary input. In an example of the technology driving computational development that yields biological insights, the high genomic resolution enabled by the SNP array has facilitated a corresponding dramatic increase in geographic resolution in ancestry inference. A principal components analysis-based method, termed EIGENSTRAT (38), facilitates the processing of large data sets to detect population stratification with greater sensitivity. This is an important application, as even subtle population stratification can inflate P-values in association studies. The initial EIGENSTRAT publication used data from the 100K array, and the authors have subsequently demonstrated (39) that EIGENSTRAT is able to subdivide individuals with European ancestry into subgroups of northwest European, southeast European and Ashkenazi Jewish ancestries.
One may regard ancestry as a gross measure of distant relatedness. SNP genotypes also provide information sufficient to reveal relatedness on a finer scale, for example identifying siblings or even distant cousins. Like population stratification, cryptic relatedness can also be a confounder in association studies. An extreme case of two samples being related is the samples’ being from the same individual. It is not uncommon that the same individual's DNA is inadvertently collected by two different centers for the same study, for example. To guard against such replication of samples, which can result in false positive results in association studies, researchers can easily screen the samples by comparing genotypes in a pairwise manner. Two arrays run on the same individual will match at nearly all genotypes (allowing some differences due to genotype error), and one group (40) identified an optimal panel of 34 SNPs from the Affymetrix 50K array to ‘bar code’ samples. Therefore, SNP array genotypes allow researchers to infer relatedness across the entire spectrum, from similarities in ancestry to individually unique DNA fingerprinting.
Applications: pooled DNA and allele-specific expression
Although the SNP array's manufacturers designed the technology to genotype genomic DNA, using one individual per array, the academic community has extended the array's genotyping capabilities to wider applications. For example, rather than treating SNP genotype as a categorical variable, researchers have demonstrated (41) that the array signals may be used to measure SNP allele frequencies, a continuous variable, in pooled DNA from hundreds of individuals. Using such an approach, a study may analyze data from pooled disease cases and pooled controls in batches to assess differences in allele frequencies, which may signal associations between genetic variants and disease susceptibility.
Another unforeseen application is the genotyping of RNA rather than DNA to assess allele-specific expression. Recently, studies have used both the Affymetrix (42) and Illumina (43) platforms to detect genes for which one of the parental alleles is expressed at a higher level than the other. The authors of the studies were able to detect allele-specific expression by using the arrays to genotype transcribed SNPs that are heterozygous in the individual's genomic DNA, then algorithmically assess the relative abundance of each of the two SNP alleles in the RNA. These studies reveal a surprising number of genes that show an imbalance in allelic expression.
LINKAGE DISEQUILIBRIUM AND GENOME-WIDE ASSOCIATION STUDIES
The dramatic increase in the density of SNP arrays has facilitated (and been driven by) a corresponding explosion of studies aiming to find inherited genomic variants associated with human disease. Armed with the ability to query DNA genome-wide, researchers may proceed agnostically, without any a priori expectations as to the associated variants’ functions or genomic locations. SNP arrays genotype far fewer than >10 million known human SNPs. How, then, is the platform able to capture a large proportion of DNA-level variation by genotyping only one million (or fewer) SNPs? This can be explained by linkage disequilibrium.
Linkage disequilibrium (LD) and phasing
LD occurs where alleles at two or more loci appear together in the same individual more often than would be expected by chance. LD in humans primarily manifests itself in loci on the same chromosome that have limited historical recombination between them. Mathematically, LD between two SNPs on the same chromosome can be quantified as correlation between alleles across population chromosomes. The standard measures of this correlation are D, D′ and r2, all of which may be expressed as functions of the allele frequencies of the two SNPs (44). Technically speaking, D is simply the traditional statistical covariance of the two binary random variables representing haploid genotypes, and r2 is the square of the statistical correlation coefficient.
