Abstract
With the ever-increasing number of studies in human functional brain mapping, an abundance of data has been generated that is ready to be synthesized and modeled on a large scale. The BrainMap database archives peak coordinates from published neuroimaging studies, along with the corresponding metadata that summarize the experimental design. BrainMap was designed to facilitate quantitative meta-analysis of neuroimaging results reported in the literature and supports the use of the activation likelihood estimation (ALE) method. In this paper, we present a discussion of the potential analyses that are possible using the BrainMap database and coordinate-based ALE meta-analyses, along with some examples of how these tools can be applied to create a probabilistic atlas and ontological system of describing function–structure correspondences.
Keywords: BrainMap, meta-analysis, activation likelihood estimation, ontology, functional atlas
Introduction
Over the last three decades, neuroimaging research has produced an enormous amount of data localizing the neural effects of specific mental operations in both healthy and diseased populations. Community-wide standards of spatial normalization and the reporting of peak activation locations in stereotactic coordinates allow researchers to compare results across studies when the primary data are unavailable or difficult to obtain. Due to the nearly universal adherence to these standards, the BrainMap project was designed to create tools for large-scale data mining and meta-analysis of the brain mapping literature (Fox and Lancaster, 2002; Laird et al., 2005a).
BrainMap is a community accessible database1 that allows a user to relate behavioral functions to specific brain locations through retrieval and visualization of peak coordinates and their associated metadata. These metadata allow each coordinate to be linked with how the observed activation was experimentally derived, a formulation that lends itself to very rich data mining. BrainMap was originally conceived by Peter Fox in 1987 and received its original funding from the James S. McDonnell Foundation (1988–1990). Continued BrainMap development was funded by the Office of Naval Research (1991–1992), the EJLB Foundation (1992–1996), and the National Library of Medicine (2000–2003). BrainMap is currently funded by the Human Brain Project of the National Institute of Mental Health.
BrainMap Software
There are three desktop applications (Scribe, Sleuth, and GingerALE) and one web application (BrainMapWeb) that allow interaction with the BrainMap database, all of which are coded in Java. The desktop applications run in the Java Runtime Environment on Macintosh, Windows, Linux, and Unix operating systems, while the web application uses Java server-side technologies. Scribe2 is used to code papers for entry into BrainMap. Peer-reviewed publications can be submitted to the database by the original authors (uncommon) or by investigators performing a meta-analysis (very common). Most data fields have candidate responses presented in scrollable lists. Coordinate tables of peak locations can be imported from a tab-delimited file or entered by hand. Upon insertion into the database, each x,y,z, coordinate is assigned an anatomical location using the Talairach Daemon3 (Lancaster et al., 2000). All entries are reviewed for quality control by BrainMap staff and faculty before being entered into the database to ensure the accuracy and consistency of coding. The Sleuth application4 allows a user to search the BrainMap database and retrieve data, which can then be filtered and visualized on a standard Talairach atlas image. Specific locations may be searched for according to user-defined, coordinate-based regions of interest (defined by Talairach or MNI coordinates) or anatomical labels from the Talairach Daemon nomenclature. BrainMap queries may also be implemented via an internet browser using BrainMapWeb5, which includes query functions that are similar to those of Sleuth, but lack 3D brain visualizations. Data view and manipulation capabilities are much more restricted than in Sleuth, which can output data in publication-ready graphics, text files of Talairach or MNI coordinates, or workspaces that specify search rules and filters for meta-analyses. By archiving coordinates of activation locations rather than raw image data, BrainMap focuses on discoveries derived from coordinate-based meta-analyses of functional neuroimaging data. The last BrainMap application, GingerALE6, is used for performing activation likelihood estimation (ALE) meta-analyses on sets of coordinates extracted from the database in Talairach or MNI space. Scribe, Sleuth, and GingerALE are closely integrated to transition seamlessly from database submission to search refinement to meta-analysis results (Figure 1).
Figure 1.
BrainMap software applications. There are three desktop applications that allow interaction with the BrainMap database: Scribe (data entry), Sleuth (search and retrieval), and GingerALE (meta-analysis). These Java applications are freely downloadable from the BrainMap website (http://brainmap.org). Database statistics are current as of 04/01/09.
BrainMap Coding Scheme
To summarize the experimental design and results of a published study for inclusion in the database, BrainMap utilizes a rigorous taxonomy that is composed chiefly of structured keywords. BrainMap entries include descriptions on the scanned subjects and experimental conditions, including the presented stimuli, instructions, and responses. A complete metadata listing can be seen in Figure 2. To facilitate meta-analysis, several hierarchically structured keywords have been developed that categorize the nature of each experimental contrast to allow rapid, comprehensive retrieval of results. “Context” broadly categorizes the purpose of the work; for example, normal mapping (the comparison of different experimental conditions in a group of healthy subjects), age effects, disease effects, or drug effects. “Behavioral Domain” classifies the research in terms of the neural systems studied according to six main categories and their related subcategories: cognition, action, perception, emotion, interoception, or pharmacology (Figure 3). “Paradigm Class” categorizes the challenge presented, preferably in the jargon of the field, such as anti-saccades, Stroop, delayed match to sample, or mental rotation tasks (Figure 4).
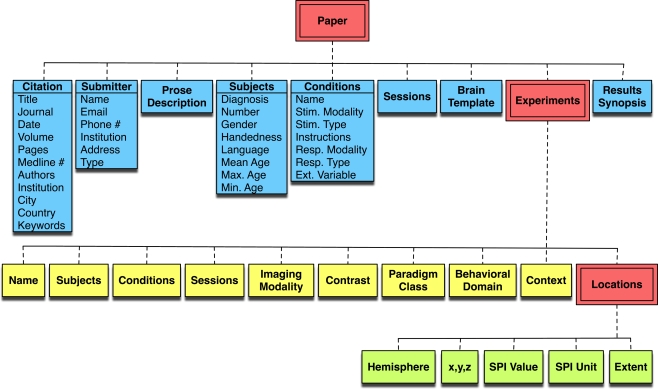
Figure 2.
The BrainMap coding scheme. The BrainMap metadata coding scheme follows a hierarchy naturally occurring within the brain mapping literature. Every Paper reports experimental results drawn from one or more subject groups, the members of which have been functionally imaged during one or more behavioral Conditions. In BrainMap, an Experiment is defined by the production of a statistical parametric image. From each Experiment one or more functional activations (Locations) are extracted in the form of peak coordinates.
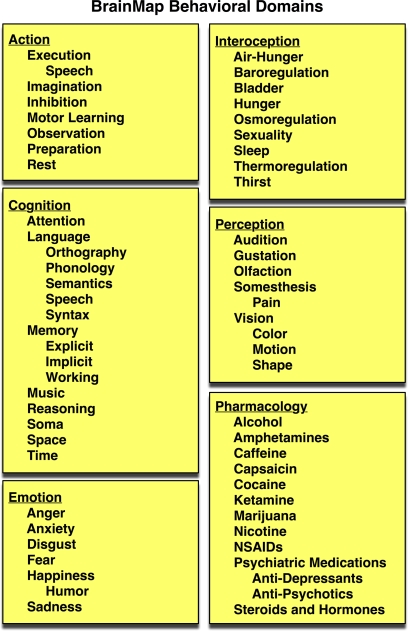
Figure 3.
