Abstract
Current DNA methylation assays are limited in the flexibility and efficiency of characterizing a large number of genomic targets. We report a method to specifically capture an arbitrary subset of genomic targets for single-molecule bisulfite sequencing for digital quantification of DNA methylation at single-nucleotide resolution. A set of ~30,000 padlock probes was designed to assess methylation of ~66,000 CpG sites within 2,020 CpG islands on human chromosome 12, chromosome 20, and 34 selected regions. To investigate epigenetic differences associated with dedifferentiation, we compared methylation in three human fibroblast lines and eight human pluripotent stem cell lines. Chromosome-wide methylation patterns were similar among all lines studied, but cytosine methylation was slightly more prevalent in the pluripotent cells than in the fibroblasts. Induced pluripotent stem (iPS) cells appeared to display more methylation than embryonic stem cells. We found 288 regions methylated differently in fibroblasts and pluripotent cells. This targeted approach should be particularly useful for analyzing DNA methylation in large genomes.
DNA methylation is a primary epigenetic mechanism for transcriptional regulation during normal development and goes awry in many diseases, including cancers. Genome-scale patterns of DNA methylation have been characterized by microarray hybridization or bisulfite sequencing1. Microarray methods have enabled methylation to be quantified at 1,536 discrete CpG sites in the human genome with the GoldenGate assay2,3. They have also been coupled with methylated DNA immunoprecipitation or methyl-specific restriction enzyme digestion to quantify relative levels of DNA methylation, although the read-outs of such approaches are only averages of the levels of methylation of multiple adjacent CpG sites4–6.
More recently, next-generation sequencing has enabled absolute quantification of DNA methylation with single-nucleotide resolution on a larger scale than previously possible. These efforts include bisulfite sequencing of PCR amplicons from human tissues and cancer cell lines7–9, single-molecule sequencing of reduced representation libraries from mouse embryonic stem cells10,11 and whole-genome bisulfite sequencing of Arabidopsis thaliana12,13. Although whole-genome bisulfite sequencing of a mammalian genome should be technically feasible, the large genome sizes pose a considerable challenge14.
Selection or enrichment of genomic targets prior to sequencing would substantially reduce sequencing cost. PCR-based target selection is highly specific, but cannot be multiplexed easily for genome-wide assays. In comparison, bisulfite sequencing of reduced representation libraries is more scalable in that sample preparation can be performed in a series of single-tube reactions11. However, this assay is restricted to CpG dinucleotides adjacent to the recognition sites of certain restriction enzymes. Here we have addressed this limitation by using padlock probes for highly parallel capture of an arbitrary set of sequencing targets from bisulfite-converted DNA. Patterns of DNA methylation across ~66,000 CpG sites were characterized in pluripotent stem cells reprogrammed from human fibroblasts. Comparison of iPS cells, derived using three different methods, with matched fibroblasts, hybrid stem cells and ES cells identified localized changes of DNA methylation that are associated with nuclear reprogramming.
RESULTS
Parallel target capture with padlock probes
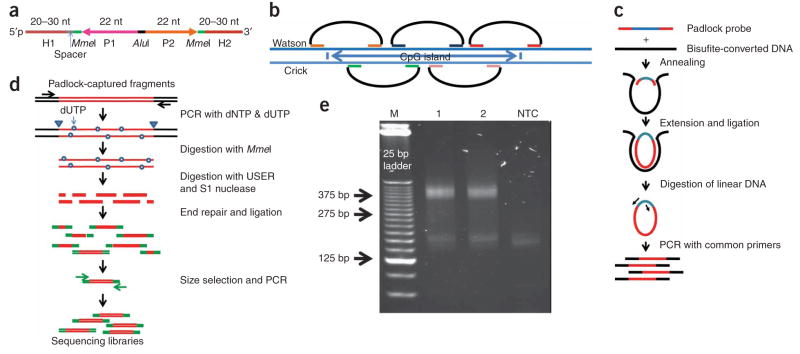
Padlock probes were previously used for exon capture and resequencing15. As in the eMIP method, our approach to targeted bisulfite sequencing involves the in situ synthesis of long (~150 nt) oligonucleotides on programmable microarrays, followed by their cleavage and enzymatic conversion into padlock probes. A library of padlock probes is annealed to the template DNA, circularized, and amplified by PCR before shotgun sequencing (Fig. 1a–c).
Figure 1.
Targeted bisulfite sequencing with padlock probes. (a) Each padlock probe has a common linker sequence flanked by two target-specific capturing arms (H1 and H2). H1 and H2 are melting temperature–normalized, and a spacer sequence is included to normalize probe lengths. The linker sequence contains priming sites (AP1 and AP2) for universal primers, two MmeI sites and a central AluI recognition site. (b) A CpG island (or other target regions) is covered by multiple padlock probes targeting partially overlapped regions on alternating strands. (c) A library of padlock probes is annealed to bisulfite-converted genomic DNA (black) and the 3′ ends are extended and ligated with the 5′ end. After removal of linear DNAs with exonucleases, all circularized padlock probes are PCR-amplified using a pair of common primers. (d) To generate a shotgun sequencing library, amplicons were reamplified in the presence of dUTP, digested with MmeI, and then with USER and S1 nuclease. Digested amplicons are end repaired and ligated with Solexa sequencing adaptors (green). Ligated products are then selected by size and amplified by PCR to generate the shotgun sequencing library. (e) Gel electrophoresis analysis of the padlock-captured products from two independent capturing reactions (1 and 2) and a no-template control. Expected amplicon size is in the range of 344–394 bp, which includes capturing targets (175–225 bp), capturing arms (58 bp) and amplification primers (111 bp). NTC, no-template control.