Two SNPs that are in strong LD may serve as proxies for one another. That is, if the correlation between the two SNPs—as measured by r2, for example—is high, genotyping one of the SNPs gives nearly complete information regarding the genotype of the other SNP. Therefore, a SNP array that genotypes 1 million SNPs effectively assays a larger proportion of human genetic variation than represented on the array. Taking advantage of this principle, the array manufacturers have been specifically designing arrays to query SNPs that correlate with, or ‘tag’, a large number of other SNPs in the human genome. The requisite knowledge of the LD structure in the genome was largely facilitated by the International HapMap Project (27), which itself uses both the Affymetrix and Illumina arrays for population genotyping along with a battery of statistical algorithms. The term HapMap is an abbreviation for ‘haplotype map’, which itself is derived from the phrase haploid genotype. The phrase implies not simply the alleles at each SNP, but the sequence of consecutive SNP alleles that occur on each chromosome. For example, suppose an individual carries heterozygous AB genotypes at each of two (genomically) consecutive SNPs. There are two possible pairs of two-SNP haplotypes for the individual's two chromosomes. Either (i) one chromosome carries the A allele at both SNPs and the other the B allele at both SNPs, or (ii) one chromosome carries the A allele at the first SNP and the B allele at the second, with the other chromosome carrying the reverse. The process of determining which of these possibilities is the reality is referred to as phasing, and the possibilities become increasingly complex with more SNPs. Phasing is necessary to determine LD structure from population genotypes, which is in turn necessary to estimate the amount of human genetic variation captured by each SNP. Although there are many algorithms for computational phasing (45), PHASE (46) is the algorithm used by the HapMap Project. Here we again witness the computation-biology-technology cycle at work. The HapMap Project uses SNP arrays to help provide genotypes in a high-throughput manner. Computational analysis of the results reveals the biological reality of LD structure. The cycle is completed as the LD structure informs the design of the next generation of SNP arrays.
The lower the LD between SNPs, the more independent information they represent. If the array can physically accommodate only one million SNPs, then the goal is to choose the SNPs that capture the largest proportion of genetic variation measured by known SNPs. A convenient metric to measure an array's ability to capture common human genetic variation is the proportion of known human SNPs captured, above a fixed r2 threshold, by array SNPs. Product literature from Affymetrix and Illumina reports these metrics, and a study by Pe’er et al. (47) found that some 80% of common (in Caucasians) human SNPs are captured (at r2 > 0.7) by the markers on the Affymetrix 500K and Illumina HumanHap300 arrays. When this study was published, Illumina's SNP selection was performed to optimally capture human genetic variation, as assessed by correlations measured using HapMap Caucasian genotypes, while Affymetrix selected SNPs that performed best with regard to genotyping accuracy. This explains the HumanHap's ability to capture a similar amount of genetic variation with fewer SNPs.
Genome wide association studies (GWAS): linking disease to DNA sequence
The power of SNP arrays to interrogate a significant proportion of human genetic variation has facilitated hundreds of GWAS, and many more are underway. The goal in GWAS is to find the variants that are statistically more prevalent in individuals with a disease than in individuals free of the disease. A study typically entails collecting large numbers of affected (cases) and unaffected (controls) individuals, and running the DNA of all individuals on SNP arrays. The researchers then mine the resulting data for statistically significant differences in allele frequencies between the two groups. The associated variant is then putatively either a disease predisposition allele, or in LD with such an allele. Pinpointing the predisposition allele can lead to genetic tests, treatment options, and insight into the disease biology.
In recent years, GWAS have used both the Affymetrix and Illumina platforms. A considerable limiting factor for doing these studies is the cost of genotyping enormous numbers of cases and controls. Large numbers are necessary because the high density of the arrays, while allowing hundreds of thousands of variants to be tested, results in hundreds of thousands of tests. In order to avert huge numbers of false positive associations, the P-value threshold for statistical significance must be very stringent—typically 10−6 or lower—to accommodate the huge multiple testing burden. This problem is exemplified by the study performed by the Wellcome Trust Case Control Consortium (48), wherein the authors performed GWAS on seven different diseases, ∼2000 cases for each disease. Rather than having separate panels of control individuals for each disease, the study shared 3000 unaffected controls across the diseases. All 17 000 individuals were genotyped on the Affymetrix 500K array. The density of the array allowed the researchers to test an enormous number of SNPs, but the multiple tests meant that only associations with P-values less than 5 × 10−7 were reported as bona fide. The large sample sizes were therefore necessary for power sufficient to obtain such low P-values.
DETECTING SOMATIC CHANGES IN CANCER CELLS
For decades, cancer biologists have known that chromosomal instability is typical of human cancers (49). The sporadic amplifications and deletions of genomic segments are an area of intense research interest, as genes in amplified regions represent candidate oncogenes, and those deleted represent candidate tumor suppressor genes. Almost immediately after SNP arrays were developed, researchers adapted them to query the tumor genome, drafting the emerging technology for applications beyond those that the manufacturers intended.
Loss-of-heterozygosity (LOH) detection from array data
LOH is the sporadic loss of all or part of one of two parental chromosome homologs. One case is hemizygous deletion, where one homolog loses a segment while the other remains at one copy per cell. However, ‘copy-neutral’ LOH (also known as uniparental disomy or gene conversion)—wherein the retained homolog is duplicated so as to preserve two total copies per cell—is quite common in some cancers (50). By definition, LOH implies a change from a heterozygous state to a homozygous state. Since SNP arrays are specifically designed to assess such states, they are natural tools for LOH detection. Lindblad-Toh et al. (51) applied the very first Affymetrix prototype SNP array (what would now be considered a very low-density array at some 1500 SNPs) to LOH detection by comparing the genotypes in the patient's tumor DNA to those in the same individual's matched normal DNA. The authors inferred LOH at regions harboring SNPs heterozygous (AB) in the normal DNA and homozygous (AA or BB) in the tumor, as one of the parental homologs has clearly been lost. SNPs-harboring homozygous genotypes in the matched normal DNA are non-informative with regard to LOH. Regions with SNPs having heterozygous genotypes in the tumor have retained heterozygosity.