BrainMap behavioral domains. In the BrainMap coding scheme, the behavioral domain is recorded for each statistical contrast, which describes the research in terms of the neural systems studied according to 6 main categories: cognition, action, perception, emotion, interoception, or pharmacology. The yellow background represents that this field lies at the Experiment level.
Figure 4.
BrainMap paradigm classes. The paradigm class is recorded for each statistical contrast in BrainMap. This field categorizes the task or challenge presented, preferably in the jargon of the field. BrainMap currently contains 80 paradigm classes. The yellow background represents that this field lies at the Experiment level.
While a given a given paradigm class (e.g., n-back) is often immediately associated with a given behavioral domain (e.g., working memory), this is not always the case. Each experiment must be evaluated based on the conditions contrasted since many comparisons are designed to elicit processing in domains not directly linked to the paradigm class employed. For example, n-back and working memory are appropriate choices when comparing an n-back condition to a control condition, but if two n-back conditions are contrasted that differ only in the modality of stimulus presentation (e.g., visual or auditory), then additional perceptual domains should be coded for that experiment. We have found that the context, behavioral domain, and paradigm class represent the three most critical components of a functional neuroimaging study; their orthogonality fully defines and gives contextual meaning to the coordinates archived in the database.
Typically, investigators use citation indexing services such as PubMed to search the literature according to user-defined keywords. This results in the identification of a subset of desired studies that explicitly match the search criteria. BrainMap's strategy has been to design paradigm class entries in order to pool similar studies, rather than segregate them according to domain-specific keywords. Categories and sub-categories are created and refined only as needed, based on the demands of the literature and the continued development of functional brain imaging. For example, the first study entered into BrainMap that utilized the Wisconsin Card Sorting Test was initially coded under the paradigm class of “Deductive Reasoning”. This was done to classify the study in the same set as other similar papers in the database (i.e., “best fit”), a practice that the classes are rich enough to be useful. Once four papers of the same sub-category are entered into the database, a new class is created and defined (e.g., “Wisconsin Card Sorting Test”). Then all entries matching this new class are manually searched for and updated to reflect the new designation. This procedure requires continuous and labor-intensive maintenance of the database, yet yields a high-quality database full of rich metadata categories and provides an evolving and flexible tool.
Given the sheer volume of neuroimaging data that is currently being produced, it is rapidly becoming overwhelming for an investigator to reconcile new results to those previously published, particularly when studies pertain to different areas of research. Derrfuss and Mar (2009) estimated that BrainMap contains approximately one-fifth of the relevant published studies, making it the largest coordinate-based database in functional neuroimaging to date. In BrainMap, an ROI of 1 cm3 currently contains an average of 23 experiments, and includes results from 15 paradigm classes. Databases designed to simply retrieve studies reporting activations in proximate locations result in subsequent manual filtering and interpretation. While BrainMap's data entry procedure is labor-intensive (Laird et al., 2005a), the depth of the current coding strategy is what provides diverse data mining opportunities and establishes the overall value of the database. BrainMap was structured with the goal of not only retrieving studies returned by regional searches without domain-specific biases, but also allowing the results to be synthesized. Specifically, BrainMap development of neuroinformatics tools focuses on knowledge discovery that is made possible by coordinate-based function–location meta-analysis (Fox et al., 1998).
Activation Likelihood Estimation Meta-Analysis
Activation likelihood estimation (ALE) is a method of coordinate-based meta-analysis that is supported within the BrainMap software environment. It is a useful tool for integrating the neuroimaging literature wherein consistent regions of activation are identified across a collection of studies. In particular, peak coordinates are collected from studies that share a similar feature of interest, which can be a specific task (e.g., go/no-go) or a more generalized cognitive process (e.g., inhibition). In ALE, coordinates are then modeled with a Gaussian function to accommodate the spatial uncertainty associated with a reported coordinate and are analyzed for where they converge.
Since its introduction (Chein et al., 2002; Turkeltaub et al., 2002), ALE has been applied in many aspects of normal brain function (Costafreda et al., 2008; Decety and Lamm, 2007; Eickhoff et al., 2006a; Grosbras et al., 2005; Soros et al., 2009; Spreng et al., 2009), as well as in studies of neuropsychiatric and neurological disorders, such as schizophrenia (Glahn et al., 2005; Minzenberg et al., 2009; Ragland et al., 2009), obsessive-compulsive disorder (Menzies et al., 2008), depression (Fitzgerald et al., 2008), and developmental stuttering (Brown et al., 2005). Recently, ALE has been extended to voxel-based morphometry (Ellison-Wright et al., 2008; Glahn et al., 2008; Schroeter et al., 2007) and diffusion tensor imaging studies (Ellison-Wright and Bullmore, 2009).
The most interesting ALE applications do not merely merge previous results in a retrospective fashion, but instead generate or test a new hypothesis (Eickhoff et al., 2009a; Price et al., 2005), identify a previously unspecified region (Derrfuss et al., 2005), resolve conflicting views (Laird et al., 2005b; Petacchi et al., 2005), or validate a new paradigm (McMillan et al., 2007). Several studies have used ALE as a preliminary step, followed by an analysis of network co-occurrences (Lancaster et al., 2005; Neumann et al., 2005, 2008; Toro et al., 2008) or structural equation modeling (Laird et al., 2008). Each of these novel meta-analytic applications was carried out using typical ALE analysis parameters, and many of them involved the comparison of multiple meta-analyses. For example, Price et al. (2005) examined the results of a picture naming meta-analysis and found that use of a high-level baseline condition to control for speech production and perceptual processing resulted in increased sensitivity to activation in areas associated with semantic processing, visual-speech integration, and response selection. A prospective fMRI study was performed to test this hypothesis, which subsequently allowed the picture naming system to be decomposed into its perceptual, semantic, and phonological components. In a different application of the ALE method, a meta-analysis was performed on studies in which transcranial magnetic stimulation was applied to left motor cortex (Laird et al., 2008). The results of this meta-analysis were used to determine the location of regions of interest in a prospective study of TMS/PET data that examined the effective connectivity of the motor system using structural equation modeling. These examples of how meta-analysis results have been applied to guide analyses in newly acquired experimental data demonstrate the power of the ALE method and provide evidence of its efficacy beyond that of a purely retrospective tool.
Modifications to the ALE Algorithm
The ALE method was originally developed and validated by Turkeltaub et al. (2002) in a meta-analysis of single word reading. BrainMap developers obtained the algorithm from the Georgetown University CSL group, ported the code into Java, and created a graphical user interface (GingerALE). A cluster analysis script was added that identifies ALE clusters (areas of high activation likelihood) and returns the cluster extent above a user-specified threshold, x-y-z coordinates of the weighted center-of-mass and peak locations, and an anatomical label assigned by the Talairach Daemon (Lancaster et al., 2000). A coordinate conversion utility was also included to convert MNI coordinates to Talairach space (Lancaster et al., 2007). Two extensions of the original ALE method, a correction for multiple comparisons and a method for computing statistical contrasts of pairs of ALE images, were also added (Laird et al., 2005c).