There are, however, two major challenges in performing padlock capture for bisulfite sequencing. First, bisulfite treatment converts all unmethylated cytosines into uracils, resulting in marked reduction of sequence complexity. Achieving specific target capture on bisulfite-converted DNA is more difficult than on native genomic DNA. Second, we initially observed a low capturing sensitivity, high bias and random losses of alleles with the eMIP method15. Obtaining accurate and efficient quantification of DNA methylation was not possible with the existing protocol, especially with the presence of allelic drop-outs.
We designed 10,582 padlock probes, each capturing a 175- to 225-bp region, including 9,350 probes covering 2,020 CpG islands (Supplementary Table 1 online) on human chromosomes 12 and 20, 705 probes covering 237 promoters in eight ENCODE (the Encyclopedia of DNA Elements) regions, and 527 probes targeting 4-kb regions centered on the transcription start sites (TSS) of 26 genes related to development or pluripotency (Supplementary Table 2 online). The total size of captured fragments was 2.1 Mbp, representing 0.064% of the human genome. Because some probes contain CpG sites within the capturing arms, we iterated all possible C/T combinations on these CpG sites, and synthesized a total of 30,000 non-degenerate probes. We chose to perform the proof-of-concept study focusing mostly on CpG islands primarily because they represent a relatively well-defined set of genomic features in the human genome annotation. To increase the sensitivity and reduce bias, we increased the probe/target ratio by more than tenfold, extended the reaction time and added an additional five cycles of circularization compared with the published protocol15. To integrate construction of sequencing libraries with padlock capture, we developed a new method that uses a combination of uracil-specific excision reagent (USER) enzymes and S1 nuclease to create fragments with random ends (Fig. 1d).
We first validated the targeted bisulfite sequencing method by capturing bisulfite-converted Jurkat cell genomic DNA with all 30,000 padlock probes in a single-tube reaction. PCR amplicons from the circularization reactions were template specific, and PAGE analysis showed the expected size distribution (Fig. 1e). We estimated the specificity of the capturing reaction by ligating captured DNA fragments to a sequencing vector, cloning these into Escherichia coli, and sequencing 96 clones. Of 89 high-quality Sanger sequencing reads obtained, 80 were from the targeted regions, indicating a specificity of 90%. We then used an Illumina Genome Analyzer to sequence the ends of the captured fragments. We mapped 5.5 millions reads to 10,364 of 10,582 targets, which translate to a sensitivity of 98%. These results indicate that padlock probes can specifically extract a large set of genomic targets for single-molecule bisulfite sequencing.
Normalization of padlock-captured DNA fragments
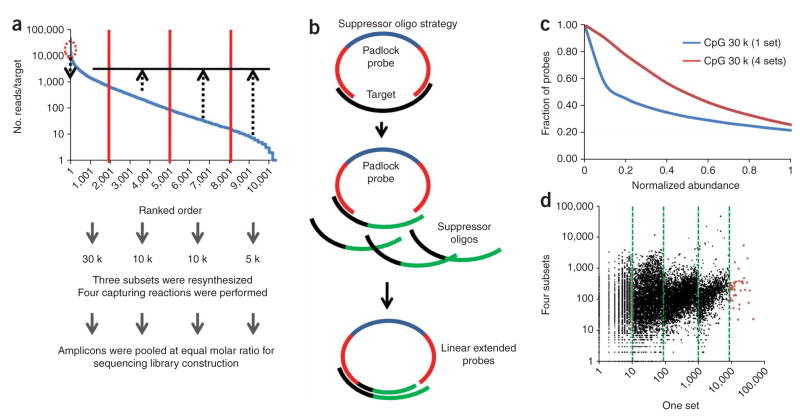
Although 98% of the targets were observed at least once in the end-sequencing analysis, the abundance of different captured fragments varied across a 10,000-fold range. Analysis of variance revealed that the bias resulted from a combination of factors, including GC content and length of the ligation arms, and the size of the targets to be captured (Supplementary Fig. 1 online). To normalize the relative abundance among different DNA fragments, we used a combination of two strategies: ‘subsetting’ and ‘suppressor oligos’ (Fig. 2). All 30,000 padlock probes were ranked based on the capturing efficiency determined by end sequencing, and divided into four subsets, two containing 5,000, and two containing 10,000, oligos. The three less efficient subsets were resynthesized. For each DNA sample, four capturing reactions were performed separately using probes from the original set of 30,000 and the three resynthesized subsets. The PCR amplicons from the capturing reactions were pooled in equal molar ratios before constructing a shotgun sequencing library (Fig. 2a). This subsetting strategy increased the relative abundance of less efficient targets by orders of magnitude. For reasons that we still do not fully understand, a very small number of probes were extremely efficient. For example, the top 48 (0.016%) most efficient probes account for 13.3% of mappable reads in the end-sequencing analysis.
Figure 2.
Normalization of padlock-capturing efficiency. (a) The ‘subsetting’ strategy. The 30,000 probes were divided into four sets (5 k, 10 k, 10 k, 5 k). The three less efficient sets were resynthesized. We reused the original 30,000-probe set because it was dominated by the most efficient 5,000 probes. (b) The ‘suppressor oligo’ strategy. (c) Distribution of normalized abundance for all captured targets with one 30,000-probe set and with four probe sets. The x-axis is the normalized abundance of each captured target, which is calculated by dividing the counts of the target by the average counts of all targets. The y-axis is the fraction of probes with the coverage equal to or greater than the normalized coverage. (d) Comparison of relative abundance for each target before and after normalization. The green vertical dash lines indicate the clear separation of four subsets of targets, as well as the fifth set normalized with the suppressor oligos.