Lin et al. (52) extended this idea to infer stretches of LOH (notwithstanding non-informative SNPs) using a hidden Markov model (HMM) approach. HMMs have a long history (53) in the speech recognition literature, and were first used by computational biologists for DNA sequence alignment (54). The HMM structure turns out to be ideally suited for various SNP array analyses, owing to the fact that the SNPs on the array can represent observational units in a Markov chain when ordered according to genomic position on a chromosome. In the LOH setting, the two hidden states are ‘loss’ or ‘retention’, and the observed data are the paired normal/tumor genotypes at each SNP. The authors in the Lin et al. study implemented the methodology in the popular dChip software as dChipSNP (http://www.dchip.org). Although it initially required the presence of matched normal genotypes (which are not necessarily available), the authors subsequently modified the HMM to allow LOH detection without paired normal DNA (55). The modified HMM is based on the observation that LOH is characterized by expanded stretches of homozygous SNP genotypes in the tumor, regardless of the matched normal genotypes. The observed values now consist solely of the tumor genotypes. As such, the hidden states increase from two to four with the addition of the (unobserved) heterozygosity status of the patient's normal DNA at each SNP.
It is important to point out that long stretches of homozygosity may have causes other than tumor LOH. For example, de novo deletions of certain chromosomal regions have been associated with a variety of neuropsychiatric (56) and mental retardation (57) disorders. At the SNP level, these deletions will manifest as homozygous genotypes in the entire region. Furthermore, it is becoming apparent that stretches of homozygosity, due to inheritance of haplotypes identical-by-descent from both parents, are longer and more common than previously believed (58). Distinguishing such autozygosity from somatic LOH events is a challenging problem.
Somatic copy number lesions
As described above, a key feature of probe hybridization-based array technology is that expected probe intensity increases with increased quantity of DNA harboring the region interrogated by the probe. While expression microarrays have exploited this feature for years, the first paper (59) describing an algorithm to detect DNA copy number changes from SNP array data appeared much later. Many other algorithms followed, but all broadly follow the same basic steps: summarization followed by smoothing/segmentation. The summarization step entails converting between 2 and 40 (depending on array platform and version) probe-level intensity values into a single measure of ‘raw’ copy number at each SNP, though the newest versions of both manufacturers’ arrays also have many thousands of singleton non-polymorphic probes that do not require summarization. Almost all methods infer raw copy number by comparing a summary measure for the sample's probe intensities to that from a panel of normal samples. These raw copy numbers are rough measures of the true underlying copy number. The smoothing/segmentation step has spawned an entire subfield of applied computational methodology, often borrowing from more well-established applications in fields such as signal processing. The goal is to infer chromosomal segments of locally constant (true) copy number from the noisy raw copy number measurements. Again, HMM approaches have proved useful for both the Affymetrix (59) and Illumina (60) data, although dozens of other methods have been proposed (61), most borrowing from established statistical methodology.
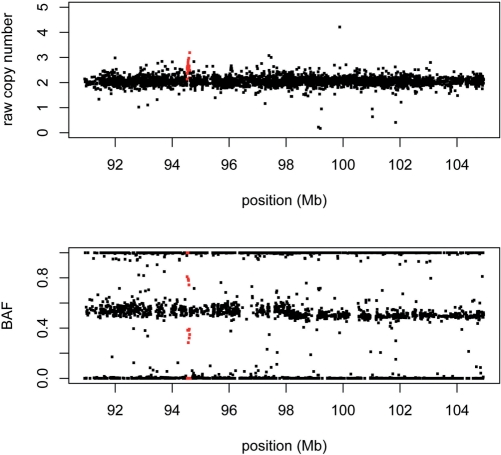
In the HMM framework for copy number inference, the hidden states are the true integer copy number, and the observed data is the raw copy number produced by the summarization step. Other pieces of the model are informed by the underlying biology. The dChipSNP software implements a copy number HMM for the Affymetrix arrays, and was demonstrated for the 10K (59) and 100K (62) versions. Soon thereafter, Peiffer et al. (63) noted that another source of information—the ‘B allele frequency’ (BAF)—can be obtained from array data, and demonstrated its informativeness using the Illumina array. The BAF is simply the B allele signal divided by the sum of the A and B signals, and is particularly straightforward to extract from the Illumina platform since the (post-normalized) data consists of precisely these two measures at each SNP. Interestingly, the BAF is reminiscent of the quantity that was clustered to determine genotype in the Affymetrix 10K array protocol (see above). Table 1 specifies the information gained by considering both raw copy number and BAF for the range between 0 and 4 copies. Figure 3 demonstrates the utility of both measures with a real example.
