Abstract
Spinal cord injury reduces the rate of skeletal muscle protein synthesis and increases protein breakdown, resulting in rapid muscle loss. The purpose of this study was to determine whether long-term paraplegia would eventually result in a downregulation of muscle mRNA and protein expression associated with both protein synthesis and breakdown. After 10 weeks of spinal cord transection, soleus muscle from 12 rats (6 sham-control, 6 paraplegic) was studied for mRNAs and proteins associated with protein synthesis and breakdown using real-time polymerase chain reaction and immunoblotting techniques. Protein kinase B (PKB/Akt), ribosomal S6 kinase 1 (S6K1), and myogenin mRNA were downregulated, whereas muscle ring finger 1 (MuRF1) and phospho-forkhead transcription factor 4 (FoxO4) protein were increased in paraplegic rats. We conclude that gene and protein expression of pathways associated with protein synthesis are reduced, whereas some markers of protein breakdown remain elevated following chronic paraplegia. Clinical interventions designed to increase muscle protein synthesis may be helpful in preventing excessive muscle loss during long-term paraplegia.
Keywords: chronic atrophy, FoxO, mTOR, MuRF1, spinal cord injury
Skeletal atrophy is a complex series of events involving an imbalance of protein synthesis and breakdown. Simplistically, atrophy can be described as a decrease in muscle protein synthesis and an increase in muscle protein breakdown, resulting in a net negative muscle protein balance. Muscle atrophy is readily apparent in conditions of disease, such as uncontrolled diabetes, cachexia, acquired immunodeficiency syndrome (AIDS), spinal cord injury, immobilization, spaceflight, and disuse. Animal and human experimental models, such as hindlimb unloading,11 denervation,33 spinal cord isolation,17,18,33 and paraplegia,15 have been used to study skeletal muscle atrophy.
Researchers have extensively studied the cellular and molecular mechanisms involved with muscle protein turnover following muscle atrophy. It is clear that upregulation and downregulation of molecular signals and morphological and functional changes to skeletal muscle occur rapidly (within days) in the aforementioned muscle-wasting conditions.40 Various studies.17,18,33 have indicated that acute atrophy proceeds initially by a rapid phase followed by a slow/steady-state phase of adaptation. Recent reports have identified insulin-like growth factor-1 (IGF-1) and protein kinase B (PKB)/Akt as central targets in the control of muscle protein synthesis and breakdown. For instance, an upregulation of IGF-1 and Akt promotes muscle protein synthesis by activation of the mammalian target of rapamycin (mTOR) and its downstream effector ribosomal S6 kinase 1 (S6K1),8,32 two key regulators of translation initiation and protein synthesis,22,31 and by blocking a major muscle protein breakdown pathway involving forkhead transcription factor (FoxO) 1, 3, and 4.14,16,36,37 Conversely, a decrease in the activation of IGF-1 and Akt leads to a decrease in protein synthesis and, subsequently, muscle loss.8 Further, reduced IGF-1 and Akt activity relieves inhibition of FoxO (hypophosphorylation), thereby allowing FoxO to assume its role as a nuclear transcription factor6,9,23 and promote the activation of several key genes of the ubiquitin-proteasome pathway such as muscle atrophy F-box (MAFbx)/atrogin-1 and muscle ring finger 1 (MuRF1).34,36
With the advancement of pharmacological treatments and rehabilitative strategies, individuals who have experienced paralyzing effects of paraplegia are living longer. Therefore, it is important to further clarify the molecular mechanisms of chronic muscle loss in order to improve the quality of life for these individuals. We have recently shown that several proteins of the muscle mTOR signaling pathway are downregulated after chronic paraplegia in rats.15 However, it is not known whether the downregulation of this key muscle protein synthesis pathway is the primary mechanism for muscle loss associated with chronic paraplegia. It is possible that the reduction in muscle protein synthesis and elevated muscle protein breakdown detected during the early stages after spinal cord injury gradually diminishes, resulting in a new steady-state condition of reduced muscle protein turnover (synthesis and breakdown) in which the muscle does not continue to atrophy. Therefore, we hypothesized that chronic paraplegia would reduce mRNA [IGF-1, mechano-growth factor (MGF), Akt, mTOR, S6K1, myogenin] and protein expression (myogenin) associated with muscle protein synthesis and would also reduce mRNA (FoxO1, MAFbx, MuRF1, myostatin) and protein (phospho-FoxO, MuRF1) expression linked with protein breakdown.