Although the subsetting strategy allowed us to adjust relative abundance among several relatively large subsets of probes, we also needed a method to specifically reduce the efficiency of a small number of probes in a library. For this, we designed a set of 48 suppressor oligos, which contained chimeric sequences: the 5′ region was reverse complementary to the extension arm H2, and the 3′ region contained a short sequence unrelated to the ligation arm H1. When these suppressor oligos were mixed with padlock probes in a high molar ratio (100-fold molar excess of suppressor oligos), the 48 most efficient probes tended to anneal to the suppressor oligos, were extended from the 3′ ends and yielded linear-extended sequences that were removed in the subsequent exonuclease digestion (Fig. 2b). We tested this normalization strategy on the same bisulfite-converted Jurkat cell DNA, performed end sequencing on the captured DNA fragments and obtained 2.2 million mappable reads. The effect of normalization was obvious: the fraction of probes with at least half of the average abundance increased from 31% to 49%; the average efficiency for the 48 most abundant probes was reduced by fivefold (Fig. 2c,d).
Accuracy of methylation quantification
To validate the measurement accuracy of our method, we took advantage of the built-in redundancy in our probe design. Each CpG island was covered by multiple probes targeting partially overlapping DNA fragments on alternating strands (Fig. 1b). The CpG sites in the overlapping regions were captured independently from two DNA strands with different probes. Because the sequencing reads were mapped in a strand-specific manner and CpG methylation is symmetric on the two DNA strands13, the accuracy of the assay can be determined by comparing the methylation level of these CpG sites on the two strands. For 2,697 such CpG sites that were covered by >50 sequencing reads, the Pearson correlation coefficient (R) was 0.987 (Supplementary Fig. 2a online). To confirm the measurement accuracy with an independent method, we quantified the methylation levels of 182 randomly selected CpG sites with conventional bisulfite Sanger sequencing. Correlation between the two assays was high (R = 0.975; Supplementary Fig. 2b). Finally, we compared the methylation measurements from two batches of IMR90 fibroblast cultures. Excellent correlation was observed between the biological replicates (R = 0.970; Supplementary Fig. 2c). Taken together, these three validation experiments indicate that our assay is highly robust.
The CpG sites characterized in this study were based on the human reference genome (UCSC hg18). A small fraction of such sites could be different in our samples owing to the presence of genetic variations. Particularly, C → T transitions at CpG sites cannot be distinguished from unmethylated CpG sites after bisulfite conversion. Such transitions are detectable, however, if the sequences of the reverse complementary strand are available. We would expect to see asymmetric dinucleotides: TG on one strand and TA on the other. We searched for sites with such an asymmetric pattern in the subset of regions that were captured and sequenced from both strands, and found from 11 to 38 transitions in each sample (Supplementary Table 3 online). The majority (57–90%) of such transitions are known C/T polymorphisms in the NCBI single-nucleotide polymorphism (SNP) database. Because Hues6-BJ-Hybrid1 is a tetraploid, the number of transitions in this hybrid line is roughly twice as many as in other cell lines. As C → T transitions represent only 0.13–0.32% of CpG sites, the resulting artifacts are negligible.
Padlock capture yields relatively short products (insert sizes of 175–225 bp). As such small DNA fragments cannot be further fragmented using conventional DNA shearing methods, such as nebulization or hydra-shearing, we developed an enzymatic fragmentation method for constructing shotgun sequencing libraries. To validate this method, we compared enzymatic fragmentation with adaptive focused sonication16. Two sequencing libraries were made from the same padlock-captured DNA (Hues6-BJ-Hybrid1) using the two methods, and sequenced. The methylation levels are highly consistent between the two libraries (Pearson R = 0.994 for the 22,937 CpG sites covered with>50 sequencing reads in both libraries; Supplementary Fig. 3a online). We also characterized the average sequencing coverage across the capturing targets. Both methods exhibit bias toward the center of the sequences, which was commonly observed in fragmentation of DNA with defined sizes. However, our enzymatic fragmentation protocol produced more even coverage across the captured DNA fragments (Supplementary Fig. 3b).
Single-molecule analysis of DNA methylation in iPS cells
To demonstrate the utility of targeted bisulfite sequencing, we characterized the changes of chromosome-wide methylation status during reprogramming of human fibroblasts to pluripotent cells. We performed the methylation assay on three sets of fibroblasts and iPS cells from three laboratories: IMR/IMR90-iPS17 reprogrammed with four factors (Oct4, Sox2, Nanog and Lin28); hFib2/hFib2-iPS18 reprogrammed with a different set of factors (Oct4, Sox2, Klf4 and Myc); BJ/BJ-iPS11/BJ-iPS12 (ref. 19) reprogrammed with the five factors (Oct4, Sox2, Nanog, Klf4 and Myc) controlled by an inducible promoter. We also characterized a line of hybrid stem cells (BJHues6-Hybrid1)20, which were reprogrammed by fusing the human fibroblasts (BJ) with hES cells (Hues6), as well as three hES cell lines (Hues12, Hues42, Hues63). We performed bisulfite conversion, padlock capture and construction of shotgun sequencing libraries on each DNA sample. We sequenced each library in one lane in the flow cell of Illumina Genome Analyzer, and obtained 2–3 million reads that were mapped to the targeted regions (Table 1). The bisulfite conversion rates were >98.5%. To avoid stochastic sampling drift, we removed CpG sites that were covered by <10 reads from the following analyses.
