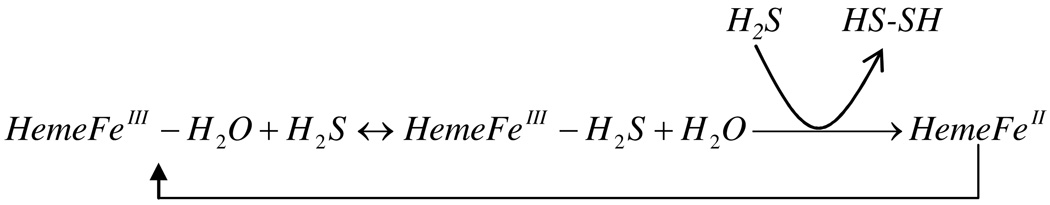Abstract
Hemoglobin I (HbI) from the clam Lucina pectinata is an intriguing hemeprotein that binds and transports H2S to sulfide-oxidizing chemoautotrophic bacteria to maintain a symbiotic relationship and to protect the mollusk from H2S toxicity. Single point mutations at E7, B10 and E11 positions were introduced in the HbI heme pocket to define the reactivity of sulfide with hemeproteins. The functional and structural properties of mutant and wild type recombinant proteins were first evaluated using the well-known ferrous CO and O2 derivatives. The effects of these mutations on the ferric environment were then studied in the metaquo and hydrogen sulfide derivatives. The results obtained with the ferrous HbI mutants show that all the E7 substitutions and the PheB10Tyr mutation influence directly CO and O2 binding and stability while the B10 and E11 substitutions induce distal structural rearrangements that affect ligand entry and escape indirectly. For the metaquo-GlnE7His, PheB10Val, PheB10Leu and the E11 variants, two individual distal structures are suggested, one of which is associated with H-bonding interactions between the E7 residues and the bound water. Similar H-bonding interactions are invoked for these HbI-H2S mutant derivatives and the rHbI, altering in turn sulfide reactivity within these protein samples. This is evident in the resonance Raman spectra of these HbI-H2S complexes, which show reduction of heme iron as judged by the appearance of the ν4 oxidation state marker at 1356 cm−1, indicative of heme-FeII species. This reduction process depends strongly on distal mutations showing faster reduction for those HbI mutants exhibiting strongest H-bonding interactions. Overall, the results presented here show that: a. H2S association is regulated by steric constraints; b. H2S release is controlled by two competing reactions involving simple sulfide dissociation and heme reduction; c. at high H2S concentrations, reduction of the ferric center dominates; d. reduction of the heme is also enhanced in those HbI mutants having polar distal environments.
Keywords: HbI, hemoglobin I; Mb, myoglobin; H2S, hydrogen sulfide; CO, carbon monoxide; O2, oxygen; RR resonance Raman; MD, molecular dynamics
Hydrogen sulfide (H2S) is a well known poisonous gas whose cytotoxic effects have been studied for more than 300 years (1). Interestingly, it has been found recently that H2S is produced in mammalian tissues and that, similar to nitric oxide (NO) and carbon monoxide (CO), it may function as a neuromodulator, neuroprotector and/or smooth muscle relaxant/constrictor (2–5). Endogenous H2S is produced at high concentrations in the brain (100 µM) by cystathionine beta-synthase enzyme (CBS, EC 4.2.1.22), where it acts as neuromodulator and neuroprotector, and in the peripheral tissues by cystathionine alpha-lyase enzyme (CSE, EC4.4.1.1) where it function as a relaxant/constrictor. As a neuromodulator, H2S enhances the activity of the N-methyl-Daspartate (NMDA) receptor and activates Ca2+ channels regulating synaptic transmission in neurons. As a neuroprotector, H2S enhances the activity of the enzyme responsible to produce glutathione, activates K+ATP and Cl− channels and acts against oxidative stress. The muscle relaxant activity of H2S is believed to be dictated by mediation of K+ATP channels in the ileum, portal vein and thoracic aorta tissues while its vasoconstrictor action is believed to be in part through inhibition of eNOS activity (2–5).
As for NO and CO, there is not a single H2S receptor responsible for all the biological activities found until now. In fact, it has been suggested that H2S can bind to hemeproteins inducing different responses that in turn modulate its cytotoxic and cytoprotective activities (6). For instance, H2S reacts with mitochondrial cytochrome c oxidase in a concentration-dependent manner to either completely inhibit the enzyme, producing cytotoxic effects or to reversibly modulate its activity, resulting in induction of a suspended animation like-state that can stimulate cytoprotective effects (7). In this regard, it has been observed that interaction of H2S with the resting enzyme at low sulfide levels (at a 1:1 stoichiometry), instead of stimulating irreversible inhibition, reduces the heme a3 cytochrome center in a reaction that results in the formation of a ferrous intermediate, O2 uptake and conversion to a low-spin ferric derivative (8, 9). This low-spin form of the heme a3 center is unable to bind oxygen, reducing its uptake and thereby down-regulating cellular respiration. It is at this level of regulation that H2S is suggested to exert protective effects in mammals (6, 7). At a higher stoichiometry however, (3 moles of H2S per mol of enzyme), the toxicity of H2S becomes apparent by the reduction of the other metal centers with its concomitant ligation to the heme a3 group, producing the final, non-functional enzyme (7–10).
On the other hand, reactions of H2S with myoglobin and hemoglobin, in the presence of O2 result in covalent modification of one of the pyrrole rings of the heme, generating the so-called sulfmyoglobin and sulfhemoglobin derivatives (11). These compounds are proposed to protect the cell from H2S toxicity (6). Others have argued against a protective role of these sulfheme compounds, since they have been found in a number of human diseases involving poisoning with sulfide containing drugs and not with H2S itself (12, 13). More recently, a study concerning the interaction of human neuroglobin with H2S suggested that this ligand binds tightly to the protein heme active site and as a consequence, it can have cytoprotective effects in the brain when levels of H2S increase (14).
Evidently, H2S exerts different responses on hemeproteins and consequently, the cell. Nevertheless, the factors controlling heme-H2S interactions required to produce all these biological responses are still unknown. Understanding these factors is therefore crucial before exploiting the potential of H2S in therapeutic interventions. Intriguingly, interactions of H2S with hemeproteins have been recognized and studied for many years in marine invertebrate organisms that live in sulfide-rich environments (15). These organisms have evolved strategies to avoid sulfide toxicity, including the binding and oxidation of H2S by hemoglobins and symbiotic bacteria, respectively. In this context, the giant tubeworm Riftia pachyptila, which lives in the deep sea hydrothermal vents, supplies O2 and H2S to the endosymbionts by binding both ligands simultaneously at two different sites on their extracellular hemoglobins (13, 15). The bacteria living inside the tubeworm oxidize H2S in the presence of O2 and utilize this energy to synthesize organic nutrients for the invertebrate.
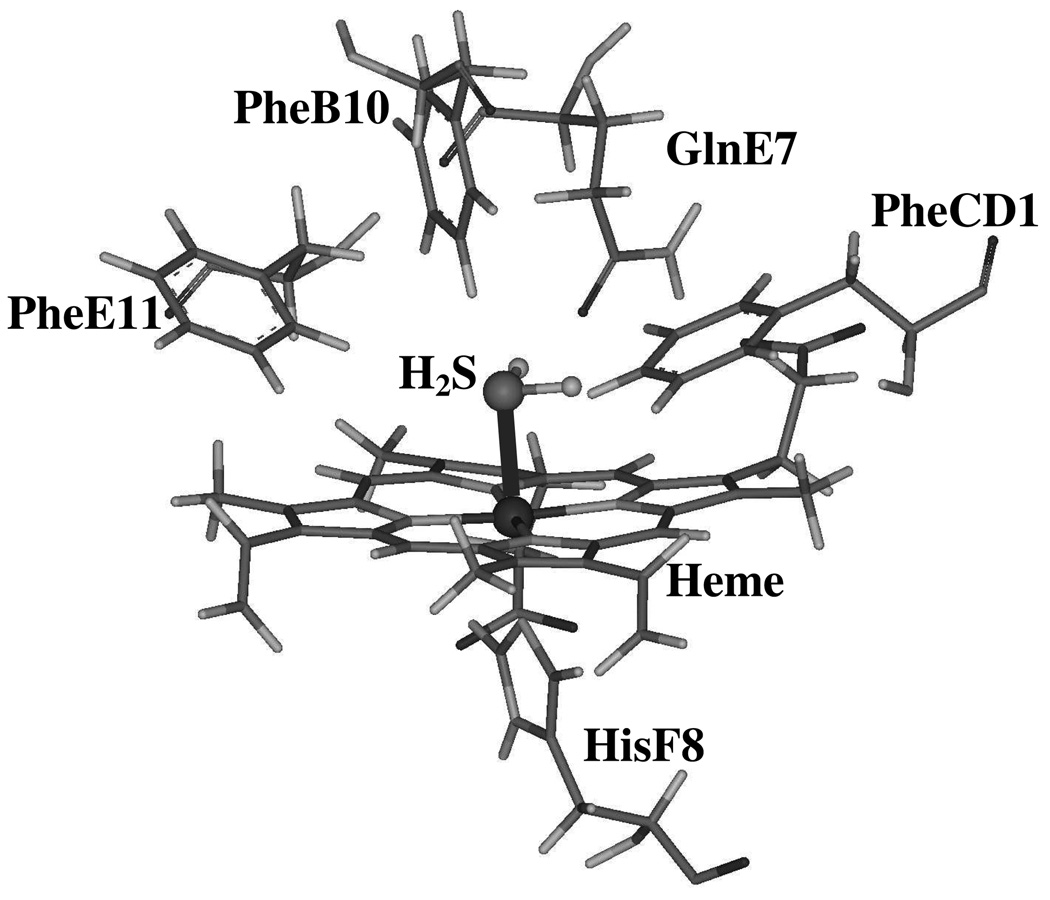
The clam Lucina pectinata is another interesting invertebrate that lives in sulfide-rich mangroves and is also characterized by the presence of symbiotic sulfide-oxidizing bacteria that need to be supplied with both H2S and O2 (16, 17). The protein responsible for delivering H2S to the bacteria is a hemeprotein called hemoglobin I (HbI). HbI is one of the few known H2S carriers in organisms that have been implicated in physiologically binding this molecule in the ferric heme iron center so as to maintain the symbiotic relationship with the bacteria as well as to protect the clam from H2S cytotoxicity. The affinity of hydrogen sulfide for ferric HbI is exceptionally high and is believed to be achieved through fast association (kon = 2.3 × 105 M−1s−1) and very slow dissociation processes (koff = 0.22 × 10−3 s−1). Structural studies of the HbI active site have shown that HbI has a glutamine (Gln) residue at the distal E7 position instead of the typical histidine (His) found in mammalian myoglobins and hemoglobins (18). In addition to GlnE7, HbI has phenylalanine residues (Phe) at the B10 and E11distal positions generating what is known as the “Phe-cage”. Figure 1 shows the peculiar amino acid composition of the HbI distal ligand binding site, which has been suggested to be responsible for the high H2S affinity. In contrast, the other two hemoglobins found in Lucina petinata (HbII and HbIII) have a tyrosine at the B10 position, which prevents H2S binding in these two hemeproteins (19–21). Indeed, spectroscopic studies of HbI with various ligands (22, 23) have demonstrated a flexibility of the GlnE7 residue that controls ligand access into the HbI heme pocket, while hydrogen bonding and multipole interactions of this residue and the Phe-cage have been suggested to be involved in H2S stability and release. Despite all these significant findings, a comprehensive understanding of H2S binding, stability, and release to and from HbI has not been established yet. Thus, a study aimed at defining the role of the heme pocket amino acids in the kinetics and dynamics of H2S binding and release to and from the heme active site of HbI and the structural aspects associated with these processes, is necessary to unravel the relationship between structure, function, and dynamics of the HbI-H2S moiety. Furthermore, these results can provide an overall picture of the role of the heme distal structural environment in hemeprotein-H2S interaction.
Figure 1.
Representation of key residues in the heme pocket of HbI from L. pectinata, taken from reference 35.