In the original implementation of the ALE method, several limitations were known to exist: (1) the size of the modeled Gaussian distribution was user-specified and therefore subjective, (2) the permutation test for significance was not anatomically constrained, leading to some modeled activation in white matter, and (3) the analysis tested for above-chance clustering of individual coordinates (a fixed-effects analysis), preventing the generalization of results that is possible in a random-effects analysis (Wager et al., 2007, 2009). Recent advances in the ALE technique have overcome these limitations to provide a more valid and statistically reliable meta-analysis framework (Eickhoff et al., 2009b). Rather than relying on user-dependent Gaussian distributions, quantitative estimates of the between-subject and between-template variability were empirically determined in order to more explicitly model the spatial uncertainty associated with each coordinate (a correction that also includes a weighting of each study by the number of included subjects). In addition, the permutation test was limited to regions of gray matter and modified to test for the above-chance clustering between experiments, resulting in a transition from a fixed-effects to a random-effects method of statistical inference. By progressing from an analysis based on the clustering across coordinates to the clustering across experiments, ALE results no longer may potentially be driven by a single study. The new ALE formulation was validated against the classical algorithm and experimental data (Grefkes et al., 2008) and found to increase the specificity of results without losing the sensitivity of the original approach. These improvements have been implemented in the most recent version of GingerALE, which is currently available for beta testing on the BrainMap website.
Function-Based Meta-Analyses
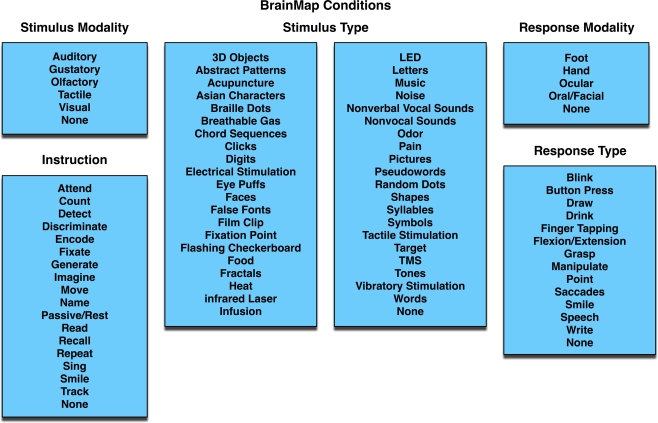
The ALE meta-analysis method can be applied in a variety of ways to answer specific research questions. Most frequently, ALE is applied to sets of neuroimaging studies that share some similar aspect of evoked brain function. These function-based meta-analyses usually involve pooling studies with similar experimental designs, and these studies may also be segregated into different collections to evaluate the functional specificity of the task or process being investigated. In the BrainMap coding scheme, experimental conditions are described according to the presented stimuli, instructions given, and response requested. Each of these fields has candidate entries to choose from, which collectively capture the essence of the scanned conditions (Figure 5). These three condition axes (stimulus, response, and instructions) provide a structure in which differential activation patterns can be systematically probed for variations of a given task.
Figure 5.
BrainMap conditions. In each paper archived in BrainMap, the scanned experimental conditions can be segregated into components based on the stimulus presented (modality and type), instructions given, and required response (modality and type). The values in the BrainMap metadata scheme are standardized to as few options as possible, so as to group similar studies. The blue background represents that this field lies at the Paper level.
For example, we performed an ALE meta-analysis of all studies in BrainMap that were coded with a paradigm class of “word generation” (66 papers, 197 experiments, 1552 coordinates), a widely used test of neuropsychological function. The meta-analytic results revealed extensive convergence in large portions of the left inferior frontal gyrus, centering on Brodmann area 44/45 (Broca's area), and the left dorsolateral prefrontal cortex (DLPFC, BA 46/9), regions commonly known to be associated with word retrieval and executive function. ALE clusters were also observed in the bilateral insula, anterior cingulate cortex (ACC, BA 32), supplementary motor area (SMA, BA6), precuneus (BA 7), posterior cingulate cortex (PCC, BA 31), left posterior temporal cortex (Wernicke's area, BA 22), left inferior parietal cortex (BA 40), right posterior cerebellum, and left thalamus. These areas are generally understood to be involved with the production of language and executive processing that is characteristic of verbal fluency tasks (Heim et al., 2008; Petersen et al., 1988; Warburton et al., 1996; Wise et al., 1991).
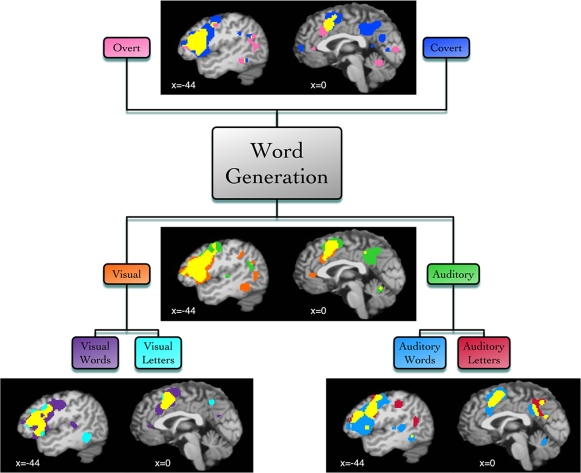
We then examined BrainMap metadata for these studies and found that the imaged tasks varied according to the modality of responses (covert or overt), the modality of stimulus presentation (visual or auditory), or the stimulus type (words or letters). Supplemental ALE meta-analyses were then performed according to each of these task variations, yielding differential patterns of activation likelihood (Figure 6). Covert word generation yielded more extensive engagement of lateral and medial prefrontal areas (similar to Basho et al., 2007), precuneus, and posterior cingulate cortex, while analysis of overt tasks revealed distinct concordance in the right cerebellum. ALE results of studies using visual stimuli were observed in visual cortex (BA 17/18) and the fusiform gyrus (BA 37), while auditory stimuli were localized to left auditory cortex (BA 41), Wernicke's area (BA 22), and precuneus. Generation of words in response to a presented word (visual or auditory) was associated with the middle frontal gyrus (BA 9). Semantic verbal fluency was also associated with the ventral portion of the left inferior frontal gyrus, in agreement with previous meta-analysis results (Costafreda et al., 2006), as well as BA 45 (Amunts et al., 2004). The observation that Wernicke's area was preferentially involved auditory stimulation, particularly with words, likely reflects this region's role in auditory processing and the comprehension of spoken words. This word generation meta-analysis highlights the rich data mining that is possible using the BrainMap coding scheme.
Figure 6.