Table 1.
Targeted bisulfite sequencing of three human fibroblast lines, four iPS cell lines, one hybrid stem cell line and three hES cell lines.
| Cell line | Raw reads | Mapped reads | CpG sites covered ≥ 1 read | CpG sites covered ≥ 10 reads | Mean coverage | Non-CpG cytosines (%) |
|---|---|---|---|---|---|---|
| BJ | 10,853,272 | 2,057,753 | 98,307 | 67,470 | 71 | 0.010 |
| BJ_iPS11 | 12,276,017 | 2,240,958 | 100,336 | 69,454 | 77 | 0.012 |
| BJ_iPS12 | 10,539,126 | 2,179,019 | 100,708 | 70,741 | 74 | 0.011 |
| hFib2 | 14,408,007 | 2,540,405 | 88,432 | 60,007 | 96 | 0.011 |
| hFib2_iPS | 14,805,000 | 2,173,844 | 88,641 | 58,531 | 80 | 0.014 |
| IMR90 | 14,795,564 | 2,896,881 | 88,369 | 59,950 | 113 | 0.011 |
| IMR90.2 | 9,892,238 | 2,051,461 | 85,741 | 55, 966 | 72 | 0.011 |
| IMR90_iPS | 10,985,469 | 2,473,997 | 99,190 | 66,424 | 58 | 0.013 |
| Hues6-BJ-Hybrid1 | 9,693,455 | 2,913,639 | 101,020 | 68,849 | 65 | 0.014 |
| Hues12 | 11,360,235 | 3,760,194 | 98,061 | 72,463 | 118 | 0.015 |
| Hues42 | 11,185,368 | 3,671,564 | 92,544 | 67,926 | 125 | 0.012 |
| Hues63 | 14,315,301 | 2,472,893 | 101,957 | 73,196 | 97 | 0.013 |
DNA methylation in 2,020 CpG islands
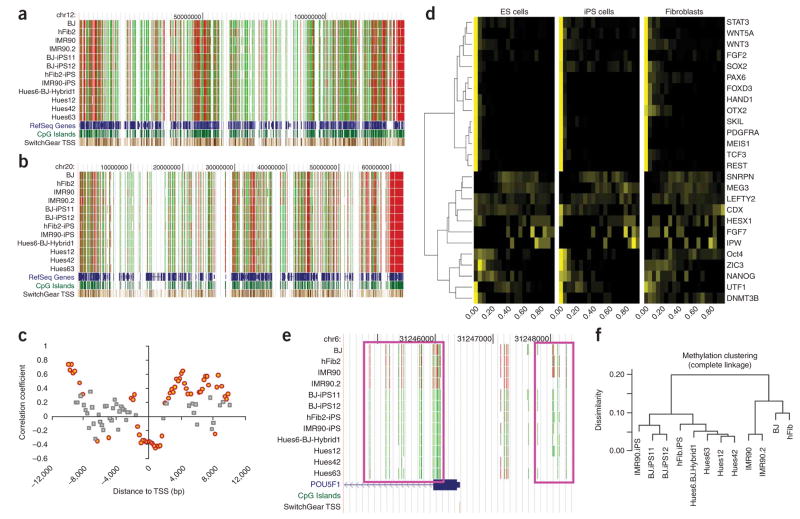
The global methylation patterns in all 12 samples (11 cell lines plus a biological replicate on IMR90 fibroblasts) were visualized using the UCSC Genome Browser. The chromosome-wide patterns of CpG island methylation were highly similar among all the cell lines (Fig. 3a,b). Globally, the methylation level of CpG dinucleotides followed similar bimodal distribution: 67% were weakly methylated (<20% methylation), 22% were highly methylated (>80% methylation) and the remaining 11% have intermediate levels of methylation (Supplementary Fig. 4 online). To distinguish CpG islands with different methylation patterns, we generated a histogram (bin size = 0.05) for the distribution of methylation on all CpG sites within a CpG island. Treating such histograms as 20-component vectors, we performed hierarchical clustering to partition the CpG islands, and divided all CpG islands into three clusters based on the similarity of distribution between pluripotent and fibroblast lines (Supplementary Fig. 5 online): in cluster 1, the CpG islands (1,451; 77.3%) have similar distributions in the two cell types (R > 0.5); in cluster 2, CpG islands (252; 13.4%) have less similar distributions (0.5 ≥ R > 0); in cluster 3, CpG islands (173; 9.2%) are anti-correlated (R ≤ 0). Therefore, only a small fraction of CpG islands show cell-type-specific methylation.
Figure 3.
Patterns of CpG methylation in fibroblasts and pluripotent cells. (a,b) Patterns of CpG island methylation on chromosomes 12 and 20 in twelve samples. Red indicates highly methylated CpG, and green indicates weakly methylated CpG. (c) Correlation between DNA methylation and gene expression. Each circle or square represents a 500-bp window. The windows in which average methylation levels are significantly correlated (P < 0.01) with gene expression are highlighted as orange circles. Gray squares indicate that the correlation is not significant. (d) Clustering of 26 selected genes based on the distribution of CpG methylation in the 4-kb TSS flanking regions. The 20 columns per cell line represent the fraction of probes within the TSS region where each probe exhibits a methylation frequency such that column header value ≤ methylation frequency < column header value + 0.05. For example, unmethylated TSS regions will have the highest probe fraction in the lowest value-labeled (left) column. Roughly half of the genes (14) were close to completely unmethylated in all cell types. Five genes (OCT4, ZIC3, NANOG, UTF1, DNMT3B) that show cell-type specific changes in methylation were grouped as a cluster. (e) Methylation pattern in the TSS flanking regions of OCT4. Two differentially methylated regions were marked by purple rectangles. (f) Hierarchical clustering of pluripotent cells and fibroblasts. The dissimilarity matrix was calculated based on Pearson correlation coefficients.