The functional aspects of many hemeproteins have been systematically examined by site-directed mutagenesis and kinetic analyses with various gaseous ligands (24, 25). In addition, the structural implications of such kinetic studies are in many instances, correlated with molecular dynamics simulations (MD) and resonance Raman (RR) analyses (26, 27).
Herein we have extended our investigations on H2S binding, stability, and release to and from hemeproteins by evaluating the effect of heme pocket mutations on the HbI-H2S interaction using kinetic, MD simulations and RR approaches. The results presented here show that: a. H2S association is dictated by steric constraints; b. H2S release is controlled by two competing reactions involving simple sulfide dissociation and heme reduction; c. at high H2S concentrations, reduction of the ferric center dominates; d. reduction of the heme is also enhanced in those HbI mutants having polar distal environments. On this basis, several factors controlling H2S reactivity with hemeproteins were identified as followed: (i.) accessibility of H2S into the heme cavity, (ii.) H2S concentration, which stimulates electron transfer (iii.) the polarity of the distal environment surrounding the bound H2S and (iv.) the stereo orientation of the distal side residues.
MATERIAL AND METHODS
Sample Preparation
Recombinant (rHbI) and mutant HbI constructs were prepared, expressed and purified as described previously (28). Briefly, HbI mutants were obtained by introducing single amino acid substitutions into the HbI coding region that was obtained by RT-PCR amplification and cloned into the pET28(a+) vector using the Quick Change Mutagenesis kit (Stratagene, La Jolla, CA, USA). The HbI mutants and rHbI were expressed in E. coli Bli5 cells transformed with the constructs described above. Expression of the samples after induction with 1mM IPTG yielded dark red cell pellets that were lysed and centrifuged to separate the soluble protein from the insoluble cell fractions as described (28). The soluble protein fractions were used for purification of the recombinant HbI proteins in Co+2 affinity columns (Talon, Invitrogen, Carlsbad, CA). Further purification of the samples was achieved by FPLC in a Hi Load 26/60 Superdex 200 gel filtration column.
The carbon monoxide and oxygen derivatives were prepared by adding a slight excess of sodium dithionite (∼50x) to the protein samples under anaerobic conditions followed by exposure to a CO or O2 atmosphere. The identity of the CO and O2 complexes was verified by their characteristic UV-vis spectra with distinct Soret bands at 420 and 416 nm, respectively (16). Carbon monoxide isotopic derivatives were formed in the same manner using 13C18O (Isotec). To form the metaquo complexes a 10% excess of potassium ferricyanide was added to the protein samples. The HbI-H2S derivatives were obtained by titrating small amounts of a 10 mM sodium sulfide solution with a gas tight syringe into the deoxygenated metaquo HbI samples (1–2 mM), until complex formation was attained. Typically 3–10 molar excess of H2S were needed to form the complexes. The sodium sulfide solution was prepared by adding the salt, previously purged with N2, to degassed and deoxygenated buffer so as to avoid oxygen contamination. The H2S concentration of the sulfide solution was determined using the Apollo 4000 free radical analyzer. The UV-Vis spectra of the samples were used to monitor formation of the H2S derivatives, which show Soret and Q bands at 426 nm and 544 nm, respectively. The optical spectra of HbI-CO, HbI-O2, metaquo and HbI-H2S derivatives were recorded using Shimadzu UV-2101PC and Agilent 8453 spectrophotometers.
Kinetic Measurements
The CO and O2 association reactions were measured with a flash photolysis system (Quantel, France) by photodissociating the HbI-CO or HbI-O2 sample derivatives using 10 ns YAG laser pulses that provided 160 mJ at 532 nm. A low intensity light source (50-watt quartz halogen lamp filtered at 436 nm) was used as the probe beam, for samples in 1 mm cuvettes. All measurements were made at 25°C, in 50 mM phosphate buffer, pH 7.4. Kinetic curves were averaged with at least three traces recorded on a LeCroy 9400 oscilloscope. Oxygen dissociation rates (koff) for all the HbI samples were measured by CO replacement techniques, as previously described (29). Samples with a slight excess of O2 were mixed with buffer containing a high concentration of the competing ligand. Alternately the oxygen koff could be obtained from the observed replacement kinetics using the flash photolysis technique for samples under a mixed O2/CO atmosphere. Analysis of the kinetics for the O2 dissociation rates were conducted using simulations of double exponential reactions to determine the kinetic constants.
The H2S association rate constants (kon) were obtained by measuring the reactions of the metaquo HbI samples with H2S solutions under anaerobic conditions, using a stopped-flow rapid scanning monochromator spectrophotometer (Olis-On-line Instrument Systems, Bogart, GA) (30). The reactions were followed under pseudo first order conditions ([H2S] >>[HbI]) in which deoxygenated solutions of the metaquo HbI samples at pH 6, were mixed rapidly with degassed solutions of sodium sulfide at different concentrations ranging from 0 to 4 mM. Data for the binding reactions were collected at 426 nm, the maximum in the static absorption of the HbI-H2S spectrum, at 1000 scans/sec. Dissociation of H2S from the rHbI protein was first evaluated under equilibrium conditions. Initially, a ∼3 µM deoxygenated and degassed metaquo rHbI sample (1 mL) was mixed with a small amount of a 10 mM sulfide stock solution to obtain a final H2S concentration of 10 µM, which is the minimum H2S concentration required to form the HbI-H2S complex (3 molar excess). The sample was equilibrated and then the absorption spectra associated with H2S dissociation were recorded as a function of time for 5 hours. The procedure was repeated using sulfide solutions with concentrations ranging from 20 µM (6 molar excess) to 5 mM (∼2000 H2S molar excess) to prepare the HbI-H2S derivative. Hydrogen sulfide dissociation kinetics were then determined for rHbI and all the HbI mutants under anaerobic and aerobic conditions by monitoring the decay of the HbI-H2S absorption signal for all the protein samples at 426 nm, using a UV-Vis spectrophotometer (Agilent 8453). For the anaerobic measurements, small volumes (5 µL) of HbI-H2S solutions (0.8–1.2 mM) were mixed rapidly with a large volume of deoxygenated buffer(1mL) at pH 6.5 and the kinetic traces as well as spectra associated with the kinetics acquired for a period of 24 hours. The H2S dissociation kinetics under aerobic conditions were measured by mixing the HbI-H2S samples with buffer saturated with O2 instead.
Molecular Dynamics Simulations
Molecular dynamics simulations were performed for unligated ferric HbI and five point mutations at positions B10 and E11: PheB10Leu, PheB10Tyr, PheB10Val, PheE11Tyr and PHeE11Val. These were carried out with the AMBER 7.0 software package (31). Initial coordinates for HbI were taken from X-ray structures, Protein Data Bank code 1b0b. All mutants were generated with the SCWRL 3.0 software package (32). In all the MD calculations the all-hydrogen topology with AMBER 94 (33) force field parameters and the TIP3P water model (34) were employed. Parameters around the iron and the charge set for heme in its FeIII state were the same as in an earlier study on sulfide-binding hemoglobins (35). Each protein was solvated with explicit water molecules in a rectangular periodic box large enough to contain the protein and 10 Å of solvent on all sides. A 1400-step steepest-descent minimization was followed by a 1400-step conjugate gradient minimization, and the systems were then heated for 50 ps to a final temperature of 300K. During the heating, a harmonic constraint of 200 kcal/Å2-mol was applied to the protein atoms. For all the mutants, the residue at the point mutation position was not constrained. The time step was 2 fs and the SHAKE algorithm was employed to constrain bonds involving hydrogen atoms. A cutoff of 10 Å was applied to nonbonded interactions. All the systems were equilibrated for 500 ps before starting each 1-ns trajectory. Data were collected at 1.0 ps intervals. During the simulations, the temperature was held constant al 300K by the weak-coupling algorithm (36) with a time constant for heat bath coupling of τ = 2.0ps.
Resonance Raman Measurements
The RR spectra of the CO, metaquo, and H2S rHbI and HbI mutant proteins (∼100 µM heme concentration) were obtained using a 413.1 nm excitation line from a Kr+ ion laser with a power of 10 mW (Spectra Physics). The laser was focused to a ∼30 µm spot on the rotating cell to prevent photodamaging. A back illuminated CCD detector,(800 × 2000 pixels), coupled to a modified Spex 1401 and notch filters centered at 410 nm, were used to record the RR spectra. For the CO and metaquo samples, three spectra were collected and averaged, each composed of 60 accumulations of 10 s. On the other hand, 30 single spectra consisting of 6 accumulations at 10 s each were obtained for the HbI-H2S derivatives. The Raman shift was calibrated using indene for both the low and high frequency regions.
RESULTS
Functional and Structural Features of the Ferrous Derivatives
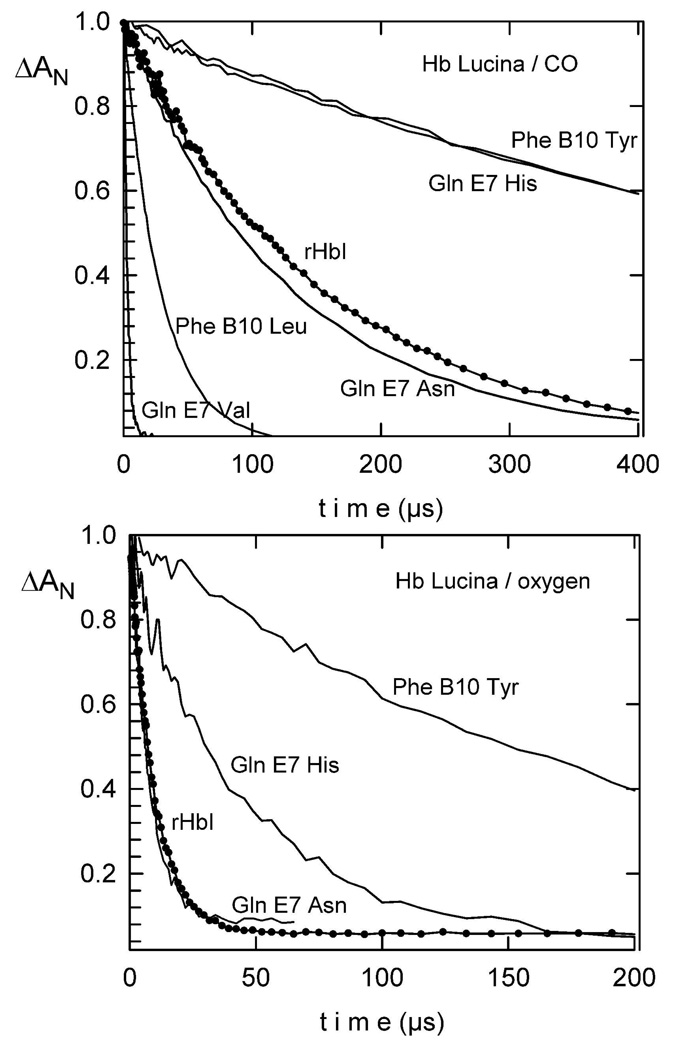
To define the reactivity of H2S with HbI as well as the HbI mutated systems, and to extrapolate the results to other hemeproteins, it is necessary to have an initial perspective on how the structure and electronic environment of these pockets behave in the presence of common ligands such as CO and O2. Thus, the kinetics and structural analyses of each HbI mutant as well as the rHbI were first evaluated with CO and O2. The microsecond bimolecular time courses for CO and O2 binding to rHbI and HbI mutants followed simple pseudo-first order kinetics and the traces were therefore fitted with single exponential functions. Figure 2 shows the comparison of CO binding for rHbI and several HbI mutants. Substitution of GlnE7 by smaller residues such as Val or Asn, increases the kon by a factor of 20 and 4, respectively. An opposite effect is observed for the bulkier and polar GlnE7His mutant, in which a 15-fold decrease of the CO association constant is observed. The results demonstrate a direct correlation between the size of the E7 residue and CO binding to the heme center. For the B10 mutations, single Phe substitution by the polar aromatic acid, Tyr, decreased the CO association constant from 7.2×106 to 6.5×105 M−1s−1. This result can be explained by fact that in this mutant, the tyrosine residue is in close proximity to the heme iron (19) thus obstructing CO entry. Mutation of PheB10 by Val or Leu however, did not affect CO association constant significantly despite their differences in size and volume (23.2 Å for the Phe-Leu and 49.9 Å for Phe-Val) (37). This suggests that CO entry is not directly modulated by the size of the B10 residue. Intriguingly, replacing PheE11 by either Val or Tyr, which differ among each other in size and polarity, provoked similar increases in the CO binding constants (16–17 × 106 M−1s−1). Consequently, this indicates that these mutations may induce analogous structural rearrangements within the distal pocket that indirectly modulate CO entry with respect to the rHbI, as judged by the two fold increase in the kon value. For SWMb, mutations of either LeuB10 and ValE11 by Phe reduces the CO kon by a factor of 2, implying that the binding of the ligand is sensitive to the size of the residues occupying those positions (24). The observation that replacement of PheB10 and PheE11 in HbI does not affect directly CO association while a direct correlation is observed for SWMb accentuates the different local structure of their heme pocket.