METHODS
study Design
All surgical procedures were reviewed and approved by the Institutional Animal Care and Use Committee at Wayne State University School of Medicine. Twelve age- and weight-matched male Sprague-Dawley rats (512 ± 16 g) were separated into two groups of six: complete spinal cord transection between thoracic level 4 and 5 (T4 and T5; paraplegic or sham spinal cord transection control). These rats are a subset from a previous study.15 After 10 weeks of paralysis induced by spinal cord transaction or sham-operated free living, the rats were killed at 24 weeks of age, and the soleus muscle was isolated, freeze-clamped, immediately frozen in liquid nitrogen and stored at -80°C until analyzed. The soleus muscle was selected for study because this muscle is primarily affected by models that induce muscle loss.40
Induced Paraplegia
All surgical procedures were performed using aseptic techniques. Rats were anesthetized with pentobarbital sodium (45 mg/kg, intraperitoneally), and supplemental doses (10-20 mg/kg, intraperitoneally) were administered if rats regained the blink reflex or responded during the surgical procedure. After anesthesia, rats were positioned prone over a thoracic roll, resulting in a moderately flexed spine. The T4 and T5 vertebrae were exposed via a midline dorsal incision. The underlying spinal cord between T4 and T5 was completely transected through the intervertebral space as previously described.12,13 The control animals had the spinal cord exposed in an identical procedure, but it was not transected. This approach allowed for minimal impact on the stability of the vertebral column. During the first week of recovery, all rats were handled several times daily. During the handling periods, visual inspection and physical manipulations were performed to prevent pressure ulcers. In addition, bladder voiding was accomplished by manual compression, after which all animals were weighed. After the first week of recovery, handling was reduced to one time each day, and bladders no longer required manual compression for voiding. At day 7, rats underwent a motor activity score using previously described criteria.42 Briefly, the motor activity score was determined by placing the animal on a table and observing hindlimb movement for 1 minute. Scores ranged from 0 to 5, with 5 indicating normal walking and 0 indicating zero weight-bearing or voluntary movement specific to the hindlimbs. All paralyzed rats had a score of 0.
RNA Extraction and cDNA Synthesis
Total RNA was isolated by homogenizing 30-40 mg of tissue with a homogenizing dispenser (T10 Basic Ultra Turrax; IKA, Wilmington, North Carolina) in a solution containing 1.0 ml of Tri-reagent and 4 μl of polyacryl carrier (Molecular Research Center, Cincinnati, Ohio). The RNA was separated into an aqueous phase using 0.2 ml of chloroform and precipitated from the aqueous phase using 0.50 ml of isopropanol. Extracted RNA was washed with 1 ml of 75% ethanol, dried, and then suspended in a known amount (1.5 μl/mg tissue) of nuclease-free water. RNA was quantified spectrophotometrically (Smart-Spec; BioRad, Hercules, California) at a wavelength of 260 nm. RNA concentration was calculated on the basis of total RNA yield. RNA quality was assessed by RNA agarose gel electrophoresis followed by visualization of the 18S and 28S ribosomal RNA bands under ultraviolet light. RNA was treated for DNA contaminates using a commercially available kit (DNA-free; Ambion, Austin, Texas). Total RNA (1 μg) was reverse transcribed into cDNA according to the manufacturer's directions (iScript; BioRad, Hercules, California). Briefly, a 20-μl reaction mixture was constructed consisting of 1 μg of total RNA, 4 μlof5× iScript reaction mix, 1 μl of iScript reverse transcriptase, and a known amount of nuclease-free water. RNA was reverse transcribed into cDNA using a thermocycler (IQ5 Real-Time PCR Cycler; BioRad, Hercules, CA) following a temperature/time protocol of 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min. All isolated RNA and cDNA samples were stored at -80°C until further analysis.
Primer pairs were customized using Beacon Designer 2.0 software (Premier Biosoft International, Palo Alto, California). Custom-designed primers are shown in Table 1. IGF-1 primer pairs have been used previously in rats.1 All primers were checked for specificity to our genes of interest by conducting a Blast analysis. Primers efficiencies were optimized, and the polymerase chain reaction (PCR) product was verified by melt analysis and a single DNA product as identified by a DNA agarose gel. Primer dimers were minimized by, first, conducting a melt analysis that consisted of continuous monitoring of the SYBR green fluorescence throughout the temperature ramp from 70°C to 90°C after each PCR run and, second, running the PCR product on a DNA agarose gel to determine if the product of interest was amplified.
Table 1.