Function-based meta-analysis of word generation. Studies utilizing a word generation paradigm were downloaded from the BrainMap database. Papers were segregated according to stimulus and response features. Separate ALE meta-analyses were performed for each set of coordinates: overt speech responses (30 papers, 78 experiments, 604 coordinates), covert speech responses (35 papers, 92 experiments, 729 coordinates), visual stimuli (39 papers, 115 experiments, 906 coordinates), auditory stimuli (32 papers, 86 experiments, 621 coordinates), visual words (25 papers, 88 experiments, 638 coordinates), visual letters (14 papers, 38 experiments, 373 coordinates), auditory words (39 papers, 124 experiments, 1001 coordinates), and auditory letters (14 papers, 35 experiments, 279 coordinates). Results are displayed at P < 0.05, corrected. Left lateral (x = −44) and medial (x = 0) sagittal slices display the comparative meta-analytic results. In all images, the overlap between meta-analysis results is shown in yellow.
Structure-Based Meta-Analyses
While function-based meta-analyses tend to dominate the literature, structure-based meta-analyses offer an alternative meta-analytic strategy. Instead of pooling studies that share a common experimental design, structure-based meta-analyses focus on a specific anatomical region and look for global coactivation patterns across a diverse range of tasks. The theory behind this type of meta-analysis is that groups of coordinates that coactivate across experiments can be pooled to identify functionally connected networks in the brain. Like other methods of analyzing functional connectivity (Cordes et al., 2000; Rogers et al., 2007; Xiong et al., 1999), structure-based meta-analyses are based on the co-occurrence of spatially separate neurophysiological events. Koski and Paus (2000) used this technique to study the meta-analytic connectivity of the anterior cingulate cortex, although their analysis was limited to the frontal lobe. In their study, the authors manually collected and filtered data from 107 studies tasks to examine regional co-occurrences and found evidence for functional heterogeneity within the ACC. A similar meta-analysis was performed on basal ganglia activation using 126 published studies to determine the functional connectivity between cortex and striatum (Postuma and Dagher, 2006).
Increasing both the size and diversity of structure-based meta-analyses adds to the generalizability of the results. When used in conjunction with the BrainMap database, the procedure is more automated, resulting in larger meta-analyses that include decades of neuroimaging data from a diverse range of paradigms and behavioral domains. Recently, a large-scale meta-analysis of the functional connectivity of the amygdala was carried out in which the ROIs for left and right amygdala were defined according the Harvard-Oxford structural probability atlas distributed with FSL (Smith et al., 2004) and seeded in BrainMap (Robinson et al., 2009). This anatomically defined meta-analysis of 240 papers (326 experiments with 3842 coordinates) revealed that the amygdala plays a integrative role in both emotion and cognition, and was validated according to the non-human primate database, CoCoMac (Stephan et al., 2001).
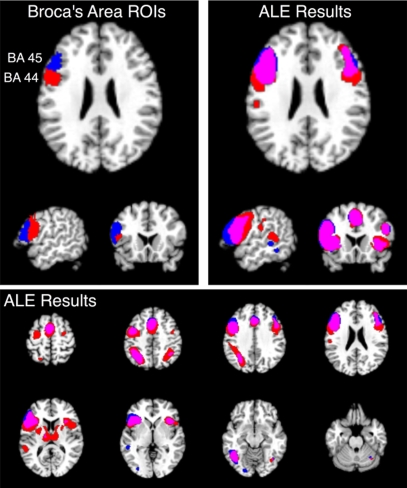
To illustrate the use of structure-based meta-analyses, we note that the function-based meta-analysis of verbal fluency studies (Figure 6) was characterized by extensive activation of the left inferior frontal gyrus, centered on Broca's area in BA 44 and 45. To determine what connectivity differences exist between these two cytoarchitectonic regions, a location query was performed within BrainMap for studies activating these regions, using ROIs of the cytoarchitectonically defined areas (Amunts et al., 1999) as distributed with the SPM Anatomy Toolbox (Eickhoff et al., 2005, 2006b). The returned studies were then analyzed in a structure-based meta-analysis of both left BA 44 and BA 45 (Figure 7). For both regions, extensive bilateral connectivity was observed across the inferior frontal gyrus, precentral gyrus, inferior parietal lobule, fusiform gyrus, and insula, as well as medial ACC and SMA. Joint connectivity was also observed in the left thalamus and right cerebellum. In contrast to the word generation meta-analysis, no connectivity was observed in the precuneus or posterior cingulate cortex, likely reflecting a memory retrieval component (Cabeza and Nyberg, 2000) of verbal fluency processing. Much overlap was observed between the two images; however, comparison of the maps for BA 44 and BA 45 revealed strong dissociation in subcortical regions. BA 44 exhibited extensive connectivity with bilateral thalamus, caudate, and putamen in agreement with Eickhoff et al. (2009a), while only the left thalamus was observed for BA 45. In addition, only the BA 44 map returned connectivity in Wernicke's area, reflecting preferential engagement of this region in comparison to BA 45. Structure-based meta-analyses can therefore find distinct differences in functionally connected networks even for regions that lie very close to each other, such as BA 44 and 45.
Figure 7.
Structure-based meta-analysis of Brodmann areas 44 and 45. ALE meta-analyses were performed for all experiments in BrainMap that reported activation in either BA 44 or BA 45 to determine the meta-analytic functional connectivity of these cytoarchitectonically-defined regions. Results are displayed at P < 0.05, corrected. The top slices (left and right) are centered at x = −54, y = 18, z = 24. The upper left slices display the ROIs used to search BrainMap for BA 44 (red) and BA 45 (blue), which were obtained from the SPM Anatomy Toolbox (Eickhoff et al., 2005; Eickhoff et al., 2006b). ALE results for these regions are shown on the top right in axial, sagittal, and coronal slices, as well as on the bottom panel in axial slices from z = 58 to z = −26. The overlap between meta-analyses for BA 44 and 45 is shown in purple.
Toro et al. (2008) expanded upon the idea of structure-based meta-analyses and developed an algorithm to test the likelihood of a functional connection between regions, yielding a 3D meta-coactivation map for every voxel in the brain. In their analysis of 825 papers (3402 experiments with 27,909 coordinates), Toro et al. observed distinct and recognizable functional networks that are commonly associated with processes such as attention, motor function, and the resting state. Given that meaningful networks were extracted from the coordinates contained in BrainMap via a coactivation analysis, a recent study proposed that known functional networks of the brain during explicit activation could be derived using independent component analysis (ICA) of BrainMap data (Smith et al., 2009). These networks, when compared to resting state networks (RSNs) obtained by ICA of resting state fMRI data (Damoiseaux et al., 2006) were virtually identical. That is, the set of major covarying activation networks identified from a massive-scale meta-analysis (1687 papers, 7342 experiments, 58,620 coordinates) matched the set of networks that are present in the resting brain. These results provide strong evidence that RSNs reflect functional neural networks, and that these dynamic networks are engaged even at rest (Fox and Raichle, 2007). Given the independent nature of these two analyses on fundamentally different types of data, as well as the heterogeneity of data contained in BrainMap due to differences in subjects, scanners, analyses, and paradigms, it is remarkable that such strong correspondence was observed between the resting state and meta-analytic results. In sum, this study supports the validity of using BrainMap and coordinate-based meta-analyses to identify functional neural networks on a large scale.