Because CpG islands are not defined in a functional manner, we divided the CpG islands into three categories. The first comprises CpG islands in the regions from 2 kb upstream to 500 bp downstream of TSS. These ‘upstream regions’ often include promoter regions. The second class (‘gene body CpG islands’) comprises CpG islands in the regions from 500 bp downstream of TSS to the ends of the last exons. The final category comprises CpG islands outside of gene body and promoter regions. CpG islands in each category were further divided into three groups according to CpG density. Consistent with previous findings, most (91.8%) CpG islands in promoter regions were weakly methylated (<20% methylation), 3.4% were highly methylated (>80% methylation) and the remaining 4.8% showed an intermediate level of methylation (20–80% methylation). The distributions are quite similar among the three groups with different CpG densities (Supplementary Fig. 6a online). In contrast, only 45.2% of CpG islands in the gene body were weakly methylated, whereas roughly one-third of them (37.7%) were highly methylated. Methylated CpGs tend to locate in islands with low CpG density (Supplementary Fig. 6b). In regions outside of gene body and promoter regions, we found more weakly methylated CpG islands (58.9%) than highly methylated CpG islands (26.6%) (Supplementary Fig. 6c). Similarly, CpG islands with low CpG density were more methylated. There were 80 genes in our data set that contained both promoter-region CpG islands and gene-body CpG islands. Sixty-two of these genes were weakly methylated in promoter regions. Among these, 48.4% were highly methylated in the gene body and 29.0% displayed weak gene-body methylation.
CpG island methylation and gene expression
As methylation at CpG dinucleotides is considered an important transcriptional regulatory mechanism, we sought to characterize the effects of methylation at different CpG sites on gene expression. We used the Illumina HumanRef BeadArray to profile gene expression of nine cell lines (hFib, hFib-iPS, BJ, BJ-iPS12, IMR90, IMR90-iPS, Hues12, Hues42, Hues63) and then investigated the distribution of functional methylation sites relative to gene structures. For both chromosomes 12 and 20, we computed the Spearman’s rank correlation between the average methylation level of all CpG sites in 500-bp windows and the expression level of its corresponding genes. The 500-bp windows were moved relative to the TSS of the corresponding genes with a step size of 200 bp from 10 kb upstream to 10 kb downstream, excluding genes with multiple TSS. Windows that overlap with adjacent genes in the upstream or downstream regions were also removed from the analysis. To exclude artifacts resulting from uneven sampling of CpG sites only from CpG islands, we permutated the gene expression values to establish the empirical distribution of background correlation coefficients. These were then used to estimate the significance of correlation between DNA methylation and gene expression. We found that methylation in a 2.4-kb region (TSS upstream 1.0 kb to downstream 1.4 kb, P < 1−10) showed the strongest negative correlation with gene expression (Fig. 3c). The methylation level and gene expression became positively correlated in the 2- to 10-kb region downstream of TSS, suggesting that active genes tend to be unmethylated around the TSS, but methylated in the gene body. This is consistent with the observation of gene body methylation in actively transcribed genes21–23. We also observed significant correlation (P < 0.01) between gene expression and methylation in other regions, which could be related to the chromatin structure of active genes or anti-sense gene expressions (Fig. 3c).
Differentially methylated regions associated with reprogramming
We next sought to identify regions that exhibit methylation differences between fibroblasts and pluripotent cells. The weakly or negatively correlated CpG island clusters identified above were based on the overall distribution of CpG methylation in CpG islands. The single-nucleotide resolution data obtained in this study allowed us to search for regions with specific changes independent of the definition of CpG island. We started by examining the methylation levels in the TSS flanking regions associated with the 26 selected genes. Most of these genes’ overall methylation frequencies were similar between pluripotent cells and fibroblasts but there were a few genes (e.g., OCT4 (also known as POU5F1), DNMT3B, and NANOG) for which methylation frequencies were noticeably different between the two cell types (Fig. 3d). We found that these genes contained differentially methylated regions (DMRs) that were separated by stretches of CpG sites with little methylation difference (Fig. 3e). To search for DMRs with similar patterns in all the regions included in this study, we performed K-mean clustering (k = 2) on the 12 data sets based on the methylation level of all CpG sites in 400-bp windows sliding along the chromosomes with the step size of 100 bp. Excluding windows that contain <3 CpG sites or >30% missing values, we examined a total of 18,434 windows. We reasoned that, for a true DMR, the eight data sets from pluripotent lines and four sets from fibroblast lines should be distinguished based on the methylation of all CpG sites in the region. Therefore, windows in which partitioning is consistent with the phenotypic difference were considered as candidate DMRs. We also required that the difference of median methylation should be at least 0.1 between stem cells and fibroblasts. A total of 1,273 partially overlapping DMRs were found.