Figure 2.
Top panel: Bimolecular kinetics for CO rebinding to rHbI and several Hb site-directed mutants. Samples of 10 µM heme-protein were monitored at 436 nm. The conditions for all reactions were 50 mM sodium phosphate at pH 7.4, 25°C under air or 1atm (1000 µM) CO. Lower panel: Bimolecular kinetics for O2 recombination after photolysis of rHbI and the distal GlnE7His, PheB10Tyr and PheE7Asn mutants, for samples under air.
Oxygen association kinetics of rHbI and various site-directed mutants are shown in the lower panel of Figure 2. The O2 kon of the E7 variants follow the trend observed for CO binding in which, an increase in the kon is observed for the GlnE7Val and GlnE7Asn mutants while, a 6-fold decrease in this value is obtained for GlnE7His. Similar to CO, a substantial decrease of the O2 kon, from 1.9×108 M−1s−1 to 6.8×106 M−1s−1, is also observed for the PheB10Tyr mutant. Substitutions of PheB10 to Val or Leu and PheE11 to Val or Tyr caused only minor changes in O2 association rate constants. For instance, a slight increase in O2 kon from 1.9 × 108 M−1s−1 to 2.3 × 108 M−1s−1 is observed for PheB10Leu mutant while similar constants are obtained for the PheB10Val, PheE11Val and PheE11Tyr variants (near 1.1 × 108 M−1s−1). Since the association kinetics of both ligands are affected in a similar fashion, a simple steric model seems appropriate in which higher rates are observed for smaller residues at position E7: GlnE7Val>GlnE7Asn>rHbI>GlnE7His. Accordingly, these results suggest that CO and O2 binding to the ferrous HbI mutants is controlled mainly by steric constraints rather than electronic, in which GlnE7His reduces the free available space in the HbI distal pocket delaying in turn, ligand binding. Mutating PheB10 to Tyr also diminishes the ligand association due to steric hindrance. In contrast, the CO and O2 association constants for the PheB10Leu, PheB10Val and the E11 mutations do not systematically depend on the size or the polarity of the residues occupying those positions. This implies that these B10 and E11 substitutions may stimulate structural displacements that can in turn affect CO and/or O2 binding relative to rHbI.
Table 1 summarizes the oxygen dissociation rate constants for rHbI and several HbI mutants (23). In this regard, it has been shown that the dissociation of O2 from the HbI mutants depends strongly on the hydrogen bonding interaction between the E7 residue and the bound ligand (21, 23). In brief, the 72% and 63% increases in the O2 koff observed for the GlnE7Val and GlE7Asn mutants were attributed to a complete or partial removal of the hydrogen bond between the bound ligand and these residues, respectively, while the decrease of this value by a factor of 47 in the GlnE7His variant was ascribed to a strong hydrogen bond (21). A strong H-bond between the bound O2 and the PheB10Tyr was also suggested in this study, which decreased the dissociation rate by a factor of 233. Both, the His and Tyr residues of the GlnE7His and PheB10Tyr mutants were implicated in a strong O2 stabilization mechanism, while a moderate stabilization role was attributed to GlnE7 in rHbI (21). The results collected here show that that replacing PheB10 and PheE11 by a smaller and non-polar residue such as Val induces a 57–65% increase of the O2 dissociation rate (from 140 s−1 to 325–400 s−1). The faster O2 dissociation behavior observed in these mutants can be rationalized in principle by the loss of multipole interactions generated by the Phe cage (22, 23). However, the energy associated with these aromatic-electrostatic interactions (∼1.5 kcal/mol) (18) appears to be insufficient to provoke these 57–65% increases in O2 release, which are similar to that obtained for the partial removal of the H-bond in the GlnE7Asn mutant (63%). Thus, the data suggest that a weakening of the hydrogen bonding interaction between GlnE7 and O2 may also occur in these mutants due to the movement of the GlnE7 amide group away from the binding. This displacement, induced by the PheB10 and PheE11 substitutions, support the idea that distal structural rearrangements occur in these mutations. Consequently, these results indicate that both factors affect O2 dissociation, which is consistent with previous reports (23, 38) in that not only GlnE7 is responsible for modulating O2 stabilization in HbI but that PheB10 and PheE11 are involved in this process as well. The apparent role of PheB10 and PheE11 in O2 stabilization has also been suggested before in a SWMb triple mutant that mimics the HbI active center (HisE7Gln, LeuB10Phe, ValB10Phe) (39). In this Mb mutant a 4 fold decrease in O2 koff was observed when compared to the wtSWMb, indicating that PheB10 and PheE11 are responsible for ligand stabilization and release in this variant. The distal structural changes suggested in the CO and O2 kinetic analyses described above for the PheB10Leu, PheB10Val, and the E11 mutants, and the stabilization roles of residues E7, B10, and E11 proposed in the dissociation studies may be important factors in determining the extent of H2S interactions with hemeproteins. Therefore, to corroborate these kinetic interpretations, the structural properties of the rHbI-CO and the HbI-CO mutated systems were examined by measuring their RR scattering responses.
Table 1.
The CO, O2 and H2S association rate constants of the rHbI and the HbI mutants. The O2 dissociation is also shown.
| Protein | CO | O2 | H2S | |
|---|---|---|---|---|
| kon (106 M−1s−1) | kon (106 M−1s−1) | koff (s−1) | kon (103 M−1s−1 | |
| WtHbIa | 0.5 | 100–200 | 61 | 27.30c |
| rHbI | 7.2 | 190 | 140b | 24.30c |
| ValE7 | 160 | 490 | 500b | 276.78 |
| AsnE7 | 31 | 230 | 375b | 39.15 |
| HisE7 | 0.47 | 31 | 3b | 65.80 |
| LeuB10 | 7.4 | 230 | 300b | 61.75 |
| ValB10 | 9.0 | 110 | 400 | nd |
| TyrB10 | 0.65 | 6.8 | 0.6b | 3.94 |
| ValE11 | 16 | 120 | 325 | 16.95 |
| TyrE11 | 17 | 110 | 40 | 11.46 |
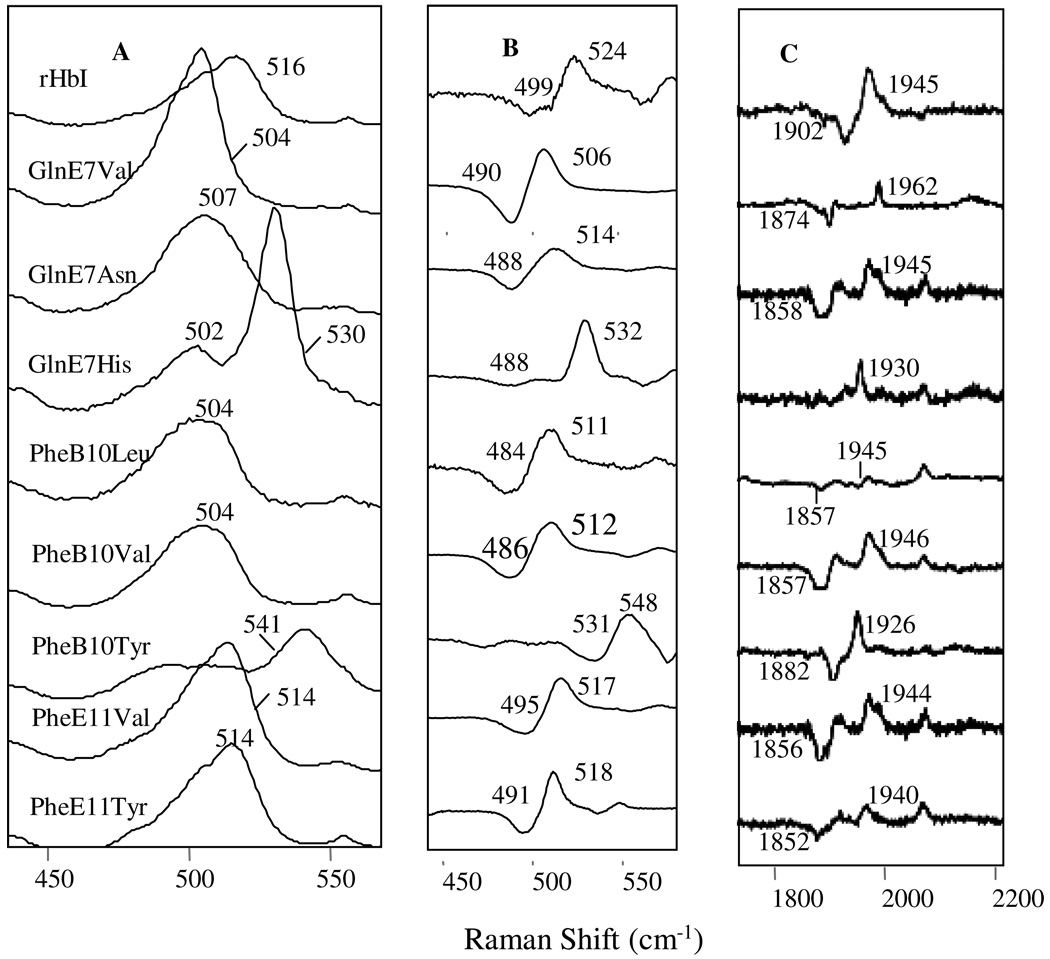
Figure 3 shows the RR spectra of the CO-bound rHbI and mutant derivatives. The low frequency spectrum of rHbI (top trace in Figure 3A) demonstrates a band at 516 cm−1, which was formerly attributed to the Fe-C stretching mode (νFe-C) of the wtHbI-CO complex (38). Panel B in Figure 3 shows that this band shifts to lower frequency upon isotope substitution (12C16O vs 13C18O), thus confirming the previous assignment. At higher frequencies, the spectrum of rHbI shows an additional isotope sensitive band at 1945 cm−1 as demonstrated in the top trace of Figure 3C, and is assigned here to the C≡O stretching mode (νC≡O). Both νFe-C and νC≡O in the low and high frequency regions, respectively, are significantly affected with distal site mutations as indicted in Figure 3. Replacing GlnE7 by Val induces a shift of the νFe-C low frequency mode to 504 cm−1 as well as a displacement of the νC≡O high frequency mode to 1962 cm−1. Mutating GlnE7 by Asn also provokes a shift of νFe-C to 507 cm−1 while the νC≡O mode is observed in the high frequency region of the spectrum at 1946 cm−1. In contrast, Figure 3 shows that the GlnE7His variant shows two isotope sensitive lines for νFe-C and νC≡O modes at 502 cm−1 and 1965 cm−1 and at 530 cm−1and 1930 cm−1, respectively, with the latter pair dominating the isotope difference spectra. The data thus far show that replacing GlnE7 by non-polar residues decreases the νFe-C and increases the νC≡O mode, respectively, while the opposite effect is observed for more polar substitutions.
Figure 3.