Mapping Function–Structure Relationships in the Brain
One of the broad goals of functional neuroimaging research is to determine function–structure relationships in the brain. A concrete deliverable of this aim is a probabilistic functional atlas, in which specific mental operations are mapped to discrete networks of brain regions. Price and Friston (2005) point out that the relationship between a brain region and a mental function is not a one-to-one mapping. Instead this relationship is a many-to-many mapping, as a single region can be involved in many cognitive processes, and a single process usually activates multiple regions. Evaluating these mappings will require collating the immense amount of neuroimaging data that has been acquired thus far and continuing the development of advanced meta-analytic techniques in order to efficiently and effectively synthesize all of these data.
As the development of comprehensive neuroinformatics tools progresses, the need for comprehensive data ontologies increases. An ontology is a machine-interpretable description of concepts and their relationships with the purpose of sharing of ideas and information in a manner facilitated by semantic interoperability (Stevens et al., 2000). Until a foundational ontology for neuroimaging is established and adopted, the communication within and between databases will be limited, hindering the creation of a functional brain atlas. In the field of neuroimaging, ontology development is proceeding rapidly in the domains of representing neuroanatomical findings [e.g., NeuroNames, Bowden and Dubach, 2003; Bowden and Martin, 1995 and the Foundational Model of Anatomy (FMA), Rosse and Mejino, 2003], describing imaging acquisition strategies (e.g., RadLex, Langlotz, 2006; Rubin, 2008), and identifying clinical assessments [e.g., the Systematized Nomenclature of Medicine (SNOMED), Coté and Robboy, 1980]. In addition, the Neuroscience Information Framework Standardized (NIFSTD) Ontology, developed by BIRN and the NIF, is a collection of these and other neuroscience ontologies (Bug et al., 2008).
There is currently no accepted ontology for describing the range of mental operations performed by the human brain, although need for such an ontology is increasingly being discussed (Binder et al., 2009; Poldrack, 2006; Price and Friston, 2005; Toga, 2002). A research question such as “find the data examining the relationship between hippocampal volume in Alzheimer's disease” requires knowledge about clinical diagnosis, neuroanatomy, and MR scan acquisition. As mentioned previously, there are ongoing ontological efforts designed to curate knowledge in these domains. However, a query such as “find the data examining neural activations observed during sustained attention” involves knowledge about cognitive processing, for which no ontology exists to date. Creating a realistic functional brain atlas will require a systematic description of mental operations that are reported in the literature so that informative and standardized labels can be applied to different brain networks.
Investigators frequently utilize alternate and sometimes competitive terminologies when referring to the cognitive processes elicited by specific tasks. When different words are employed to represent the same concepts, the grouping of related ideas across different resources is impeded. For example, a semantically interoperable ontology will allow the linking of data designated with terms such as “declarative” and “explicit” memory types and be capable of relating “working memory” to either “memory” or “executive processing”. Cognition represents the most difficult domain to explicate as it contains the most intricate of all concepts, such as language, attention, and memory. But while much imaging research focuses on cognitive processing, important results are also being published in the areas of perception, action, emotion, and autonomic functions. To comprehensively describe the many-to-many mappings of structure and function being investigated in neuroimaging research, a complete mental ontology must be developed.
Poldrack (2008) suggests that the many-to-many mapping dilemma may be complicated by our current understanding of what cognitive processes exist and how they are defined across functional neuroimaging experiments. Optimally, an appropriate and useful cognitive ontology will not merely be a catalogue of various mental operations parsed down to very fine detail in accord with current theories of cognitive psychology. While the consideration of competing theories often results in new knowledge discovery, the development of such a top-down ontology would be so continuously and vehemently debated that it could never reach a sufficient degree of consensus in order to be considered adoptable by the neuroimaging community. In contrast, a biologically based ontology that is driven by the way we observe the brain to operate in imaging experiments may reveal a cognitive architecture that has not previously been considered.
BrainMap's Future Role in Atlas and Ontology Development
The synergy that exists between the BrainMap database and the ALE meta-analysis method was designed to facilitate the creation of a functional brain atlas. BrainMap's search capabilities can support different types of queries, such as “for a given function, what regions are typically engaged?”, “for a given region, what tasks elicit activation?”, or “for a given region, what other regions are coactivated?” These questions highlight the value of meta-analytic results in comparison to results from individual studies. We believe that these correspondences (function-to-regions, region-to-tasks, or region-to-network) must be constructed according to a bottom-up strategy, using knowledge gleaned from data-driven analyses. Current probabilistic structural atlases (e.g., the Harvard-Oxford structural probability atlas, Smith et al., 2004, or the Jülich cytoarchitectonic atlas, Eickhoff et al., 2005, 2006a) have proven to be useful in testing region-to-tasks or region-to-network associations, as shown in Figure 7. Theoretically, any method of determining regions of interest can be used to query the BrainMap database, whether structurally or functionally defined.
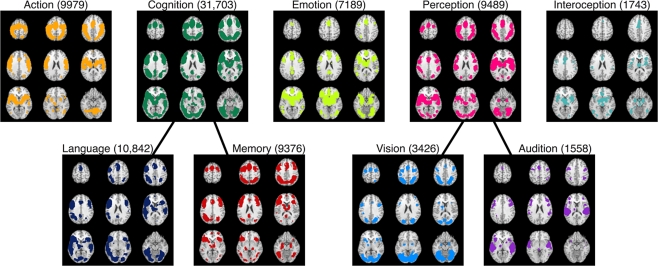
BrainMap is also capable of generating function-to-regions associations, albeit at a coarse resolution. Whole brain meta-analytic maps can be created for each behavioral domain category in BrainMap, which can then be decomposed into sub-networks based on different levels of the domain hierarchy. To illustrate, ALE meta-analyses were performed on nine different behavioral domain categories and sub-categories: action, cognition, emotion, perception, interoception, language, memory, vision, and audition (Figure 8). Each ALE image provides a unique mapping of the neural network associated with the relevant domain. Many regions are observed in multiple domain maps, and some maps are very similar, but none are identical. However, a more detailed domain structure is needed to fully characterize the range of human cognition. We propose that applying high-level filters from the entire BrainMap coding scheme to these meta-maps, in the way condition-based filters were applied in the word generation meta-analysis (Figure 6), can be an effective strategy for refining the spatial specificity of these images. Thus, while paradigm class and behavioral domain are important metadata fields in the BrainMap coding scheme, all fields have the potential to assist in unraveling the brain's systems and their interactions.
Figure 8.
BrainMap's functional atlas development strategy. ALE meta-analyses were performed on nine arbitrarily selected behavioral domain categories and sub-categories: action, cognition, emotion, perception, interoception, language, memory, vision, and audition. These results illustrate the mapping of function-to-regions correspondences that are currently capable using the BrainMap database. Refinement of these maps can be accomplished through high-level filtering and mining of the database. Results are displayed at P < 0.05, corrected. The total number of coordinates in each meta-analysis is listed after the domain heading for each map in parentheses.