Table 1.
Copy number state information from SNP array data
| CNV type | Possible SNP genotypes | Expected A+B signal | Expected BAF |
|---|---|---|---|
| Homozygous gain | AAAA | 4 | 0 |
| AAAB | 4 | 0.25 | |
| AABB | 4 | 0.5 | |
| ABBB | 4 | 0.75 | |
| BBBB | 4 | 1 | |
| Hemizygous gain | AAA | 3 | 0 |
| AAB | 3 | 0.33 | |
| ABB | 3 | 0.67 | |
| BBB | 3 | 1 | |
| Normal | AA | 2 | 0 |
| AB | 2 | 0.5 | |
| BB | 2 | 1 | |
| Hemizygous loss | A_ | 1 | 0 |
| B_ | 1 | 1 | |
| Homozygous loss | _ _ | 0 | Undefined |
Figure 3.
Two sources of information from SNP arrays. The raw copy number (top panel) and BAF (bottom panel) are plotted for a 14 Mb region on chromosome 9. Both views of the data, from a custom Illumina array, provide evidence for a focal gain (in red). Note that the gain manifests itself in the BAF plot as clusters of points intermediary between 0.5 and 0 or 1, as expected from the values in Table 1.
Although these copy number inference methods perform well, technological and biological issues can have a negative impact. As a consequence of the complexity-reduction step of the array protocols (12,13), the SNPs are harbored on DNA fragments that vary with regard to both length and G+C content. Two studies (64,65) observed that these parameters can affect PCR amplification kinetics, thereby leading to biases in downstream copy number inferences. To ameliorate this effect, both studies added terms in a linear regression model to correct for both of these artifacts.
Another problem that emerges from the biological reality is that a primary tumor samples can have contamination from normal cells, or even consist of several different subclones, each with different sets of lesions. This motivated the Ogawa group to modify their copy number inference algorithm CNAG (65) to call LOH from cancer samples with up to 70–80% contaminating normal cells (66) from array data. Assié et al. (67) and Li et al. (68) subsequently extended this to allow copy number inference in the presence of normal cells, and were even able to estimate the proportion of cells harboring a particular lesion when the sample consists of various subclones.
Since the SNP array provides both copy number and SNP allelic information, we and others sought to measure SNP allele-specific copy number (69). From the copy number of the SNP alleles, one can determine the chromosomal homolog(s) harboring each amplification and deletion event. We performed our inferences by modeling probe intensity as a function of allele-specific copy number, for which we adopted a generalized linear model. In another example of novel computational methodology yielding biological insight when applied to array data, the study showed that somatic amplification in lung cancer appears to be strictly a monoallelic phenomenon (70). That is, where amplification occurs, it affects only one of the two parental homologs.
GERMLINE COPY NUMBER VARIATION
Until recently, the consensus view was that SNPs comprise the vast majority of human genetic variation. This view began to change in 2004 with the publication of two landmark articles (2,3) that uncovered inherited CNVs on a widespread scale. A detailed comparison (71) between one individual's maternal and paternal chromosomes underscored the importance of non-SNP variation in DNA-level differences. That study's authors found that although SNPs accounted for 78% of all discrete differences between the two chromosomal homologs, they only accounted for 26% of the total nucleotide differences. That is, the number of bases at which two individuals’ DNA sequences differ due to SNPs is likely fewer than the number of bases at which they differ due to CNVs. The inferences from the study were partially facilitated by the Affymetrix 500K and Illumina HumanHap650Y arrays. Both manufacturers have influenced, and been influenced by, the germline CNV field over the last few years.
Methods to mine array data for CNVs
Two studies published in early 2006 (72,73) exploited erroneous SNP genotype calls to infer deletions at clusters of calls that violated Mendelian inheritance or had high ‘No Call’ rates. Mendelian inheritance is violated when the genotype of a child is inconsistent with that of its parents. The violations occur, however, as a result of the (biallelic) assumption of three possible genotypes (AA, AB or BB) at each SNP site. Under this assumption, individuals with a deletion on one homolog would likely be assigned a homozygote call, which would often result in apparent Mendelian inconsistency. For example, suppose that a child inherits a deletion from a mother and has a father with a true AA genotype at a SNP in the region. It will appear that the child has an AA genotype. If the mother harbors a B allele at the SNP on her non-deleted homolog, she will appear to have a BB genotype, yielding apparent Mendelian inconsistency for the trio. If the deletion is common and large enough, there will be a cluster of SNPs showing the violation across multiple father–mother–child trios. Similarly, individuals with deletions at the same locus on both chromosomes will often yield ‘No Calls’, and clusters of these null genotypes may also indicate deletion.