Resonance Raman spectra of the HbI-CO protein samples. Figures 3A corresponds to the low frequency region while Figure 3B and 3C demonstrates the 12C16O – 13C18O isotope difference spectra of the low and high frequency regions, respectively. Some of the spectra on 3C show flat peaks due to the subtraction process.
This trend is analogous to that observed in many hemeproteins (Figure 4) including Mb mutants (26), in which replacement of HisE7 by non-polar residues shifts the νFe-C (512 cm−1) to lower frequencies while displacing the νC≡O (1944 cm−1) to higher values (27). Furthermore, substitution of HisE7 in Mb by more polar residues had the opposite effect. This phenomenon has been very well established and it indicates loss or gain of H-bonding interactions, respectively that could stabilize the bound CO. Hence, the displacements of the νFe-C and the νC≡O modes observed in the GlnE7Val and GlnE7Asn variants to lower and higher frequencies, respectively, imply loss or weakening of H-bonding interaction between the E7 residue and the CO ligand, confirming the role of GlnE7 in the stabilization of ligands (Figure 4). On the other hand, the shift of these modes to higher and lower frequencies in GlnE7His indicates a stronger ligand stabilization mechanism in comparison with the rHbI. The HbI PheB10 mutants show a similar tendency. For instance, substitution of PheB10 by non-polar Leu or Val in HbI produced the same spectral changes showing νFe-C at 504 cm−1 and νC≡O modes at ∼1944 and 1946 cm−1, respectively, implying loss of a positive distal site environment. The data also confirm the importance of PheB10 in stabilizing the bound ligand through the aromatic multipoles interactions. Moreover, the substantial shift of the νFe-C mode (504 cm−1 for the mutants and 516 cm−1 for rHbI) and the identical spectral features of the PheB10Leu and PheB10Val variants, support the notion that these mutations provoke analogous conformational changes, involving movement of the GlnE7 amide group, that weaken further the H-bonding interaction with the bound ligand. On the other hand, the polar PheB10Tyr mutant produced shifts of the νFe-C and νC≡O modes to 541 cm−1 and 1925 cm−1, for the low and high frequency regions, respectively, indicating a strong positive field in the vicinity of the ligated CO.
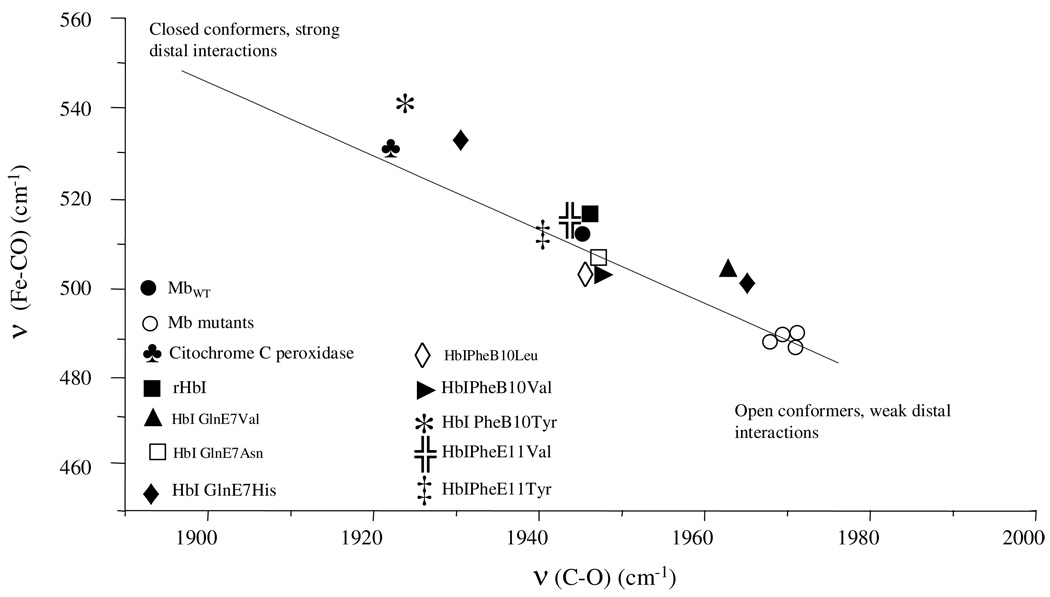
Figure 4.
The νFe-C versus νC≡O inverse correlation curve for Mb, Mb mutants, cytochrome C peroxidase and the HbI proteins.
Interestingly, this trend of increasing νFe-C while decreasing νC≡O upon polar residue replacement is not observed in the HbI PheE11 variants. As Figure 3 and Figure 4 show, substitution of PheE11 to non-polar Val or to polar Tyr residues yields similar low frequency νFe-C modes at 514 cm−1 and high frequency νC≡O modes at 1944 cm−1 and 1940 cm−1, implying that these modes are less sensitive to the polarity of the residues occupying this position. Nonetheless, as in the PheB10Val and PheB10Leu mutants, the similarities in spectral behavior of these E11 variants and the small shift of both modes when compared to rHbI (514 cm−1versus 516 cm−1), suggest related structural changes that induce a slight loss of the positive field surrounding bound CO. Taken together, these RR results support the idea drawn from the kinetic analyses in that: (i.) GlnE7His and PheB10Tyr exert stronger stabilization mechanisms with bound ligands, (ii.) a distal structural rearrangement, involving GlnE7, occurs in the PheB10Leu, PheB10Val and E11 mutations and (iii.) that GlnE7, PheB10 and PheE11 contribute synergistically to the stabilization of ligand in deoxyHbI. These results along with those obtained in the association kinetic study, provide the structural information associated with the HbI mutants that can in turn affect the interaction of HbI and its site-directed mutants with H2S in a ferric heme pocket environment.
Structural Aspects of the Metaquo Derivatives
The direct steric constraint suggested in the CO and O2 association kinetics for the GlnE7His and the PheB10Tyr mutants, relative to rHbI, and the strong stabilization mechanisms invoked for these variants in the ferrous dissociation and RR studies, are important factors that can affect the nature of H2S binding and release in HbI and consequently in hemeproteins. Moreover, the distal conformational changes inferred from the ferrous kinetic and RR analyses upon B10 and E11 substitutions can not only alter H2S interaction but it can also help to define the different responses exerted by this ligand in hemeproteins. Hence, to identify the structural properties of the HbI mutants in the ferric environment and to estimate how they will influence H2S stability and release, the resonance Raman scattering of several metaquo-HbI mutant derivatives was measured in the high frequency region. The high frequency region of the metaquo RR spectra provides information on the overall heme environment and it was used here to characterize the ferric structural features of the HbI mutant complexes in comparison with the wtHbI previously reported (38).
Figure 5 shows the high frequency RR spectra of the metaquo-GlnE7Val, PheB10Tyr, GlnE7His, and PheB10Val HbI mutated systems. Similar to the wtHbI (38), the oxidation state marker band, ν4, for the mutated systems presented in the Figure appears at 1371 cm−1, demonstrating the presence of heme-FeIII derivatives. The RR scattering of rHbI, GlnE7Val and PheB10Tyr show that the ν3 and ν2 modes at 1480 cm−1 and 1560 cm−1 dominate the spectra. A small contribution of the low-spin, six-coordinated metaquo species is also observed in these protein samples as indicated by ν2 mode at 1580 cm−1. Such mixtures, involving the presence of a small population of the low-spin state, are typical of metaquo-complexes with the water molecule occupying the six coordination position. The spectra of the PheB10Val and GlnE7His mutants also display the ν3 and ν2 modes at 1480 cm−1 and 1560 cm−1, indicative of high-spin, six-coordinated metaquo species as well. However, substantial increases of the ν3 and ν2 markers at 1505 cm−1 and 1580 cm−1, respectively, are observed, suggesting a stronger contribution of the low-spin, six-coordinated metaquo species in these variants. These results suggest that in PheB10Val and GlnE7His, the water molecule is coordinated to the heme distal site with an equal mixture of high and low-spin configurations. Resonance Raman studies of various metaquohemoglobin derivatives showing high contribution of the low-spin state, such as the truncated Hb from the protozoan P. caudatum (trHbP) (27) and a HisE7Arg SWMb mutant (40), have indicated that the low-spin character of the mixture arises from H-bonds between heme-bound water and the distal residues. These H-bonding interactions increase the ligand field strength, increasing in turn the low-spin state of the complexes. Therefore, the high contribution of the low-spin markers in the GlnE7His variant can then be rationalized by the formation of a strong hydrogen bond between the imidazole side chain and the water-bound molecule, which generates a strong ligand field and increases the low spin character of this derivative. In the PheB10Val mutant, the best explanation for the strong low-spin contribution is that movement of the GlnE7 towards the ferric iron may induce the generation of a strong H-bond between this residue and H2O that stabilizes the low-spin state of this complex. Similar H-bonding interaction can be predicted in these Hb-H2S mutant derivatives, which may alter in turn sulfide stabilization and release. In addition, these results demonstrate that, in agreement with the ferrous analyses, the PheB10Val mutation stimulates structural changes within the heme distal site that can affect ligand dissociation. These alterations are also expected to occur in the PheB10Leu and the E11 mutations. To assert this hypothesis, molecular dynamics simulations of these ferric HbI mutants were conducted.
Figure 5.
High frequency resonance Raman spectra of the metaquo derivatives for some of the HbI mutants, showing the ν3 and ν2 modes.
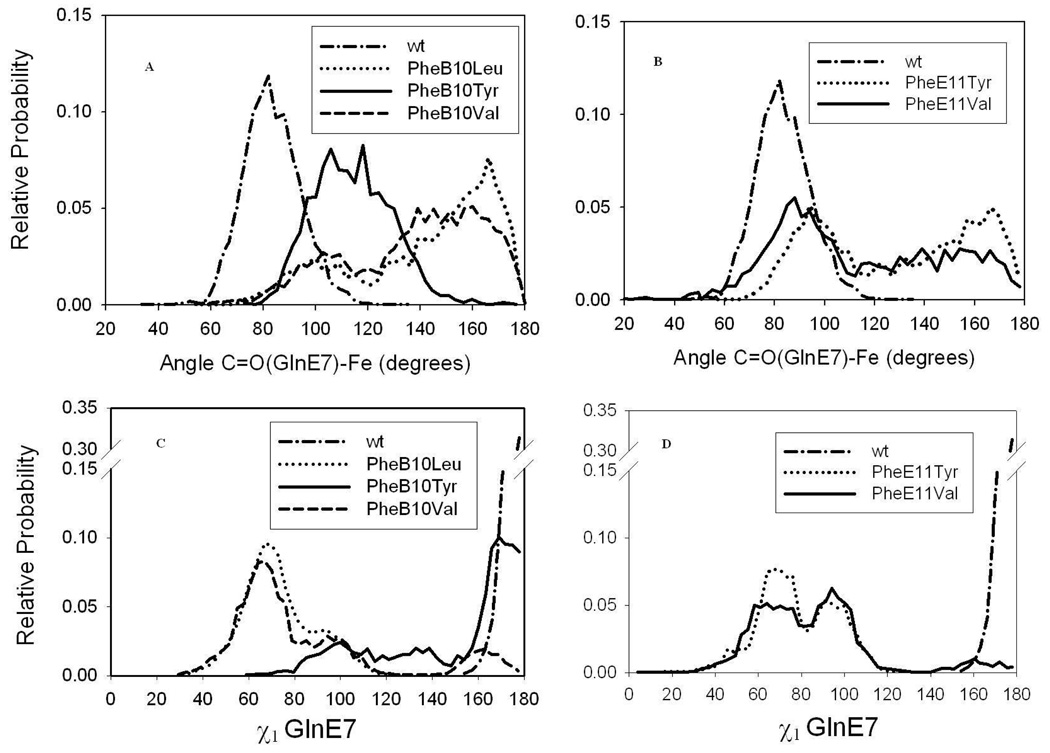
The MD results demonstrate that in the PheB10 and PheE11 mutations a structural rearrangement of the GlnE7 occurs. As Figure 6C and D show, in wtHbI the GlnE7 dihedral angle χ1 is found to be at 180°, while a 90° rotation about this angle is predicted for the PheB10 and the PheE11 mutants. The consequence of this rotation would be a reorientation of the GlnE7 C=O side chain. For instance, Figure 6A and B show that the distribution of the GlnE7 carbonyl group angle with respect to the ferric iron center varies substantially in these B10 and E11 mutants. The C=O(GlnE7)-Fe angle changes from ∼ 90° to 180° in the PheB10Val and PheB10Leu mutants with the C=O pointing towards the ferric iron. Although less dramatic, similar changes are observed for the PheE11Val and PheE11Tyr variants. This is in good agreement with the previous suggestion, in which these B10 and E11 mutants alter the HbI distal structural environment affecting ligand entry and release from the heme pocket. Furthermore, the data also set the stage to define how these perturbations induced by the mutations, can affect the interaction of H2S with the ferric heme center.