To evaluate the robustness of this procedure, we randomly shuffled the 12 data sets, and applied the same algorithm on the permutated data where at least three samples were placed into incorrect categories. We found an average of 6.8 DMRs in 100 permutations, suggesting a false-positive rate of 0.53%. After combining multiple overlapping DMRs, we found 288 discontinuous DMRs (44 from ENCODE promoters, 16 from selected genes, 228 from CpG islands) that were associated with reprogramming. To identify associated genes, we mapped these DMRs to regions from 10 kb upstream to 10 kb downstream of the TSS of RefSeq genes. We mapped 132 DMRs to 155 genes, including 10 from the 26 selected genes, 49 in the ENCODE regions, and 96 on chromosomes 12 and 20 (Supplementary Table 4 online). We then looked for the enrichment of genes in certain functional categories. Because some targets were chosen based on known functions, we focused on a subset of 125 genes on chromosomes 12 and 20, and five ENCODE regions (ENm004, ENm005, ENm007, ENr123, ENr333) to avoid sampling bias. We performed gene ontology analysis using DAVID tools24. At a false-discovery rate of < 0.05, we found six enriched functional categories related to ion transport for the 94 genes associated with an elevated level of methylation in pluripotent cells. The other 31 genes associated with reduced methylation in pluripotent cells were enriched for eight functional categories related to transcription, metabolic and developmental regulations (Supplementary Table 5 online).
hES cells have unique gene expression and methylation signatures3,25. To test whether we can distinguish pluripotent cells based on the methylation profiles established in this study, we performed hierarchical clustering on the methylation level on all CpG sites with at least 50× sequencing coverage. Fibroblasts and iPS/ES cell lines were clearly grouped into two different clades (Fig. 3f). In addition, the iPS and hES cell lines had subtle yet noticeable differences, with the Hues6-BJ hybrid line more closely resembling hES cells. Interestingly, the similarity between hES cell lines and fibroblasts were slightly higher than that between iPS cell lines and fibroblasts (see Supplementary Table 6 online for the similarity matrix). Globally, both iPS and hES cell lines were more methylated than fibroblasts; and iPS cell lines were more methylated than hES cell lines (Supplementary Fig. 7 online). Locally, key pluripotent genes were less methylated in pluripotent cells (Fig. 3d,e). These results suggest that iPS cells were reprogrammed into an epigenetic state that made them less similar to fibroblasts than hES cells are. Possibly, the persistent overexpression of transcription factors from integrated viral transgenes accounts for the more substantial shift in the overall epigenetic state (Supplementary Fig. 8 online). Alternatively, such a difference could be due to the minor variations in iPS derivation and hES culture among different laboratories. Further characterization of a larger panel of iPS/hES cell lines with similar passage numbers under standard culture conditions is required to reach a generalized conclusion on whether the epigenetic states of iPS cells differ systematically from those of hES cells.
DISCUSSION
We have demonstrated that padlock probes can specifically extract a large number of genomic regions in single-tube reactions for bisulfite sequencing analysis. The degree of multiplexity is at least four orders of magnitude greater than that possible with conventional PCR-based bisulfite sequencing. The high capturing specificity is contributed by the cooperative annealing of the two capturing arms on the target molecules in proper orientation and distance, the selectivity of DNA polymerase and ligase, and the removal of linear DNA with exonuclease. Although padlock probes have been successfully applied to exon capturing15 and SNP genotyping26, we demonstrate their use with bisulfite-converted DNA with highly skewed nucleotide composition and low sequence complexity. Recent studies showed that most methylation changes are restricted to a very small fraction of the genome outside of CpG islands11,23,27. Padlock capture is more efficient than full-genome bisulfite sequencing12,13 for quantifying DNA methylation, as it allows for focused sequencing on the most informative genomic regions. It also provides a much greater flexibility than reduced representation bisulfite sequencing11 in the selection of genomic targets, because the latter method is limited to genomic regions closely adjacent to the recognition sites of restriction enzymes, such as MspI.
One current limitation associated with padlock probes is the uneven capturing efficiencies among different probes. We observed a >10,000-fold difference between the most efficient and least efficient probes. Although we have identified some DNA sequence features (that is, melting temperature, gap length, GC compositions) that are statistically correlated with the efficiency, they explained only 18% of the total variation. Other major factors still remain elusive. Further theoretical and experimental analyses are required to achieve a better understanding of padlock formation. Using a combination of two normalization strategies, we have dramatically reduced the capturing bias, which is only slightly higher than hybrid selection of genomic fragments28. However, we think there is room for further improvement, especially in better understanding the annealing thermodynamics of padlock probes, characterizing potential sequence-dependent bias of DNA polymerase or ligase, and post-capture normalization of biased libraries.
In this study we showed that 30,000 probes can capture specific sequences in a single tube. Previously, we have captured exonic targets with 55,000 probes, and SNPs with 132,000 probes (K.Z. et al., unpublished data). As we have not encountered an upper limit, it seems possible that all CpG islands in the human genome (~20 Mb in total size) or other genomic targets of similar size can be captured and sequenced in single-tube reactions. Achieving this goal would probably require further reducing representation bias. Performing multiplexed capture not only has an obvious advantage in reducing reagent cost and labor, it also reduces the amount of starting materials. In our experiments, the amount of input DNA is 200 ng per capturing reaction, which is modest compared with the amount of DNA needed for most genome-scale assays. We also have preliminary evidence that our current probe set works with as little as 50 ng of input DNA, which is equivalent to ~10,000 cells (data not shown). The observation that certain probes are 10,000× more efficient than others suggests that at least some of the targets can potentially be captured from a very small number of cells. Fourteen DMRs identified in this study were covered by the 200 most efficient probes. This would open the door for large-scale methylation analysis on clinical specimens or other samples with a very limited amount of starting materials.