In this way, these two studies were able to reveal a large collection of novel germline deletions, but did not actually take advantage of SNP array data per se, rather exploiting downstream SNP genotypes. Moreover, their methods were blind to gains. Given the extensive computational algorithm development already undertaken for detecting somatic copy number lesions in cancer from arrays, it is unsurprising that the first published CNV detection algorithm came from the cancer community. A detailed study (74) of the 270 HapMap samples applied a computational method (75) to Affymetrix 500K array data for CNV discovery. The method was adapted from the authors’ tumor DNA algorithm GIM [Genome Imbalance Map (65)]. In the new adaptation, the study compared GIM-inferred raw copy numbers pairwise across all 270 samples, inferring CNV regions at consistently aberrant raw copy number ratios. The algorithm uses cluster analysis to reveal a ‘maximal clique’ of samples that it infers to be copy number two, calling the remainder as gains or losses based on the ratios of their raw copy number to that of the copy number two samples. Soon after this study was published, the computational community developed corresponding methods for Illumina arrays. Two of the first, QuantiSNP (76) and PennCNV (60) take an HMM approach. Rather than using only raw copy number as observed emission in the Markov process, both of these methods take also advantage of the BAF measure described above.
Most recently, a pair of papers appeared (31,32) describing a new collection of algorithms, termed Birdsuite. Affymetrix in fact developed their 6.0 array in collaboration with these researchers and in tandem with these algorithms. The papers’ authors divided the CNV calling procedures into two separate methods, one for genotyping common CNVs (termed copy number polymorphisms, or CNPs), and one for identifying rare or de novo CNVs. For CNP genotyping, Birdsuite takes advantage of an existing map of CNP locations generated by McCarroll et al. (31). To assign a genotype to a sample at a CNP, the algorithm summarizes for the raw signals from the probes within the CNP boundaries, and then clusters this measure across samples to infer the individual genotypes. For rare CNV detection, Birdsuite adopts an HMM approach similar to that used by dChipSNP described above.
Combining CNVs with SNP genotypes
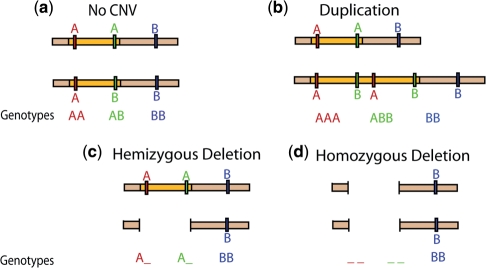
For a given SNP, genotyping has traditionally meant the classification of an individual as either AA, AB or BB. As such, the manufacturers originally designed and optimized the arrays with this goal in mind. As this review has demonstrated, this is a well-studied problem algorithmically. If the SNP lies within a copy number variable region, however, the assumption of two copies at each locus is no longer valid. Similar to allele-specific copy number in cancer described above, one can consider a generalized genotype whereby the SNP is multi-allelic when considering both base residue and copy number (Figure 4). SNP genotypes should, in theory, be tractable from array data, since allelic intensity provides a noisy measure of allelic dosage. However, this problem is computationally much more difficult than the three-class problem. Where the data is very clean, one can cluster samples by generalized genotype (Figure 5), but the noisiness of the data usually necessitates a more sophisticated approach. We presented such an approach in Macconaill et al. (77), and the Birdsuite collection mentioned above also contains a method to provide SNP allele-specific CNV calls.
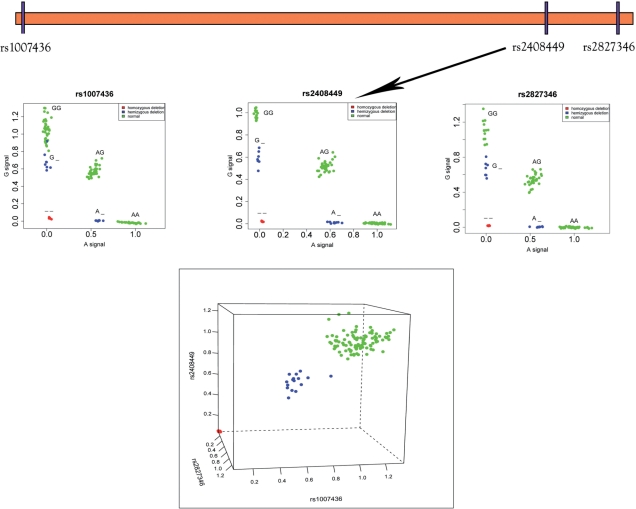
Figure 4.