As the BrainMap database increases in size, these results will evolve and grow more powerful, perhaps leading to a multi-layered, multi-modal probabilistic functional brain atlas derived from many different large-scale coordinate-based meta-analyses. This approach would likely be enhanced by the development of a standardized mental ontology. Differences in competitive terminology must be resolved to allow for the union of experimentally similar data sets. Perhaps the best strategy would be to combine all of BrainMap's data-driven methods in establishing function–structure relationships with other ontology initiatives, such at the Cognitive Atlas7 (Bilder et al., 2009), the NIFSTD ontology (Bug et al., 2008), or the Neural ElectroMagnetic Ontologies (NEMO) (Frishkoff et al., 2009). Given the complex nature of human brain function, it is reasonable to suggest that no single approach will be powerful enough to solve the fundamental challenges associated with mapping the mind, but rather a joint effort will be required.
Conflict of Interest Statement
The authors report no competing interests.
Acknowledgments
This work was supported by the NIMH (R01-MH074457-03; PI = Peter Fox and R01-MH084812-01; PI = Angela Laird and Jessica Turner), NINDS (T35-NS051166-04; PI = Peter Fox), NIDCD (F32-DC009116-02; PI = Matthew Cykowski), NCRR (U24-RR021992; PI = Steven Potkin), and the Helmholz Initiative on Systems-Biology (SBE).
Footnotes
References
- Amunts K., Schleicher A., Burgel U., Mohlberg H., Uylings H. B., Zilles K. (1999). Broca's region revisited: cytoarchitecture and intersubject variability. J. Comp. Neurol. 412, 319–341 [DOI] [PubMed] [Google Scholar]
- Amunts K., Weiss P. H., Mohlberg H., Pieperhoff P., Eickhoff S., Gurd J. M., Marshall J. C., Shah N. J., Fink G. R., Zilles K. (2004). Analysis of neural mechanisms underlying verbal fluency in cytoarchitectonically defined stereotactic space – the roles of Brodmann areas 44 and 45. Neuroimage 22, 42–56 10.1016/j.neuroimage.2003.12.031 [DOI] [PubMed] [Google Scholar]
- Basho S., Palmer E. D., Rubio M. A., Wulfeck B., Muller R. A. (2007). Effects of generation mode in fMRI adaptations of semantic fluency: paced production and overt speech. Neuropsychologia 45, 1697–1706 10.1016/j.neuropsychologia.2007.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder R. M., Sabb F. W., Parker D. S., Kalar D., Chu W. W., Fox J., Freimer N. B., Poldrack R. A. (2009). Cognitive ontologies for neuropsychiatric phenomics research. Cogn. Neuropsychiatry (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder J. R., Desai R. H., Graves W. W., Conant L. L. (2009). Where is the semantic system? A critical review and meta-analysis of 120 functional neuroimaging studies. Cereb. Cortex (in press). 10.1093/cercor/bhp055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden D. M., Dubach M. F. (2003). NeuroNames 2002. Neuroinformatics 1, 43–59 10.1385/NI:1:1:043 [DOI] [PubMed] [Google Scholar]
- Bowden D. M., Martin R. F. (1995). NeuroNames brain hierarchy. Neuroimage 2, 63–83 10.1006/nimg.1995.1009 [DOI] [PubMed] [Google Scholar]
- Brown S., Laird A. R., Ingham R. J., Ingham J. C., Fox P. T. (2005). Stuttered and fluent speech production: an ALE meta-analysis of functional neuroimaging studies. Hum. Brain Mapp. 25, 105–117 10.1002/hbm.20140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bug W. J., Ascoli G. A., Grethe J. S., Gupta A., Fennema-Notestine C., Laird A. R., Larson S. D., Rubin D., Shepherd G. M., Turner J. A., Martone M. E. (2008). The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 6, 175–194 10.1007/s12021-008-9032-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeza R., Nyberg L. (2000). Imaging cognition II: an empirical review of 275 PET and fMRI studies. J. Cogn. Neurosci. 12, 1–47 10.1162/08989290051137585 [DOI] [PubMed] [Google Scholar]
- Chein J. M., Fissell K., Jacobs S., Fiez J. A. (2002). Functional heterogeneity within Broca's area during verbal working memory. Physiol. Behav. 77, 635–639 10.1016/S0031-9384(02)00899-5 [DOI] [PubMed] [Google Scholar]
- Cordes D., Haughton V. M., Arfanakis K., Wendt G. J., Turski P. A., Moritz C. H., Quigley M. A., Meyerand M. E. (2000). Mapping functionally related regions of brain with functional connectivity MR imaging. Am. J. Neuroradiol. 21, 1636–1644 [PMC free article] [PubMed] [Google Scholar]
- Costafreda S. G., Brammer M. J., David A. S., Fu C. H. (2008). Predictors of amygdala activation during the processing of emotional stimuli: a meta-analysis of 385 PET and fMRI studies. Brain Res. Rev. 58, 57–70 10.1016/j.brainresrev.2007.10.012 [DOI] [PubMed] [Google Scholar]
- Costafreda S. G., Fu C. H. Y., Lee L., Everitt B., Brammer M. J., David A. S. (2006). A systematic review and quantitative appraisal of fMRI studies of verbal fluency: role of the left inferior frontal gyrus. Hum. Brain Mapp. 27, 799–810 10.1002/hbm.20221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coté R. A., Robboy S. (1980). Progress in medical information management. Systemized nomenclature of medicine (SNOMED). JAMA 243, 756–762 10.1001/jama.243.8.756 [DOI] [PubMed] [Google Scholar]
- Damoiseaux J. S., Rombouts S. A., Barkhof F., Scheltens P., Stam C. J., Smith S. M., Beckmann C. F. (2006). Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. U.S.A. 103, 13848–13853 10.1073/pnas.0601417103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decety J., Lamm C. (2007). The role of the right tempoparietal junction in social interaction: how low-level computational processes contribute to meta-cognition. Neuroscientist 13, 580–593 10.1177/1073858407304654 [DOI] [PubMed] [Google Scholar]
- Derrfuss J., Brass M., Neumann J., Yves von Cramon D. (2005). Involvement of the inferior frontal junction in cognitive control: meta-analyses of switching and Stroop studies. Hum. Brain Mapp. 25, 22–34 10.1002/hbm.20127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrfuss J., Mar R. A. (2009). Lost in localization: the need for a universal coordinate database. Neuroimage (in press). 10.1016/j.neuroimage.2009.01.053 [DOI] [PubMed] [Google Scholar]
- Eickhoff S., Stephan K. E., Mohlberg H., Grefkes C., Fink G. R., Amunts K., Zilles K. (2005). A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. Neuroimage 25, 1325–1335 10.1016/j.neuroimage.2004.12.034 [DOI] [PubMed] [Google Scholar]
- Eickhoff S. B., Amunts K., Mohlberg H., Zilles K. (2006a). The human parietal operculum. II. Stereotaxic maps and correlation with functional imaging results. Cereb. Cortex 16, 268–279 10.1093/cercor/bhi106 [DOI] [PubMed] [Google Scholar]
- Eickhoff S. B., Heim S., Zilles K., Amunts K. (2006b). Testing anatomically specified hypotheses in functional imaging using cytoarchitectonic maps. Neuroimage 32, 570–582 10.1016/j.neuroimage.2006.04.204 [DOI] [PubMed] [Google Scholar]