Figure 6.
Histograms for GlnE7 dihedral angle χ1 and the angle associated with the carbonyl group of GlnE7 and the heme iron. The A and B panels display the C=O(GlnE7)-Fe angle for the B10 and E11 mutants, respectively, while C and D show the GlnE7 dihedral angle χ1 of the same variants.
Interaction of H2S with the Metaquo Derivatives
The RR results of the metaquo-HbI mutants presented above suggest that the reactivity of H2S may be modulated in the GlnE7His and PheB10Val mutants relative to the rHbI, due to their differences in the distribution of the high and low-spin configurations. The data along with the MD simulations also indicates that PheB10Val, PheB10Leu and E11 mutations produce distal conformational changes that can modify H2S binding and stability as well. To identify how these findings influence H2S reactivity with metaquo-rHbI and the HbI site-directed mutants, sulfide dissociation kinetics as well as RR measurements were conducted. In addition, the effects of these mutations on H2S binding were evaluated.
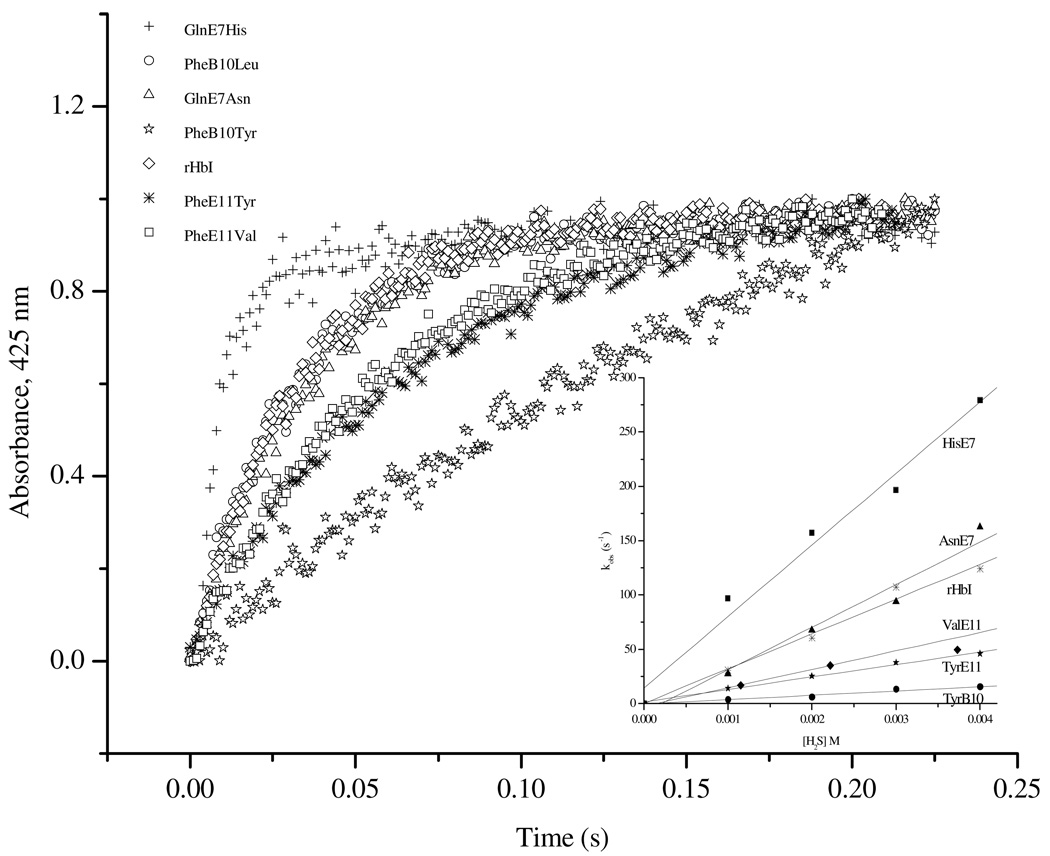
Figure 7 shows the comparison of the H2S association kinetics of the metaquo rHbI previously reported (30) and various HbI mutants studied here. All the traces followed single kinetic event indicating that the ligand reacts readily with the ferric heme center. The calculated association rate constants for each protein were obtained by fitting the data with single exponential functions and plotting the observed constants, kobs, as a function of H2S concentration as shown in the inset. The binding constants thus obtained are summarized in Table 1. Evaluation of the association constants for the E7 mutants at pH 7, demonstrates that replacing GlnE7 by the Val, Asn or His increases the association constant by a factor of 11, 2, and 2.7 respectively. As opposed to CO and O2 binding, there is not a direct correlation between the size of the E7 residue and H2S binding to the heme center, suggesting that its association may also be governed by an external kinetic barriers. In this respect, transient fluctuations of the protein may be as important as steric effects for H2S binding, probably because of the larger nature of the ligand itself. The results show that H2S binding is nearly 1000 times slower that CO and O2, suggesting that protein fluctuations are required to allow H2S access into the iron heme center. That could explain why the GlnE7His mutant seems too fast for H2S while it is the slowest E7 mutant for CO and O2 binding.
Figure 7.
Hydrogen sulfide association kinetics of the rHbI and the mutated samples. The inset displays the plot of the observed rate constants as a function of H2S concentration.
Mutation of PheB10 by Leu increases the association constant from 2.43 × 104 M−1s−1 to 6.58 × 104 M−1s−1 while substitution of this residue by Tyr decreases the kon value by a factor of 6. This suggests that once inside the protein, H2S binding may be dictated by the free accessible space in the distal site.
Interestingly, substitution of PheE11 by a much smaller non-polar residue such as Val instead of increasing the H2S association constant, as observed in the GlnE7Val mutant, decreases this value from 2.43 × 104 M−1s−1 to 1.69 × 104 M−1s−1. Similarly, the association rate of the PheE11Tyr mutant decreases 2-fold to 1.15 × 104 M−1s−1. Hence, like deoxy CO and O2, the structural displacements caused by these E11 mutations affect ligand entry into the heme distal site. Overall, our results show that H2S association is influenced by an outer kinetic barrier that requires transient motions of the protein to allow its movement into the distal pocket. Once H2S is inside the protein its binding is controlled by either electronic or steric constraints exert by nearby residues.
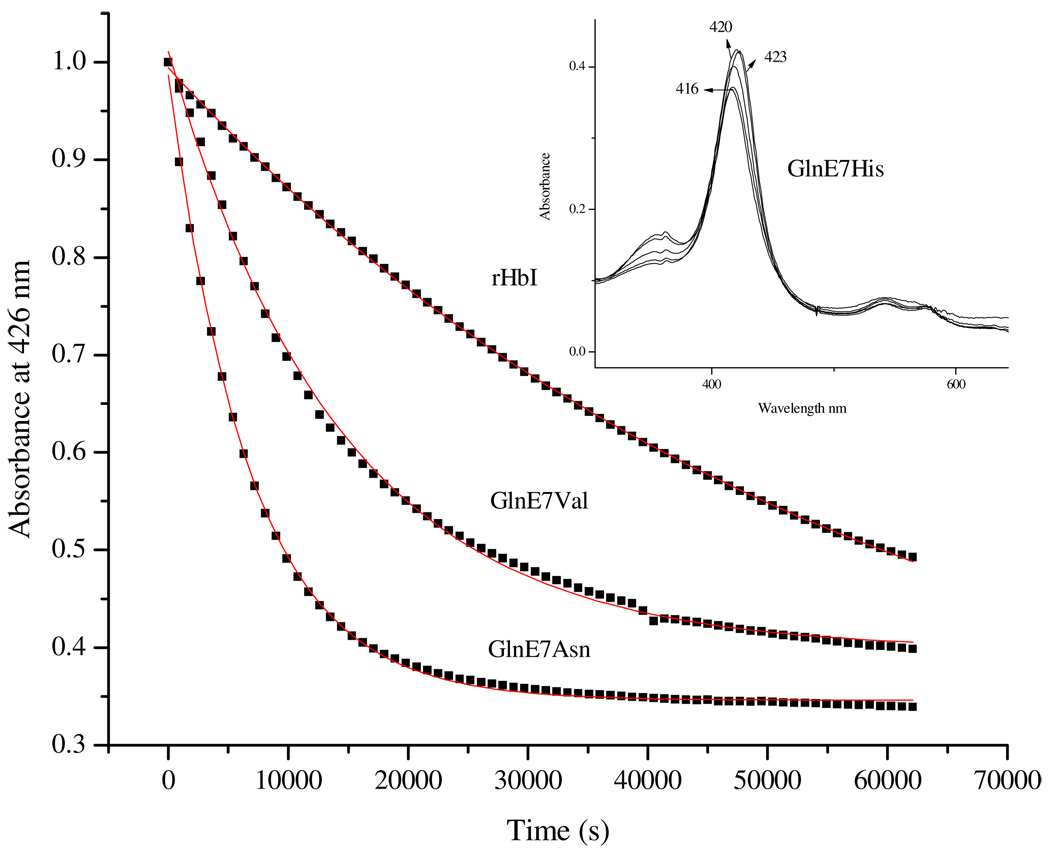
Dissociation of H2S from rHbI was studied first under equilibrium conditions using the lowest sulfide concentration required to form the complex (10 µM or 3 H2S molar excess). Under these conditions, H2S dissociation from rHbI showed a tendency for the metaquo complex formation, as judged by the 425 nm to 408 nm displacement of the Soret band. However, a broad Soret band was observed suggesting that other processes may be contributing to H2S dissociation. Similar results were observed using H2S concentrations ranging from 20 µM to 480 µM, corresponding to 6 and 160 sulfide molar excess, respectively, to form the complex. At higher sulfide concentrations, (600 µM-5 mM, equivalent to 200–2000 H2S molar excess) H2S dissociation from rHbI, at equilibrium, revealed the formation of a final deoxy-like species with Soret and Q bands at 433 nm and 560 nm respectively, indicating reduction of the heme iron center (Figure not shown). Hence, the data suggest that H2S dissociation under equilibrium conditions is dominated by two competing reactions involving simple H2S release from the ferric iron and heme iron reduction followed by H2S liberation. At low H2S concentrations the former process seems to dictate the dissociation reaction while the latter reduction process appears to dominate at higher sulfide concentrations. The H2S dissociation kinetics of the rHbI and the HbI mutants were therefore measured at the limit of complex formation (3–10 molar excess) in order to minimize the difficulties associated with competing reactions such as heme reduction.