CpG islands are defined by DNA sequence features29 that do not correlate perfectly with biological functions. Recent studies11,30 showed that CpG-rich promoters are associated with both ‘house-keeping’ genes and genes expressed during embryonic development. It is the CpG-poor promoters that are generally associated with highly tissue-specific genes. Consistent with these reports, we did not observe widespread changes of DNA methylation during reprogramming in the CpG islands on chromosomes 12 and 20. Surprisingly, we still found 288 DMRs that distinguish pluripotent cells and fibroblasts, including 228 from CpG islands. Extrapolation from these numbers suggests that, of the 28,226 CpG islands in the entire human genome, there would be ~3,186 DMRs that distinguish fibroblasts from pluripotent. In contrast, we identified 44 DMRs from the 237 ENCODE promoters included in this study. The significant (P = 0.005) enrichment of DMRs in ENCODE promoters compared with CpG islands is unsurprising because all ENCODE promoters have been experimentally validated. Focusing exclusively on CpG islands might not be the most efficient strategy to expand our targeted bisulfite sequencing efforts from two chromosomes to the full genome. It seems that whole-genome bisulfite sequencing of a carefully selected set of cell lines would be especially useful for generating a list of genomic regions, which could then be prioritized for targeted analysis in a larger number of samples.
METHODS
Padlock probe design
We developed a probe design algorithm to search for an optimal set of padlock probes covering an arbitrary set of nonrepetitive genomic targets. This algorithm weights candidate probes based on several sequence features that were previously not considered in eMIP probe design, including the melting temperature, size and word statistics (distribution of 12-mers in the bisulfite-converted genome) of the capturing arms, and gap sizes. When the capturing arms contain one or more CpG dinucleotides, we used multiple probes to iterate all possible methylation state combinations of the CpGs contained within the arms. Chromosome positions of CpG islands were retrieved from the UCSC genome browser (http://genome.ucsc.edu/) based on the hg18 annotation. All the probe sequences, annotations and their relative efficiencies are listed in Supplementary Table 7 online.
Padlock probe production
Libraries of long oligonucleotides (~150 nt) were synthesized by ink-jet printing on programmable microarray, and released (Agilent Technologies). The estimated total yield is 10 fmol per library. PCR amplification was performed in 32–96 reactions (100 μl each) with 0.1 nM template oligonucleotides, 200 μM dNTPs, 400 nM Ap1V4IU primer, 400 nM Ap2V4 primer, 0.8× SybrGreen I, 36 units JumpStart Taq polymerase in 1× JumpStart buffer (Sigma), at 94 °C for 2 min, 22 cycles of 94 °C for 30 s, 55 °C for 2 min, 72 °C for 45 s and, finally, 72 °C for 5 min. The amplicons were purified by either column purification (Zymo DNA Concentrator- 100 columns) or ethanol precipitation.
Approximately 40–60 μg of the purified PCR amplicons were digested with 40 units Lambda Exonuclease (5 U/μl; New England Biolabs (NEB)) in 1× lambda exonuclease buffer (NEB) at 37 °C for 2 h, followed by denaturing at 90 °C for 5 min, and purified with six Qiagen Qiaquick PCR purification columns. The resulting single-stranded DNA was subsequently digested with 6 units USER enzyme (1 U/μl; NEB) in 1× DpnII buffer (NEB) at 37 °C for 4 h. We added 10 μl of 100 μM DpnII_V4 guide oligo into the reaction and denatured the mixture at 95 °C for 5 min in a thermocycler, followed by a gradual decrease of temperature (0.1 °C/s) to 60 °C and a 20-min incubation at 60 °C. The mixture was digested with 100 U DpnII (50 U/μl) at 37 °C for 2 h. The single-stranded 102-nt probes were finally purified from the digestion with 6% denaturing PAGE (6% TB-Urea 2D gel; Invitrogen).
Multiplex capture on bisulfite-converted DNA
Genomic DNA was extracted from frozen pellets of fibroblast, iPS or hES cells using Qiagen DNeasy columns, and bisulfite converted with the Zymo DNA Methylation Gold Kit (Zymo Research). Padlock probes (60 nM) and 200 ng of bisulfite-converted genomic DNAwere mixed in 10 μl 1× Ampligase Buffer (Epicentre), denatured at 95 °C for 10 min, then hybridized at 55 °C for 18 h, after which 1 μl gap-filling mix (200 μM dNTPs, 2 U AmpliTaq Stoffel Fragment (ABI) and 0.5 units Ampligase (Epicentre) in 1× Ampligase buffer) was added to the reaction. For circularization, the reactions were incubated at 55 °C for 4 h, followed by five cycles of 95 °C for 1 min, and 55 °C for 4 h. To digest linear DNA after circularization, 2 μl exonuclease mix (containing 10 U/μl exonuclease I and 100 U/μl exonuclease III; USB) was added to the reaction, and the reactions were incubated at 37 °C for 2 h and then inactivated at 95 °C for 5 min.
Capture circles amplification
10-μl circularization products were amplified by PCR in 100 μl reactions with 200 nM AmpF6.2-SoL primer, 200 nM AmpR6.2-SoL primer, 0.4× SybrGreen I and 50 μl iProof High-Fidelity Master Mix (Bio-Rad) at 98 °C for 30 s, eight cycles of 98 °C for 10 s, 58 °C for 20 s, 72 °C for 20 s, 14 cycles of 98 °C for 10 s, 72 °C for 20 s and 72 °C for 3 min. The amplicons of the expected size range (344–394 bp) were purified with 6% PAGE (6% TBE gel; Invitrogen).