- Eickhoff S. B., Heim S., Zilles K., Amunts K. (2009a). A systems perspective on the effective connectivity of overt speech production. Philos. Transact. A Math. Phys. Eng. Sci. 367, 2399–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff S. B., Laird A. R., Grefkes C., Wang L. E., Zilles K., Fox P. T. (2009b). Coordinate-based activation likelihood estimation meta-analysis of neuroimaging data: a random-effects approach based on empirical estimates of spatial uncertainty. Hum. Brain Mapp. (in press). 10.1002/hbm.20718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison-Wright I., Bullmore E. (2009). Meta-analysis of diffusion tensor imaging studies in schizophrenia. Schizophr. Res. 108, 3–10 10.1016/j.schres.2008.11.021 [DOI] [PubMed] [Google Scholar]
- Ellison-Wright I., Glahn D. C., Laird A. R., Thelen S. M., Bullmore E. T. (2008). The anatomy of first-episode and chronic schizophrenia: an anatomical likelihood estimation meta-analysis. Am. J. Psychiatry 165, 1015–1023 10.1176/appi.ajp.2008.07101562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald P. B., Laird A. R., Maller J., Daskalakis Z. J. (2008). A meta-analytic study of changes in brain activation in depression. Hum. Brain Mapp. 29, 683–695 10.1002/hbm.20426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M. D., Raichle M. E. (2007). Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 8, 700–711 10.1038/nrn2201 [DOI] [PubMed] [Google Scholar]
- Fox P. T., Lancaster J. L. (2002). Mapping context and content: the BrainMap model. Nat. Rev. Neurosci. 3, 319–321 10.1038/nrn789 [DOI] [PubMed] [Google Scholar]
- Fox P. T., Parsons L. M., Lancaster J. L. (1998). Beyond the single study: function–location meta-analysis in cognitive neuroimaging. Curr. Opin. Neurobiol. 8, 178–187 10.1016/S0959-4388(98)80138-4 [DOI] [PubMed] [Google Scholar]
- Frishkoff G. A., Dou D., Frank R., LePendu P., Liu H. (2009). Development of Neural Electromagnetic Ontologies (NEMO): representation and integration of event-related brain potentials. Proc. Int. Conf. Biomed. Ontologies (in press). [Google Scholar]
- Glahn D. C., Laird A. R., Ellison-Wright I., Thelen S. M., Robinson J. L., Lancaster J. L., Bullmore E., Fox P. T. (2008). Meta-analysis of gray matter anomalies in schizophrenia: application of anatomic likelihood estimation and network analysis. Biol. Psychiatry 64, 774–781 10.1016/j.biopsych.2008.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn D. C., Ragland J. D., Abramoff A., Barrett J., Laird A. R., Bearden C. E., Velligan D. I. (2005). Beyond hypofrontality: a quantitative meta-analysis of functional neuroimaging studies of working memory in schizophrenia. Hum. Brain Mapp. 25, 60–69 10.1002/hbm.20138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefkes C., Eickhoff S. B., Nowak D. A., Dafotakis M., Fink G. R. (2008). Dynamic intra- and interhemispheric interactions during unilateral and bilateral hand movements assessed with fMRI and DCM. Neuroimage 41, 1382–1394 10.1016/j.neuroimage.2008.03.048 [DOI] [PubMed] [Google Scholar]
- Grosbras M.-H., Laird A. R., Paus T. (2005). Cortical regions involved in eye movements, shifts of attention, and gaze perception. Hum. Brain Mapp. 25, 140–154 10.1002/hbm.20145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim S., Eickhoff S. B., Amunts K. (2008). Specialisation in Broca's region for semantic, phonological, and syntactic fluency? Neuroimage 40, 1362–1368 10.1016/j.neuroimage.2008.01.009 [DOI] [PubMed] [Google Scholar]
- Koski L., Paus T. (2000). Functional connectivity of the anterior cingulate cortex with the human frontal lobe: a brain-mapping meta-analysis. Exp. Brain Res. 133, 55–65 10.1007/s002210000400 [DOI] [PubMed] [Google Scholar]
- Laird A. R., Lancaster J. L., Fox P. T. (2005a). BrainMap: the social evolution of a human brain mapping database. Neuroinformatics 3, 65–78 10.1385/NI:3:1:065 [DOI] [PubMed] [Google Scholar]
- Laird A. R., McMillan K. M., Lancaster J. L., Kochunov P., Turkeltaub P. E., Pardo J. V., Fox P. T. (2005b). A comparison of label-based review and activation likelihood estimation in the Stroop task. Hum. Brain Mapp. 25, 6–21 10.1002/hbm.20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A. R., Fox M., Price C. J., Glahn D. C., Uecker A. M., Lancaster J. L., Turkeltaub P. E., Kochunov P., Fox P. T. (2005c). ALE meta-analysis: controlling the false discovery rate and performing statistical contrasts. Hum. Brain Mapp. 25, 155–164 10.1002/hbm.20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A. R., Robbins J. M., Li K., Price L. R., Cykowski M. D., Narayana S., Laird R. W., Franklin C., Fox P. T. (2008). Modeling motor connectivity using TMS/PET and structural equation modeling. Neuroimage 41, 424–436 10.1016/j.neuroimage.2008.01.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J. L., Laird A. R., Fox M., Glahn D. E., Fox P. T. (2005). Automated analysis of meta-analysis networks. Hum. Brain Mapp. 25, 174–184 10.1002/hbm.20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J. L., Tordesillas-Gutierrez D., Martinez M., Salinas F., Evans A., Zilles K., Mazziotta J. C., Fox P. T. (2007). Bias between MNI and Talairach coordinates analyzed using the ICBM-152 brain template. Hum. Brain Mapp. 28, 1194–1205 10.1002/hbm.20345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J. L., Woldorff M. G., Parsons L. M., Liotti M., Freitas C. S., Rainey L., Kochunov P. V., Nickerson D., Mikiten S. A., Fox P. T. (2000). Automated Talairach atlas labels for functional brain mapping. Hum. Brain Mapp. 10, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlotz C. P. (2006). RadLex: a new method for indexing online educational materials. Radiographics 26, 1595–1597 10.1148/rg.266065168 [DOI] [PubMed] [Google Scholar]