Figure 8 shows the dissociation kinetic traces for rHbI, GlnE7Asn and GlnE7Val. The calculated dissociation constants, obtained by fitting the traces with single exponential functions, are of 0.03 × 10−3 s−1, 0.15 × 10−3 s−1 and 0.06 × 10−3 s−1 for rHbI, GlnE7Asn and GlnE7Val, respectively. The results show an increase in H2S koff by 80% in GlnE7Asn probably due to a substantial weakening of the hydrogen bonding strength between this residue and the bound ligand. An increase in the koff is also observed for the GlnE7Val mutant but not as significant as that for the Asn mutant (80% vs 50%), suggesting that the other competing reaction (heme reduction) is also contributing to the observed signals. In fact, the spectra associated with these kinetics showed a final broad Soret band centered at 409 nm supporting the above suggestion. Furthermore, the kinetic spectra of the GlnE7His mutant under aerobic conditions showed that an intermediate is formed rapidly and decays to form a new species with a characteristic Soret band at 416 nm (Inset in Figure 8). Additionally, the kinetic trace of H2S dissociation from GlnE7His shows much more complex behavior and could not be fitted with a single exponential. Similar difficult kinetic traces are observed for the B10 and E11 mutated systems studied. These data were therefore fitted with a two consecutive exponential function yielding constants for the first and second phases. The constants thus obtained do not correlate with any physical-chemical properties of the mutated residues. However, the 416 nm Soret band in the GlnE7His mutant suggests that an oxy-like species may be formed, indicating that reduction of the heme-FeIII-H2S moiety dominates in this mutant even at the minimum H2S concentration needed to form the complex (3–10 molar excess). We thus hypothesize that in rHbI and the HbI mutants both competing processes are involved in H2S dissociation and that the contribution of heme reduction depends strongly on the distal site environment. To corroborate this interpretation, resonance Raman studies of these derivatives were performed. The sensitivity of RR spectroscopy for studying heme structural properties, as opposed to UV-Vis, should yield relevant information associated with the H2S dissociation kinetics described above.
Figure 8.
Dissociation kinetic traces of rHbI, GlnE7Asn and GlnE7Val. The inset shows the spectra related to kinetics of H2S dissociation for the GlnE7His mutant under aerobic conditions.
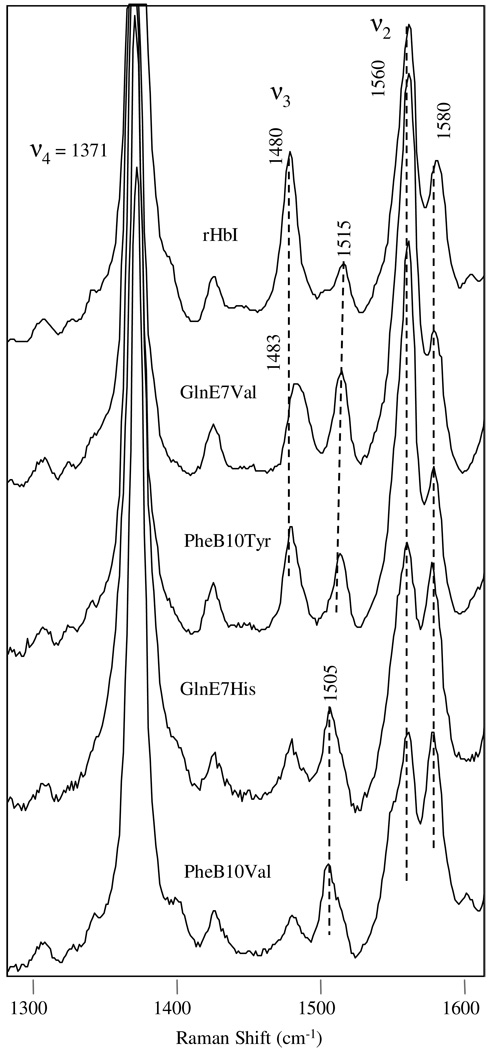
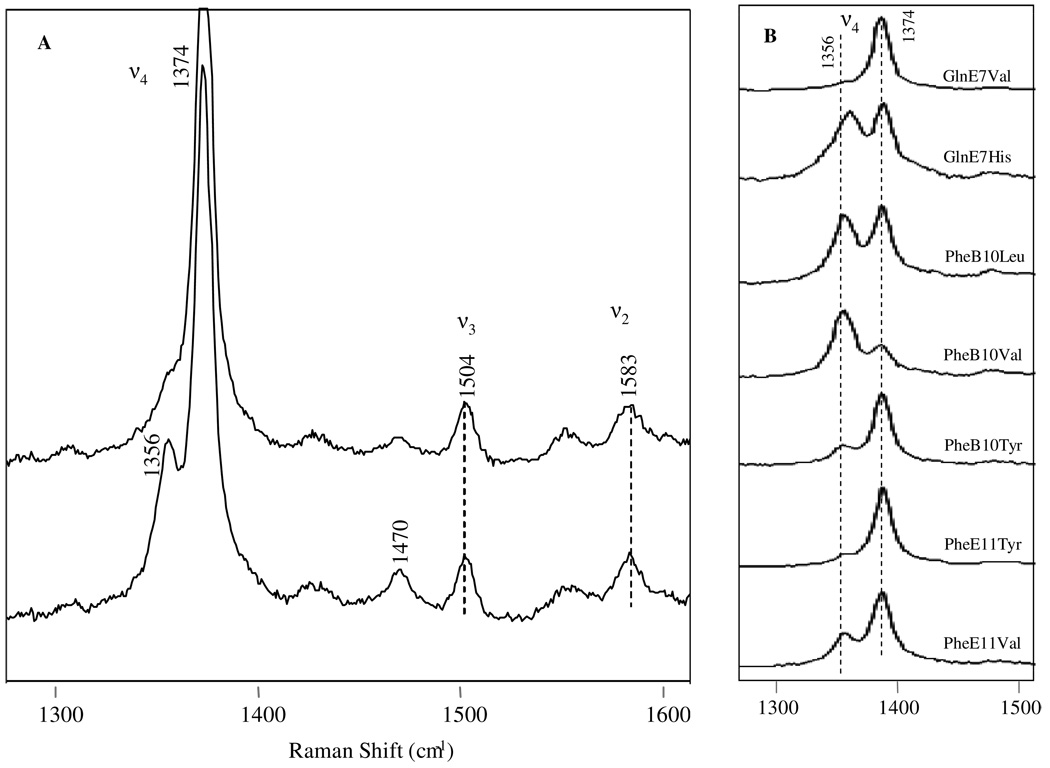
The high frequency RR spectra obtained upon reaction of H2S with metaquo-rHbI is shown in Figure 9A. The top trace shows the oxidation state marker at 1374 cm−1, characteristic of a FeIII heme protein. The presence of the ν3 and ν2modes at 1504 cm−1 and 1583 cm−1, respectively, indicates a low-spin six-coordinated metsulfide HbI complex. Interestingly, the bottom trace shows that the HbI-H2S complex changes within a few minutes indicating the presence of a mixture of a deoxy high spin species and a metsulfide derivative as evidenced by the appearance of new oxidation and spin markers at 1356 cm−1 and 1470 cm−1, respectively. Figure 9B demonstrates similar results for all the mutant proteins studied. Laser power dependence studies indicate that this reduction process is power independent (data not shown), but it depends strongly on distal site mutations. The tendency of FeIIIH2S reduction for all the site-directed mutants studied is as follow: PheB10Val > PheB10Leu ≈ GlnE7His > PheE11Val > PheB10Tyr > PheE11Tyr > rHbI >GlnE7Val. This reduction process is also confirmed by the instantaneous appearances of the νFe-His frequency in the low region at 219 cm−1 (Figure not shown) The data confirm the dissociation kinetic analyses in that reduction of the heme iron take place in HbI and the HbI mutated systems at the limit of H2S concentrations for complex formation. Furthermore, the results show that HbI distal site mutations induce different structural conformations that affect the nature of H2S reactivity as described in detail below.
Figure 9.
High frequency resonance Raman spectra of the rHbI-H2S and mutant derivatives. The top trace in Figure 9A is the first spectrum recorded upon H2S reaction with the sample, while the second one is the last spectrum taken (after 30 minutes). The ν4, ν3, and ν2 modes are highlighted. Figure 9B shows the High frequency resonance Raman spectra of various HbI-H2S mutants, showing the contribution of ν4 band at 1356 cm−1, characteristic of deoxy species.
DISCUSSION
Distinct distal structural geometry influenced H2S reactivity in hemeproteins
The kinetics and RR results obtained with the ferrous HbI mutant derivatives, which are summarized in Table 1 and Table 2, respectively, show that mutating E7, B10 and E11 provoke different heme distal structures that influence directly or indirectly the extent of CO and O2 interaction with the ferrous iron center. The data reveal that relative to rHbI and GlnE7Val, His and Tyr in the ferrous GlnE7His and PheB10Tyr variants, diminish the free available space in the HbI distal pocket that reduce CO and O2 binding. Upon CO and O2 coordination, GlnE7His, PheB10Tyr and wtHbI form H-bonding interactions with the bound ligands generating closed distal cavity configurations. On the contrary, no CO and O2 stabilization is invoked in GlnE7Val, inducing in turn an open distal structure. For the PheB10Val, the PheB10Leu and the E11 variants, the data show that they do not directly dictate ligand binding and release, instead these mutations induce distal structural rearrangement that affect indirectly both, ligand entry and release.
Table 2.
Selected RR band assignments for the CO and H2S-HbI mutant derivatives.
| Protein | CO | H2S | |||
|---|---|---|---|---|---|
| νFe-CO | νc-o | ν2 | ν3 | ν4 | |
| WtHbIa | 516 | 1945 | 1579 | 1500 | 1372 |
| rHbI | 516 | 1945 | 1583 | 1504, 1470 | 1374 (87%)b |
| 1356 (13%) | |||||
| ValE7 | 504 | 1962 | 1580 | 1501 | 1372 |
| AsnE7 | 507 | 1946 | nd | nd | nd |
| HisE7 | 502 | 1965 | 1583 | 1505, 1470 | 1373 (53%) |
| 532 | 1930 | 1356 (47%) | |||
| LeuB10 | 504 | 1944 | 1583 | 1502, 1470 | 1373 (53%) |
| 1355 (47%) | |||||
| ValB10 | 504 | 1946 | 1582, 1562 | 1500, 1470 | 1372 (32%) |
| 1355 (68%) | |||||
| TyrB10 | 541 | 1925 | 1583 | 1503. 1470 | 1372 (80%) |
| 1354 (20%) | |||||
| ValE11 | 514 | 1944 | 1581 | 1502, 1470 | 1373 (70%) |
| 1355 (30%) | |||||
| TyrE11 | 514 | 1940 | 1581 | 1502, 1470 | 1373 (86%) |
| 1356 (14%) | |||||
Data taken from reference reference 31.
The RR spectra were decomposed using peak fitting software from Grams spectra. The relative intensity of each band was calculated by the ratio of their height to the total relative height. nd, not determined.
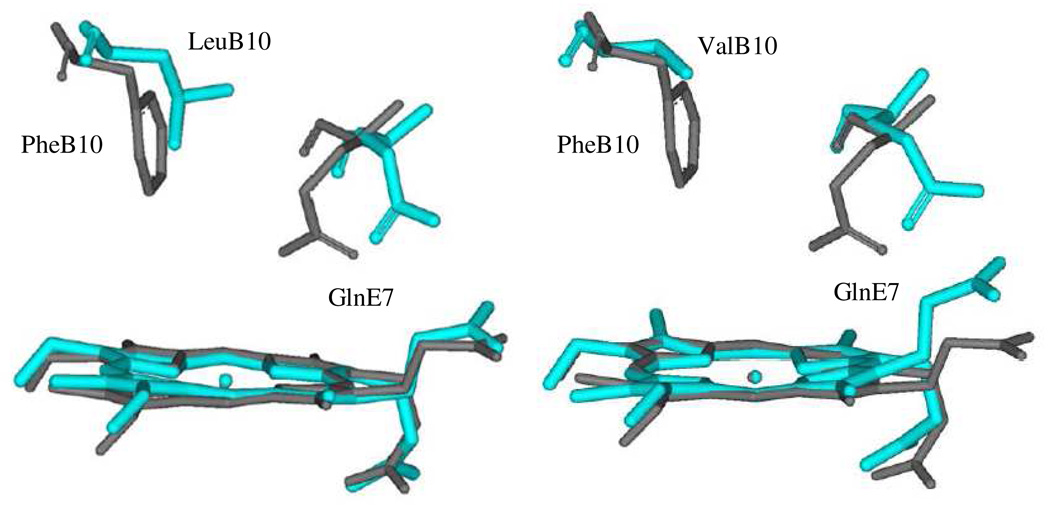
Distinct structural geometries are also observed in the ferric HbI mutants. Regarding this, the equal contributions of spin states illustrated in the RR spectra of metaquo-GlnE7His and the PheB10Val mutant derivatives suggest that two individual distal structures coexist in a conformational equilibrium relative to the metaquo-wtHbI, GlnE7Val and PheB10Tyr derivatives. One of these structures is associated with H-bonding interactions between the HisE7 and the bound water in the GlnE7His mutant and with GlnE7 and the coordinated ligand in the PheB10Val variant, with this latter interaction being induced by the Val replacement. As depicted in Figure 10, substitution of PheB10 by Val, resulted in a stereo orientation of GlnE7 different from wtHbI, where the carbonyl group points towards the heme. This GlnE7 orientation in the PheB10Val mutant can generate the structure involving H-bonding interaction with the water molecule as observed in the RR study. Changes in GlnE7 orientation were also predicted for PheB10Leu (Figure 10, right panel), and to a lesser extent, for the E11 mutants (not shown). Similar variations in distal structures can be invoked for the HbI-H2S mutant derivatives, altering in turn sulfide reactivity within these proteins samples, as judged by their responses in heme reduction.