SNP genotypes in the presence of CNVs. (a) Traditional SNP genotyping, under the assumption of two copies. (b) A chromosome harbors a duplication of the orange region, resulting in multi-allelic genotypes for the two SNPs contained in the region. (c) A chromosome harbors a deletion of the orange region. (d) This individual carries a deletion of the orange region on both chromosomes, resulting in _ _ genotypes for the two SNPs.
Figure 5.
Calling SNP/CNV alleles from raw data. All three SNPs shown here on chromosome 21 have alleles A and G. All plots show A allele and G allele intensity values from Illumina HumanHap550 data for 112 HapMap samples. The top three panels show each of the three SNPs individually along with their generalized genotypes. The bottom panel shows the total raw copy number sums (A signal + G signal) plotted, with each axis representing one of the SNPs. Note that the samples clearly separate into homozygous deletions (red), hemizygous deletions (blue), and normal (green).
As with SNP genotypes, copy number calls are ambiguous with regard to phase. Matters are further complicated when SNP genotypes are considered simultaneously with copy number. Mother–father–child trio information may sometimes allow unambiguous phase inference from the generalized SNP genotypes (77). In the absence of trios, Kato et al. (78) generalized SNP phasing techniques to infer SNP/CNV haplotypes in this multiallelic setting. The authors’ algorithm MOCSphaser (mixture of CNV and SNP phaser) is a straightforward application of the EM algorithm similar to one developed earlier for SNPs (79), and is implemented in free software (http://emu.src.riken.jp/MOCSphaser).
CONCLUSIONS AND FUTURE PROSPECTS
As this review has shown, the history of the development of SNP arrays is characterized by two recurrent themes. First, the technology has repeatedly spawned applications extending well beyond the purpose for which it was originally designed. Initially driven by computational work from the cancer community, SNP arrays were serendipitously well-positioned for the near-concurrent discovery of CNVs as a major source of human genetic variation. Second, we repeatedly witness a synergy between the array's technological evolution and advances in computational development and biological insight. As a result, the SNP array is now a central tool in biomedical research.
After regular dramatic increases in marker density throughout the first several years of the 2000s, this trend seems to have leveled off. The last arrays with substantial increases in marker density were commercially released by Affymetrix and Illumina in May 2007 and July 2007, respectively. Both manufacturers have been developing custom SNP genotyping arrays for non-human species such as mouse, dog, and cow, but plans for the human arrays are unclear. This review has focused on human SNP arrays, but applications to other species promises bring additional insights to evolution, molecular biology, and genetics.
One contributor to the lack of recent updates to SNP arrays may be the emergence of a new technology. Next-generation sequencers of the sort produced by Illumina/Solexa, Roche/454 and ABI (with others on the way) are able to produce all of the information that SNP arrays can produce, but with (theoretically) greater resolution and accuracy (80). These new machines can sequence billions of bases DNA sequence much more cheaply, and in a much shorter time than previously thought possible. For example, a large study recently used billions of paired end reads (81) to detect thousands of CNVs in a single individual, most of which would have been missed by SNP arrays. Other groups (82,83) have also recently shown that between 10 and 30 million reads are sufficient to detect somatic amplifications and deletions in tumor DNA at a resolution comparable to that of the densest SNP arrays. Furthermore, paired end reads can also uncover structural DNA changes that are invisible to SNP arrays, such as translocations and inversions (Table 2). In the first full sequencing of a cancer genome (85), DNA base-level differences—both germline (SNPs) and somatic (point mutations)—were detectable at far more loci than any array could cover. However, as Table 2 shows, cost considerations make next-generation sequencers impractical for some applications at the moment. Moreover, it is important to note that the computational methodology spawned by the SNP array community has already begun to prove useful for high-throughput sequence data. For example, the studies in refs (82,83) adopted a segmentation approach, similar to the circular binary segmentation (86) algorithm developed for array data, to infer copy number lesions from millions of sequence reads. Costs associated with next-generation sequencers will continue to decrease, and some are predicting the demise of the microarray (14). Whether this demise comes sooner or later, the insights gained from SNP arrays and the computational methodologies developed to handle the resulting data represent significant scientific advances from the last decade.
Table 2.