- McMillan K. M., Laird A. R., Witt S. T., Meyerand M. E. (2007). Self-paced working memory: validation of verbal variations of the n-back paradigm. Brain Res. 1139, 133–142 10.1016/j.brainres.2006.12.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies L. A. C., Chamberlain S. R., Laird A. R., Thelen S. M., Sahakian B. J., Bullmore E. T. (2008). Integrating evidence from neuroimaging and neuropsychological studies of obsessive compulsive disorder: the orbitofronto-striatal model revisited. Neurosci. Biobehav. Rev. 32, 525–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minzenberg M. J., Laird A. R., Thelen S. M., Carter C. S., Glahn D. C. (2009). Meta-analysis of 41 functional neuroimaging studies of executive cognition reveals dysfunction in a general-purpose cognitive control system in schizophrenia. Arch. Gen. Psychiatry (in press). 10.1002/hbm.20133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann J., Lohmann G., Derrfuss J., Yves von Cramon D. (2005). The meta-analysis of functional imaging data using replicator dynamics. Hum. Brain Mapp. 25, 165–173 10.1002/hbm.20380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann J., Yves von Cramon D., Lohmann G. (2008). Model-based clustering of meta-analytic functional imaging data. Hum. Brain Mapp. 29, 177–192 10.1002/hbm.20137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petacchi A., Laird A. R., Fox P. T., Bower J. M. (2005). Cerebellum and auditory function: an ALE meta-analysis of functional neuroimaging studies. Hum. Brain Mapp. 25, 118–128 10.1038/331585a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S. E., Fox P. T., Posner M. I., Mintun M., Raichle M. E. (1988). Positron emission tomographic studies of the cortical anatomy of single-word processing. Nature 331, 585–589 10.1016/j.tics.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Poldrack R. A. (2006). Can cognitive processes be inferred from neuroimaging data? Trends Cogn. Sci. 10, 59–63 10.1016/j.conb.2008.07.006 [DOI] [PubMed] [Google Scholar]
- Poldrack R. A. (2008). The role of fMRI in cognitive neuroscience: where do we stand? Curr. Opin. Neurobiol. 18, 223–227 10.1093/cercor/bhj088 [DOI] [PubMed] [Google Scholar]
- Postuma R. B., Dagher A. (2006). Basal ganglia functional connectivity based on a meta-analysis of 126 positron emission tomography and functional magnetic resonance imaging publications. Cereb. Cortex 16, 1508–1521 10.1002/hbm.20132 [DOI] [PubMed] [Google Scholar]
- Price C. J., Devlin J. T., Moore C. J., Morton C., Laird A. R. (2005). Meta-analyses of object naming: effect of baseline. Hum. Brain Mapp. 25, 70–82 10.1080/02643290442000095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. J., Friston K. J. (2005). Functional ontologies for cognition: the systematic definition of structure and function. Cogn. Neuropsychol. 22, 262–275 10.1176/appi.ajp.2009.08091307 [DOI] [PubMed] [Google Scholar]
- Ragland J. D., Laird A. R., Ranganath C. S., Blumenfeld R. S., Gonzales S. M., Glahn D. C. (2009). Prefrontal activation deficits during episodic memory in schizophrenia. Am. J. Psychiatry (in press). 10.1002/hbm.20854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. L., Laird A. R., Glahn D. C., Lovallo W. R., Fox P. T. (2009). Meta-analytic connectivity modeling: Delineating the functional connectivity of the human amygdala. Hum. Brain. Mapp. (in press). 10.1016/j.mri.2007.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B. P., Morgan V. L., Newton A. T., Gore J. C. (2007). Assessing functional connectivity in the human brain by fMRI. Magn. Reson. Imaging 25, 1347–1357 10.1016/j.jbi.2003.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse C., Mejino J. L. V. (2003). A reference ontology for biomedical informatics: The Foundational Model of Anatomy. J. Biomed. Inform. 36, 478–500 10.1007/s10278-007-9073-0 [DOI] [PubMed] [Google Scholar]
- Rubin D. L. (2008). Creating and curating a terminology for radiology: ontology modeling and analysis. J. Digit. Imaging 21, 355–362 10.1016/j.neuroimage.2007.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter M. L., Raczka K., Neumann J., Yves von Cramon D. (2007). Towards a nosology for frontotemporal lobar degenerations – a meta-analysis involving 267 subjects. Neuroimage 36, 497–510 [DOI] [PubMed] [Google Scholar]
- Smith S. M., Fox P. T., Miller K. L., Glahn D. C., Fox P. M., Mackay C. E., Filippini N., Watkins K. E., Toro R., Laird A. R., Beckmann C. F. (2009). The brain's functional architecture: Correspondence between rest and activation. Proc. Natl. Acad. Sci USA (in press). 10.1016/j.neuroimage.2004.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. M., Jenkinson M., Woolrich M. W., Beckmann C. F., Behrens T. E., Johansen-Berg H., Bannister P. R., De Luca M., Drobnjak I., Flitney D. E., Niazy R. K., Saunders J., Vickers J., Zhang Y., De Stefano N., Brady J. M., Matthews P. M. (2004). Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23(Suppl. 1), S208–S219 10.1002/hbm.20680 [DOI] [PubMed] [Google Scholar]
- Soros P., Inamoto Y., Martin R. E. (2009). Functional brain imaging of swallowing: an activation likelihood estimation meta-analysis. Hum. Brain Mapp. (in press). 10.1162/jocn.2008.21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreng R. N., Mar R. A., Kim A. S. (2009). The common neural basis of autobiographical memory, prospection, navigation, theory of mind and the default mode: a quantitative meta-analysis. J. Cogn. Neurosci. 21, 489–510 10.1098/rstb.2001.0908 [DOI] [PubMed] [Google Scholar]
- Stephan K. E., Kamper L., Bozkurt A., Burns G. A., Young M. P., Kötter R. (2001). Advanced database methodology for the Collation of Connectivity data on the Macaque brain (CoCoMac). Philos. Trans. R. Soc. Lond. B, Biol. Sci. 356, 1159–1186 10.1093/bib/1.4.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens R., Goble C. A., Bechhofer S. (2000). Ontology-based knowledge representation for bioinformatics. Brief. Bioinform. 1, 398–414 10.1038/nrn782 [DOI] [PubMed] [Google Scholar]
- Toga A. W. (2002). Neuroimage databases: the good, the bad, and the ugly. Nat. Rev. Neurosci. 3, 302–309 10.1093/cercor/bhn014 [DOI] [PubMed] [Google Scholar]
- Toro R., Fox P. T., Paus T. (2008). Functional coactivation map of the human brain. Cereb. Cortex 18, 2553–2559 10.1006/nimg.2002.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkeltaub P. E., Eden G. F., Jones K. M., Zeffiro T. A. (2002). Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage 16, 765–780 10.1093/scan/nsm015 [DOI] [PubMed] [Google Scholar]
- Wager T. D., Lindquist M., Kaplan L. (2007). Meta-analysis of functional neuroimaging data: current and future directions. Soc. Cogn. Affect. Neurosci. 2, 150–158 10.1016/j.neuroimage.2008.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager T. D., Lindquist M., Nichols T. E., Kober H., Van Snellenberg J. X. (2009). Evaluating the consistency and specificity of neuroimaging data using meta-analysis. Neuroimage 45, S210–S221 10.1093/brain/119.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton E., Wise R. J., Price C. J., Weiller C., Hadar U., Ramsay S., Frackowiak R. S. (1996). Noun and verb retrieval by normal subjects. Studies with PET. Brain 119, 159–179 10.1093/brain/114.4.1803 [DOI] [PubMed] [Google Scholar]
- Wise R., Chollet F., Hadar U., Friston K., Hoffner E., Frackowiak R. (1991). Distribution of cortical neural networks involved in word comprehension and word retrieval. Brain 114, 1803–1817 [DOI] [PubMed] [Google Scholar]
- Xiong J., Parsons L. M., Gao J. H., Fox P. T. (1999). Interregional connectivity to primary motor cortex revealed using MRI resting state images. Hum. Brain Mapp. 6, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]