Shotgun sequencing library construction
Purified PCR products with the four probe sets on the same template DNA were pooled in equal molar ratio, and reamplified in 4× 100 μl reactions with 4-μl template (10~15 ng/μl), 200 μM dNTPs, 20 μM dUTP, 200 nM AmpF6.3 primer, 200 nM AmpR6.3 primer, 0.4× SybrGreen I and 200 μl 2× Taq Master Mix (NEB) at 94 °C for 3 min, 8 cycles of 94 °C for 45 s, 55 °C for 45 s, 72 °C for 45 s and 72 °C for 3 min. PCR amplicons were purified with Qiaquick columns, and digested with MmeI: ~3.6 nmole purified PCR amplicons, 16 units of MmeI (2 U/μl; NEB), 100 μM SAM in 1× NEB Buffer 4 at 37 °C for 1 h. The digestions were again column purified, and digested with 3 U USER enzyme (1 U/μl) at 37 °C for 2 h, then with 10 units S1 nuclease (10 U/μl; Invitrogen) in 1× S1 nuclease buffer at 37 °C for 10 min. The fragmented DNA was column purified, and end repaired at 25 °C for 45 min in 25-μl reactions containing 2.5 μl 10× buffer, 2.5 μl dNTP mix (2.5 mMeach), 2.5 μl ATP (10 mM), 1 μl end-repair enzyme mix (Epicentre), and 15 μl DNA. Approximately 100–500 ng of the end-repaired DNA was ligated with 60 μM Solexa sequencing adaptors in 30 μl of 1× QuickLigase Buffer (NEB) with 1 μl QuickLigase for 15 min at 25 °C. Ligation products of 150~175 bp in size were size selected with 6% PAGE, and amplified by PCR in 100 μl reactions with 15 μl template, 200 nM Solexa PCR primers, 0.8× SybrGreen I and 50 μl iProof High-Fidelity Master Mix (Bio-Rad) at 98 °C for 30 s, 12 cycles of 98 °C for 10 s, 65 °C for 20 s, 72 °C for 20 and 72 °C for 3 min. The PCR amplicons were purified with Qiaquick PCR purification columns, and sequenced on Illumina Genome Analyzer. All primer sequences were listed in Supplementary Table 8 online.
Read mapping and data analysis
Mapping of bisulfite sequencing reads was performed with SOAP31 driven by a customized Perl script. An unbiased mapping strategy in which the mapping success rate is independent of the methylation status was developed. Sequences of the captured targets were extracted from the repeat-masked human genome (hg18), and both strands were ‘bisulfite converted’ in silico assuming no methylation on all CpG dinucleotides. The raw sequencing reads were also converted to ‘unmethylated reads’. To do this, the C/T and A/G ratios for each read were first compared to determine whether the reads corresponding to the bisulfite-converted strand or the reverse-complementary strand. In the latter case, the raw sequence was reverse complemented. All Cs were replaced by Ts in the resulting sequences. The unmethylated reads were then aligned to the unmethylated template sequences using SOAP. The false mapping rate that was due to the use of captured targets instead of the full human genome sequence was 0.21%. Finally, based on the mapping position, the methylation status of each CpG site was retrieved from the unconverted raw reads. Cluster analyses and statistical analyses were performed with R, Cluster3 Perl module, and in-house Perl scripts. The UCSC Genome Browser (http://genome.ucsc.edu/) and Multiexperiment Viewer (http://www.tm4.org/mev.html) was used for data visualization.
The algorithm for padlock probe design, as well as the Perl scripts for read mapping and data analysis are freely available in Supplementary Data online or at http://genome-tech.ucsd.edu/public/NBT-RA20324.
Acknowledgments
We thank George Church, Billy Jin Li, Jay Shendure for inputs related to padlock probes; Huidong Shi, Billy Jin Li and Madeleine Ball for suggestions on methylation analysis; Ruiqiang Li for suggestions on read mapping; James Sprague for assistance on gene expression profiling, Colleen Ludka for assistance on Illumina sequencing. This work was supported by the UCSD new faculty startup fund, and partially by NIH/NIDA R01-DA025779 (to K.Z.). J.D. was sponsored by a CIRM post-doctoral fellowship.
Footnotes
Accession numbers. All sequence reads and methylation data have been deposited at GEO, with accession number GSE15007.
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
K.Z. and Y.G. oversaw the project. J.D. and K.Z. designed and performed experiments related to padlock probe preparation, target capture, sequencing library construction and various validation assays. B.X. and Y.G. performed Illumina sequencing. E.M.L. provided oligonucleotide libraries. J.A.-B., D.E., N.M., I.-H.P., J.Y. G.Q.D., K.E. K.H. J.T. provided DNA/RNA from stem cells and fibroblasts. J.D., R.S., A.G. W.W., Y.G., and K.Z. performed data analysis. J.D. and K.Z wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/
References
- 1.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 2.Bibikova M, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibikova M, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Khulan B, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakyan VK, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor KH, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 10.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 15.Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 16.Quail MA, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 19.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 21.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 22.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 23.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009 29 March; doi: 10.1038/nbt.1533. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis G, Jr, et al. DAVID: database for annotation, visualization, and integrated discovery . Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 25.Müller FJ, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardenbol P, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 30.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]