SNP array/next-generation sequencer cost and feasibility comparison
| Genome-wide interrogation goal | Estimated cost per sample |
||
|---|---|---|---|
| SNP arraya | Next-generation sequencerb | References | |
| SNP genotypingc | $700 | $36 000d | (12,13) |
| SNP/point mutation discoveryc | NA | $180 000 | (84,85) |
| LOH detectione | $1400 | $72 000d | (12,13) |
| Copy number assessment | $700 | $2000 | (31,60,82,83) |
| Inversion detection | NA | $2000–$180 000f | (82,83) |
| Translocation detection | NA | $2000–$180 000f | (82,83) |
aAt flat cost of $700 per array.
bAt $2000 per experiment, each producing 1 Gb of sequence.
cAt 30X coverage for reliable SNP/mutation calling.
dAfter first performing a complexity-reduction step (similar to SNP array protocols) yielding ∼1 M fragments of average size 600 bp.
eUsing both tumor and matched normal DNA samples.
fDepending upon level of resolution desired.
N.B. All costs are very approximate, and will vary with different platforms, economies of scale and other factors.
FUNDING
National Cancer Institute (Grant number R01 CA131341). Funding for open access charge: National Cancer Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The author acknowledges Moulik Kothari, Katherine Wilkins and Russellitta Young for expert help with the figures, and thanks Mark Adams for discussions regarding next-generation sequencing.
REFERENCES
- 1.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat. Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 4.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genetics. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 5.Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, et al. APP locus duplication causes autosomal dominant early-onset alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 6.Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am. J. Hum. Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 9.Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat. Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon-Sanchez J, Scholz S, Matarin Mdel M, Fung HC, Hernandez D, Gibbs JR, Britton A, Hardy J, Singleton A. Genomewide SNP assay reveals mutations underlying Parkinson disease. Hum. Mutat. 2008;29:315–322. doi: 10.1002/humu.20626. [DOI] [PubMed] [Google Scholar]
- 11.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 12.Affymetrix. (California): Santa Clara; 2007. Affymetrix Genome-Wide Human SNP Array 6.0 data sheet. [Google Scholar]
- 13.Illumina. Genome-Wide DNA Analysis BeadChips Data Sheet. California: San Diego; 2009. [Google Scholar]
- 14.Coombs A. The sequencing shakeup. Nat. Biotechnol. 2008;26:1109–1112. doi: 10.1038/nbt1008-1109. [DOI] [PubMed] [Google Scholar]
- 15.Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 16.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21(1 Suppl):20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, et al. Large-scale genotyping of complex DNA. Nat. Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, et al. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu WM, Di X, Yang G, Matsuzaki H, Huang J, Mei R, Ryder TB, Webster TA, Dong S, Liu G, et al. Algorithms for large-scale genotyping microarrays. Bioinformatics. 2003;19:2397–2403. doi: 10.1093/bioinformatics/btg332. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman L, Rousseeuw PJ. Clustering by means of medoids. In: Dodge Y, editor. Statistical Data Analysis Based on the L1-norm and Related Methods. Amsterdam: North Holland; 1987. pp. 405–416. [Google Scholar]
- 21.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data, a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 24.Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C, Mathews DJ, Shah NA, Eichler EE, Warrington JA, et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 26.Tukey JW. Exploratory Data Analysis. Reading, Massachusetts: Addison-Wesley; 1977. [Google Scholar]
- 27.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antipova AA, Tamayo P, Golub TR. A strategy for oligonucleotide microarray probe reduction. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0073. RESEARCH0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smemo S, Borevitz JO. Redundancy in genotyping arrays. PLoS ONE. 2007;2:e287. doi: 10.1371/journal.pone.0000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen F, Huang J, Fitch KR, Truong VB, Kirby A, Chen W, Zhang J, Liu G, McCarroll SA, Jones KW, et al. Improved detection of global copy number variation using high density, non-polymorphic oligonucleotide probes. BMC Genet. 2008;9:27. doi: 10.1186/1471-2156-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy numbervariation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 32.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J. Roy. Statist. Soc. B. 1977;39:1–38. [Google Scholar]
- 34.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 35.Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat. Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 36.Zuckerman M. Some dubious premises in research and theory on racial differences. Scientific, social, and ethical issues. Am. Psychol. 1990;45:1297–1303. doi: 10.1037//0003-066x.45.12.1297. [DOI] [PubMed] [Google Scholar]
- 37.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 39.Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demichelis F, Greulich H, Macoska JA, Beroukhim R, Sellers WR, Garraway L, Rubin MA. SNP panel identification assay (SPIA): a genetic-based assay for the identification of cell lines. Nucleic Acids Res. 2008;36:2446–2456. doi: 10.1093/nar/gkn089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butcher LM, Meaburn E, Liu L, Fernandes C, Hill L, Al-Chalabi A, Plomin R, Schalkwyk L, Craig IW. Genotyping pooled DNA on microarrays: a systematic genome screen of thousands of SNPs in large samples to detect QTLs for complex traits. Behav. Genet. 2004;34:549–555. doi: 10.1023/B:BEGE.0000038493.26202.d3. [DOI] [PubMed] [Google Scholar]