Figure 10.
Representation of the B10 mutated residue and GlnE7 in the distal heme of HbI (dark line) and the PheB10Leu mutant (blue line) are shown on the left while that for HbI (dark) and the PheB10Val mutant (blue) are displaced on the right side.
Reduction of heme active center by hydrogen sulfide
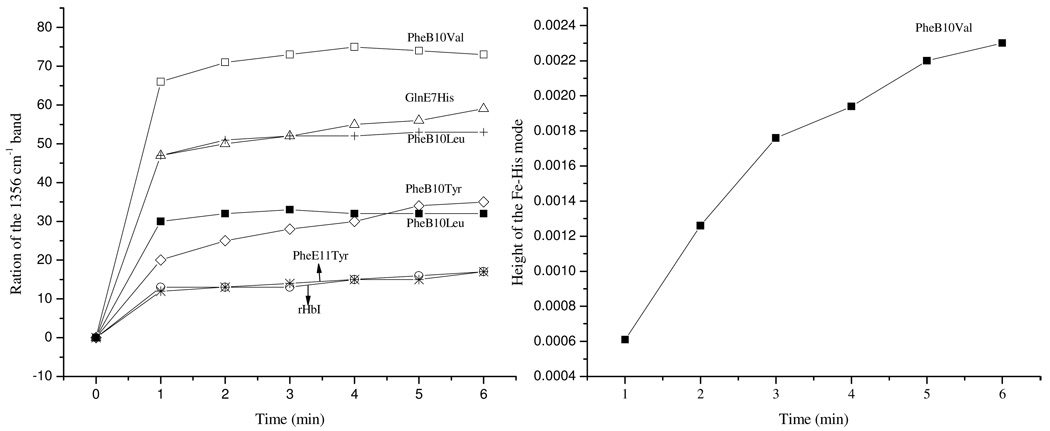
Figure 11 and Table 2 show that reduction of the HbI ferric iron center upon reaction with H2S is controlled by the residues at the distal active site as judged by the increase of the oxidation state marker band ν4 at 1356 cm−1 and the νFe-His mode at 219 cm−1, which are characteristic of deoxy like species. The relative intensity of the 1356 cm−1 band can be obtained for each mutant by calculating the ratio of its height to the total relative height of both the ν4 1356 and the 1372 cm−1 peaks. Accordingly, the faster reduction reactions for the PheB10Val, GlnE7His and PheB10Leu mutants, are evidenced by the 68% and 47% contributions of the 1356 cm−1 band, respectively. Likewise, although to a lesser extent, reaction of PheE11Val, PheB10Tyr, PheE11Tyr and rHbI with H2S produced rapid reduction of the ferric iron with 30%, 20%, 14% and 13% contributions of the 1356 cm−1 band, respectively. While no immediate reduction of the ferric iron atom was observed for the PheE7Val mutant, a small contribution of the ν4 band, at 1356 cm−1 was detected as a function of time, indicating a very slow electron transfer reaction for this variant. The dependence of the heme reduction process on distal side residues can be explained in principle by the strength and directionality of the hydrogen bond between the E7 residue and the bound H2S. In the case of GlnE7Val the lack of proton acceptor groups in close proximity to the bound ligand retards substantially the reduction of the heme iron induced by H2S. In the GlnE7His variant the N atom of the imidazole side chain forms a stronger hydrogen bond (when compared to the rHbI) with the bound H2S thus inducing a faster electron transfer to the ferric iron. For the PheB10Val and PheB10Leu mutants, movement of the GlnE7 carbonyl group towards the ferric iron may place this side chain in a new orientation so as to form a more stable and stronger H-bond with H2S, accelerating in turn the reduction and consequent formation of the unbound deoxy species, as judged by the rapid increase in the Fe-His mode (Figure 11, right panel). A similar mechanism can be invoked for the E11 mutations. However, mutation of PheE11 to either Val or Tyr changes the direction of the GlnE7 carbonyl group so that the hydrogen bond becomes weaker than the B10 mutants but stronger than the recombinant wild type protein.
Figure 11.
The relative intensity of the 1356 cm−1 band as a function of time for various HbI mutants (left panel). The intensities were obtained by calculating the ratio of its height to the total relative height of both the ν4 1356 and the 1372 cm−1 peaks. On the right, the high of the Fe-His mode for the PheB10Val mutant versus time is shown.
Factor affecting the reactivity of hydrogen sulfide with hemeproteins
The results obtained with the HbI site-directed mutants, indicate that interactions of H2S with hemeproteins are controlled by the following factors: (i.) accessibility of H2S into the heme cavity, (ii.) H2S concentration, (iii.) polarity of the distal environment surrounding the bound H2S, and (iv.) the stereo orientation of the distal side residues. At the limit of complex formation (3 to 10 H2S molar excess), hemeproteins with a rather open distal pocket will react with H2S readily forming the heme-H2S complex, as judged by the H2S bimolecular binding reactions. Sulfide release is then dictated by two competing processes involving simple H2S dissociation from the ferric adduct and heme iron reduction induced by H2S itself. The complex behavior of H2S dissociation kinetics can be related to these species. The electron transfer process is greatly enhanced at high H2S concentrations and in those variants having proton acceptor groups near the bound H2S. For instance, in GlnE7His, PheB10Val and PheB10Leu the reduction could take place without large excesses of H2S (10 or less molar excess), as evidenced by the immediate appearance of the ν4 mode in their RR spectra. In fact, the spectral changes associated with H2S dissociation from the GlnE7His mutant, which were taken at the limit of complex formation, show the generation of an oxy-like species, suggesting rapid reduction of the heme and formation of this derivative.
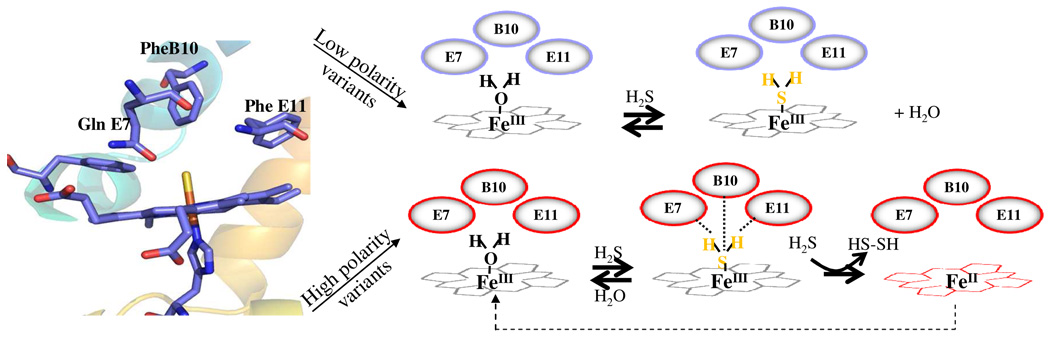
On these bases we proposed that interaction of H2S with hemeproteins can be generalized by the reactions schematized in Figure 12. On the limit of complex formation, proteins with low polar distal environment in the vicinity of the iron will react with H2S according to scheme 1.
Figure 12.
The hypothesized reactions of H2S with hemeproteins deduced from the results of the HbI site-directed mutants.
Scheme 1.
Reaction proposed for the interaction of H2S with low polar heme pocket environments.
In this context, these proteins will bind H2S and sulfide release is then dictated by simple H2S dissociation from the ferric adducts without inducing significant heme reduction. Furthermore, heme pockets with distal polar environment in which their residues are not oriented to form strong H-bonding interactions with H2S can follow the same reaction. In contrast, hemeproteins with polar side chains showing stereo orientations favorable to form strong H-bonding interactions with the bound ligand can stimulate the reduction process under the same conditions as shown in scheme 2. In these hemeproteins the H-bonding interaction between the polar residues and the bound H2S can induce rapid deprotonation of H2S stimulating in turn the formation of a FeII-SH· radical intermediate by one electron transfer from the FeIII-SH− moiety. In the presence of slight ligand excess, H2S can react with the FeII-SH· radical intermediate producing in turn the final deoxy hemeFeII and disulfide species.
Scheme 2.
Reaction proposed for the interaction of H2S with high polar heme pocket environments.
This reduction reaction is supported by the observation that interaction of H2S with other hemeproteins, such as cytochrome c oxidase (8, 10) and flavocytochrome c sulfide dehydrogenase (41), induces heme reduction and generates SH· radical intermediates and final disulfides or elemental sulfur at low and high H2S concentrations, respectively. Reduction of the heme iron center by several organic thiols with the concomitant generation SH· radical intermediates and final disulfides compounds has also been suggested to occur in myoglobin (42). Moreover, oxidation of the reduced Mb by the disulfide products was also observed in that work.
We therefore suggest that these disulfides or hydrogen persulfide species (H2S2) can in principle oxidize the reduced heme reconverting the deoxy species to the active metaquo form of the protein. Alternatively, spontaneous autoxidation of the ferrous heme can occur, resulting in the formation the functional ferric protein as indicated in scheme 2. As with other hemeproteins (24), this autoxidation process should be influenced by the properties of each mutant.
Thus, for wtHbI the flexibility of GlnE7 allows H2S to bind rapidly to the ferric heme. Stabilization of the bound H2S through H-bonding interaction with GlnE7 can in turn induce some heme reduction and H2S delivery as function of H2S concentration. The fact that the X-ray structure of HbI indicates that its heme group is more exposed to the solvent than in HbII (21), suggest that heme oxidation of the ferrous iron can occur, reconverting the deoxy specie to the metaquo form of the protein as H2S is being consumed.
In the case of Mb, HisE7 lies closed to the active center (43) and its reaction with a small excess of H2S induces not only reduction of the heme but also sulfmyoglobin formation (11, 12). This further indicates that the proper orientation of the distal residues is not only important for heme reduction induced by H2S, but that this factor is crucial for sulfhemeprotein formation. Consistent with this argument is the fact that under certain conditions, H2S provokes reduction of the ferric heme a3 in cytochrome c oxidase, which is characterized by a very polar distal heme site involving the CuB center and a Tyr at position 244, without an apparent formation of sulfheme species (9).
Interestingly, the interaction of human neuroglobin (Ngb) with H2S has been examined recently for the first time by analyzing its binding reaction with the ferric center of this new member of the globin family, which it is especially abundant in the brains of mammals (14). It was found in this study that H2S association, using 400 sulfide molar excess, followed biphasic behavior with a fast process being concentration dependent and a slower one independent of H2S concentration. The rapid reaction was ascribed to the bimolecular binding of H2S, while the slowest was not attributed to any particular process. Based on the structural properties of Ngb (44) and factors affecting H2S reactivity presented here, we suggest that heme iron reduction should be considered for the slow process observed in the H2S binding reaction with this hemeprotein. The distal heme cavity of Ngb is characterized by the presence of a HisE7 coordinated to the heme iron in the absence of exogenous ligand (45). Upon exogenous ligation HisE7 is detached from the iron but still near to the bound ligand (46). Hence, reaction of a high excess of H2S with Ngb can also induce iron reduction due to the peculiar HisE7 orientation, and it should be contemplated when analyzing the interaction of H2S with this hemeprotein.