- 42.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 43.Milani L, Lundmark A, Nordlund J, Kiialainen A, Flaegstad T, Jonmundsson G, Kanerva J, Schmiegelow K, Gunderson KL, Lönnerholm G, et al. Allele-specific gene expression patterns in primary leukemic cells reveal regulation of gene expression by CpG site methylation. Genome Res. 2009;19:1–11. doi: 10.1101/gr.083931.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 45.Niu T. Algorithms for inferring haplotypes. Genet. Epidemiol. 2004;27:334–347. doi: 10.1002/gepi.20024. [DOI] [PubMed] [Google Scholar]
- 46.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pe'er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat. Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 48.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 50.Maciejewski JP, Mufti GJ. Whole genome scanning as a cytogenetic tool in hematologic malignancies. Blood. 2008;112:965–974. doi: 10.1182/blood-2008-02-130435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindblad-Toh K, Tanenbaum DM, Daly MJ, Winchester E, Lui WO, Villapakkam A, Stanton SE, Larsson C, Hudson TJ, Johnson BE, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat. Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- 52.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 53.Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- 54.Durbin R, Eddy SR, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 55.Beroukhim R, Lin M, Park Y, Hao K, Zhao X, Garraway LA, Fox EA, Hochberg EP, Mellinghoff IK, Hofer MD, et al. Inferring loss-of-heterozygosity from unpaired tumors using high-density oligonucleotide SNP arrays. PLoS Comput. Biol. 2005;2:e41. doi: 10.1371/journal.pcbi.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook EH, Jr., Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 57.Slavotinek AM. Novel microdeletion syndromes detected by chromosome microarrays. Hum. Genet. 2008;124:1–17. doi: 10.1007/s00439-008-0513-9. [DOI] [PubMed] [Google Scholar]
- 58.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao X, Li C, Paez JG, Chin K, Jänne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 60.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lai WR, Johnson MD, Kucherlapati R, Park PJ. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao X, Weir BA, LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, et al. Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 63.Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A, Kurokawa M, Chiba S, Bailey DK, Kennedy GC, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 65.Ishikawa S, Komura D, Tsuji S, Nishimura K, Yamamoto S, Panda B, Huang J, Fukayama M, Jones KW, Aburatani H. Allelic dosage analysis with genotyping microarrays. Biochem. Biophys. Res. Commun. 2005;333:1309–1314. doi: 10.1016/j.bbrc.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am. J. Hum. Genet. 2007;81:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Assié G, LaFramboise T, Platzer P, Bertherat J, Stratakis CA, Eng C. SNP arrays in heterogeneous tissue: highly accurate collection of both germline and somatic genetic information from unpaired single tumor samples. Am. J. Hum. Genet. 2008;82:903–915. doi: 10.1016/j.ajhg.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li C, Beroukhim R, Weir BA, Winckler W, Garraway LA, Sellers WR, Meyerson M. Major copy proportion analysis of tumor samples using SNP arrays. BMC Bioinformatics. 2008;9:204. doi: 10.1186/1471-2105-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laframboise T, Harrington D, Weir BA. PLASQ: a generalized linear model-based procedure to determine allelic dosage in cancer cells from SNP array data. Biostatistics. 2007;8:323–336. doi: 10.1093/biostatistics/kxl012. [DOI] [PubMed] [Google Scholar]
- 70.LaFramboise T, Weir BA, Zhao X, Beroukhim R, Li C, Harrington D, Sellers WR, Meyerson M. Allele-specific amplification in cancer revealed by SNP array analysis. PLoS Comput. Biol. 2005;1:e65. doi: 10.1371/journal.pcbi.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 73.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 74.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Komura D, Shen F, Ishikawa S, Fitch KR, Chen W, Zhang J, Liu G, Ihara S, Nakamura H, et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Macconaill LE, Aldred MA, Lu X, Laframboise T. Toward accurate high-throughput SNP genotyping in the presence of inherited copy number variation. BMC Genomics. 2007;8:211. doi: 10.1186/1471-2164-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kato M, Nakamura Y, Tsunoda T. MOCSphaser: a haplotype inference tool from a mixture of copy number variation and single nucleotide polymorphism data. Bioinformatics. 2008;24:1645–1646. doi: 10.1093/bioinformatics/btn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 80.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 81.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-widemassi vely parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Tassell CP, Smith TP, Matukumalli LK, Taylor JF, Schnabel RD, Lawley CT, Haudenschild CD, Moore SS, Warren WC, Sonstegard TS. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat. Methods. 2008;5:247–252. doi: 10.1038/nmeth.1185. [DOI] [PubMed] [Google Scholar]
- 85.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]