Although reduction of hemeproteins by H2S has been observed before, the dependence of this process on the polarity of the distal site environment should be taken into account when considering interaction of hemeproteins as well as metalloproteins with H2S. This can explain, in principle, the different responses that H2S exerts on hemeproteins and in different tissues and how they can influence sulfide biological activities within cells.
ACKNOWLEDGEMENTS
We thank undergraduate students Abner Nieves and Frances Marie Pietri for their technical assistance during the works. R.P. also thanks the Alfred P. Sloan (NACME) and CNY-PR AGEP for their fellowship support.
Footnotes
This work was supported in part by funds from the National Science Foundation, Cellular Biology (Grant 0544250) and NIH-NIGMS/MBRS-SCORE S06GM08103-34, NCRR G12RR03051, P20RR016439 and NCMHD T37MD001477.
REFERENCES
- 1.Reiffenstein RJ, Hulbert WC, Roth SH. Toxicology of Hydrogen Sulfide. Annu. Rev. Pharmacol. Toxicol. 1992:109–134. doi: 10.1146/annurev.pa.32.040192.000545. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Moore PK. Putative biological roles of hydrogen sulfide in health and disease: a breath of not so fresh air? Trends Pharmacol Sci. 2008;29:84–90. doi: 10.1016/j.tips.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Chen CQ, Xin H, Zhu YZ. Hydrogen sulfide: third gaseous transmitter, but with great pharmacological potential. Acta Pharmacol Sin. 2007;28:1709–1716. doi: 10.1111/j.1745-7254.2007.00629.x. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Moore PK. An overview of the biological significance of endogenous gases: new roles for old molecules. Biochem Soc Trans. 2007;35:1138–1141. doi: 10.1042/BST0351138. [DOI] [PubMed] [Google Scholar]
- 5.Łowicka E, Bełtowski J. Hydrogen sulfide (H2S) – the third gas of interest for pharmacologists. Pharmacol Rep. 2007;59:4–24. [PubMed] [Google Scholar]
- 6.Szabó C. Hydrogen sulphide and its therapeutic potential. Nat Rev Drug Discov. 2007;6:917–935. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- 7.Blackstone E, Morrison M, Roth MB. H2S induces a suspended animation-like state in mice. Science. 2005;308:518. doi: 10.1126/science.1108581. [DOI] [PubMed] [Google Scholar]
- 8.Nicholls P, Kim JK. Oxidation of sulphide by cytochrome aa3. Biochim Biophys Acta. 1981;637:312–320. doi: 10.1016/0005-2728(81)90170-5. [DOI] [PubMed] [Google Scholar]
- 9.Nicholls P, Kim JK. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can J Biochem. 1982;60:613–623. doi: 10.1139/o82-076. [DOI] [PubMed] [Google Scholar]
- 10.Hill BC, Woon TC, Nicholls P, Peterson J, Greenwood C, Thomson AJ. Interactions of sulphide and other ligands with cytochrome c oxidase. An electron-paramagnetic-resonance study. Biochem J. 1984;224:591–600. doi: 10.1042/bj2240591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson EA. The reversion to haemoglobin of sulphhaemoglobin and its coordination derivatives. Biochim Biophys Acta. 1970;207:30–40. doi: 10.1016/0005-2795(70)90134-0. [DOI] [PubMed] [Google Scholar]
- 12.Berzofsky JA, Peisach J, Blumberg WE. Sulfheme proteins. I. Optical and magnetic properties of sulfmyoglobin and its derivatives. J Biol Chem. 1971;246:3367–3377. [PubMed] [Google Scholar]
- 13.Bailly X, Vinogradov S. The sulfide binding function of annelid hemoglobins: relic of an old biosystem? J Inorg Biochem. 2005;99:142–150. doi: 10.1016/j.jinorgbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Brittain T, Yosaatmadja Y, Henty K. The interaction of human neuroglobin with hydrogen sulphide. IUBMB Life. 2008;60:135–138. doi: 10.1002/iub.16. [DOI] [PubMed] [Google Scholar]
- 15.Weber RE, Vinogradov SN. Nonvertebrate hemoglobins: functions and molecular adaptations. Physiol Rev. 2001;81:569–628. doi: 10.1152/physrev.2001.81.2.569. [DOI] [PubMed] [Google Scholar]
- 16.Kraus DW, Wittenberg JB. Hemoglobins of the Lucina pectinata/bacteria symbiosis I. J. Biol. Chem. 1990;265:16043–16053. [PubMed] [Google Scholar]
- 17.Kraus DW, Wittenberrg JB. Hemoglobins of the Lucina pectinata/ bacteria symbiosis II. J. Biol. Chem. 1990;265:16054–16059. [PubMed] [Google Scholar]
- 18.Rizzi M, Wittenberg JB, Coda A, Ascenzi P, Bolognesi M. Structural bases for sulfide recognition in Lucina pectinata hemoglobin I. J. Mol. Biol. 1996;258:1–5. doi: 10.1006/jmbi.1996.0228. [DOI] [PubMed] [Google Scholar]
- 19.Pietri R, Granell L, Cruz A, De Jesús W, Lewis A, Leon R, Cadilla CL, Garriga JL. Tyrosine B10 and heme-ligand interactions of Lucina pectinata hemoglobin II: control of heme reactivity. Biochim Biophys Acta. 2005;1747:195–203. doi: 10.1016/j.bbapap.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 20.De Jesús-Bonilla W, Cruz A, Lewis A, Cerda J, Bacelo DE, Cadilla CL, López-Garriga J. Hydrogen-bonding conformations of tyrosine B10 tailor the hemeprotein reactivity of ferryl species. J Biol Inorg Chem. 2006;11:334–342. doi: 10.1007/s00775-006-0082-0. [DOI] [PubMed] [Google Scholar]
- 21.Gavira JA, Camara-Artigas A, De Jesús-Bonilla W, López-Garriga J, Lewis A, Pietri R, Yeh S-R, Cadilla CL, García-Ruiz JM. Structure and Ligand Selection of Hemoglobin II from Lucina pectinata. J Biol Chem. 2008;283:9414–9423. doi: 10.1074/jbc.M705026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen BD, Zhao X, Vyas K, La Mar GN, Lile RA, Brucker EA, Phillips JN, Olson JS, Wittenberg JB. Solution and crystal structures of a sperm whale myoglobin triple mutant that mimics the sulfide-binding hemoglobin from Lucina pectinata. J. of Biol. Chem. 1998;273:9517–9526. doi: 10.1074/jbc.273.16.9517. [DOI] [PubMed] [Google Scholar]
- 23.Pietri R, León RG, Kiger L, Marden MC, Granell LB, Cadilla CL, López-Garriga J. Hemoglobin I from Lucina pectinata: a model for distal heme-ligand control. Biochim Biophys Acta. 2006;1764:758–765. doi: 10.1016/j.bbapap.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Springer BA, Sligar SG, Olson JS, Phillips GN., Jr Mechanisms of ligand recognition in myoglobin. Chem. Rev. 1994;94:699–714. [Google Scholar]
- 25.Draghi F, Miele AE, Travaglini-Allocatelli C, Vallone B, Brunori M, Gibson QH, Olson JS. Controlling ligand binding in myoglobin by mutagenesis. J Biol Chem. 2002;277:7509–7519. doi: 10.1074/jbc.M109206200. [DOI] [PubMed] [Google Scholar]
- 26.Bikiel DE, Boechi L, Capece L, Crespo A, De Biase PM, Di Lella S, González Lebrero MC, Martí MA, Nadra AD, Perissinotti LL, Scherlis DA, Estrin DA. Modeling heme proteins using atomistic simulations. Phys Chem Chem Phys. 2006;8:5611–5628. doi: 10.1039/b611741b. [DOI] [PubMed] [Google Scholar]
- 27.Egawa T, Yeh SR. Structural and functional properties of hemoglobins from unicellular organisms as revealed by resonance Raman spectroscopy. J. Inor. Biochem. 2005;99:72–96. doi: 10.1016/j.jinorgbio.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 28.León RG, Munier-Lehmann H, Barzu O, Baudin-Creuza V, Pietri R, López-Garriga J, Cadilla CL. High-level production of recombinant sulfide-reactive hemoglobin I from Lucina pectinata in Escherichia coli. High yields of fully functional holoprotein synthesis in the BLi5 E. coli strain. Protein Expr Purif. 2004;38:184–195. doi: 10.1016/j.pep.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Kiger L, Poyart C, Marden MC. Oxygen and CO binding to triply NO and asymmetric NO/CO hemoglobin hybrids. Biophys J. 1993;65:1050–1058. doi: 10.1016/S0006-3495(93)81164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collazo E, Pietri R, De Jesús W, Ramos C, Del Toro A, Leon R, Cadilla CL, Lopez-Garriga J. Functional Characterization of the Purified Holo form of Hemoglobin I from Lucina pectinata Over-Expressed in Escherichia coli. The Protein Journal. 2004;23:239–245. doi: 10.1023/b:jopc.0000027848.40972.97. [DOI] [PubMed] [Google Scholar]
- 31.University of California at San Francisco; Available at http://www.amber.ucsf.edu. [Google Scholar]
- 32.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd generation force-field for the simulation of proteins, nucleicacids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 35.Fernandez-Alberti S, Bacelo DE, Binning RC, Jr, Echave J, Chergui M, Lopez-Garriga J. Sulfide-binding hemoglobins: Effects of mutations on the active-site flexibility. Biophys. J. 2006;91:1698–1709. doi: 10.1529/biophysj.106.081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 37.Zamyatin AA. Protein volume in solution. Prog. Biophys. Mol. Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]
- 38.Cerda J, Echevarría Y, Morales E, López-Garriga J. Resonance Raman studies of the heme-ligand active site of hemoglobin I from Lucina pectinata. Biospectroscopy. 1999;5:289–301. [Google Scholar]
- 39.Dou Y, Maillett DH, Eich RF, Olson JS. Myoglobin as a model system for designing heme protein based blood substitutes. Biophys Chem. 2002;10:127–148. doi: 10.1016/s0301-4622(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 40.Morikis D, Champion PM, Springer BA, Egebey KD, Sligar SG. Resonance Raman studies of iron spin and axial coordination in distal pocket mutants of ferric myoglobin. J Biol Chem. 1990;265:12143–12145. [PubMed] [Google Scholar]
- 41.Steudel R. Mechanism for the formation of elemental sulfur from aqueous sulfide in chemical and microbiological desulfurization processes. Ind. Eng. Chem. Res. 1996;35:1417–1423. [Google Scholar]
- 42.Romero FJ, Ordoñez I, Arduini A, Cadenas E. The reactivity of thiols and disulfides with different redox state of myoglobin. Redox and addition reactions and formation of thiyl radical intermediates. J Biol Chem. 1992;267:1680–1688. [PubMed] [Google Scholar]
- 43.Kundu S, Blouin GC, Premer SA, Sarah G, Olson JS, Hargrove MS. Tyrosine B10 inhibits stabilization of bound carbon monoxide and oxygen in soybean leghemoglobin. Biochemistry. 2004;43:6241–6252. doi: 10.1021/bi049848g. [DOI] [PubMed] [Google Scholar]
- 44.Pesce A, Dewilde S, Nardini M, Moens L, Ascenzi P, Hankeln T, Burmester T, Bolognesi M. Human brain neuroglobin structure reveals a distinct mode of controlling oxygen affinity. Structure. 2003;11:1087–1095. doi: 10.1016/s0969-2126(03)00166-7. [DOI] [PubMed] [Google Scholar]
- 45.Nienhaus K, Kriegl JM, Nienhaus GU. Structural dynamics in the active site of murine neuroglobin and its effects on ligand binding. J Biol Chem. 2004;279:22944–22952. doi: 10.1074/jbc.M401561200. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa H, Finkelstein IJ, Kim S, Kwak K, Chung JK, Wakasugi K, Massari AM, Fayer MD. Neuroglobin dynamics observed with ultrafast 2D-IR vibrational echo spectroscopy. Proc Natl Acad Sci U S A. 2007;104:16116–16121. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]