Primer sequences used for real-time PCR.
| Name | Accession no. | Primer sequence (5' to 3') | Product size (bp) | |
|---|---|---|---|---|
| MGF | X06108 | Sense | CGTCTTCACATCTCTTCTACC | 122 |
| Antisense | TGGTCCACACACGAACTG | |||
| Akt1 | NM_033230 | Sense | AGAAGGAGGTCATCGTTGC | 120 |
| Antisense | GGTCGTGGGTCTGGAATG | |||
| mTOR | NM_019906 | Sense | TCTTCTTCCAGCAAGTTCAGC | 97 |
| Antisense | GAATCAGACAGGCACGAAGG | |||
| S6K1 | NM_031985 | Sense | CGGGTACTTGGTAAAGGG | 150 |
| Antisense | CCGCTCTGCTTTTGTATG | |||
| Myogenin | NW_047395 | Sense | CAGTGAATGCAACTCCCACA | 85 |
| Antisense | CAAATGATCTCCTGGGTTGG | |||
| FoxO1 | AF247812 | Sense | CACCTTGCTATTCGTTTGC | 130 |
| Antisense | CTGTCCTGAAGTGTCTGC | |||
| Myostatin | AF019624 | Sense | GGCAGAGTATTGATGTGAAGAC | 119 |
| Antisense | TGGGAAGGTTACAGCAAGATC | |||
| MAFbx | NM_133521 | Sense | GCAAAACATAAGACTCATACG | 83 |
| Antisense | GTAGAGTGGTCTCCATTCG | |||
| MuRF1 | NM_080903 | Sense | AGGTGAAGGAGGAACTGAG | 86 |
| Antisense | AACTGCTCTCGGTACTGG | |||
| GAPDH | AF106860 | Sense | GGATGCAGGGATGATGTTCT | 116 |
| Antisense | AAGGGCTCATGACCACAGTC | |||
| β2M | NM_012512 | Sense | TCCACCCACCTCAGATAG | 149 |
| Antisense | AGCATATACATCGGTCTCG |
Semiquantitative Real-Time PCR
Determination of relative mRNA expression was performed by real-time PCR using the iQ5 Multicolor Real-Time PCR Cycler (Bio-Rad, Hercules, California). cDNA was diluted (1:8) and analyzed using SYBR green fluorescence (iQ SYBR Green Supermix; BioRad, Hercules, California). The reaction vessel contained 12.5 μl of iQ SYBR Green Supermix, 0.5-0.7-μl forward and reverse primers, 2.0 μl cDNA, and a known amount of sterile water. The total volume of the reaction tube was 25 μl. All samples were run in duplicate. An initial cycle for 5 min at 95°C was used to denature the cDNA. This was followed with 40 PCR cycles consisting of denaturation at 95°C for 20 s, and primer annealing and extension at 55°C for 30 s. Relative fold changes in mRNA within and baseline differences between groups were determined as described by Livak and Schmittgen27 after normalizing to our housekeeping genes.
Internal Controls
There were no statistical differences in glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or β2-microglobulin (β2M) mRNA expression between control and paraplegic rats. Therefore, we combined the two housekeeping genes to provide more accurate and reliable normalization of our gene-expression data.41 The average cycle threshold values of the two housekeeping genes were 22.65 ± 0.19 (control) and 22.84 ± 0.30 (paraplegic). Validation of GAPDH and β2M were calculated to determine whether the expression of these two housekeeping genes changed with paraplegia. The validation of the two housekeeping genes produced a fold change of 1.07 ± 0.16 and a slope of 0.055.
Immunoblotting
Soleus muscles were homogenized [1:9 (wt/vol) in a buffer containing 50 mM Tris-HCl, 250 mM mannitol, 50 mM NaF, 5 mM Na pyrophosphate, 1 mM ethylene-diamine tetraacetic acid (EDTA), 1 mM ethylene-glycol tetraacetic acid (EGTA), and 1% Triton X-100 (pH 7.4); we added 1 mM dithiothreitol (DTT), 1 mM benzamadine, 0.1 mM phenylmethysulfonylfluoride (PMSF), and 5 μg/ml of soybean trypsin inhibitor just prior to use. Muscle samples were centrifuged for 10 min at 4°C, and supernatant was collected. Total protein concentrations were determined using the Bradford assay (Smartspec Plus; BioRad, Hercules, California). Aliquots of homogenates were boiled at 100°C for 3 minutes in 2× sample buffer containing 125 mM Tris (pH 6.8), 25% glycerol, 2.5% sodium dodecylsulfate (SDS), 2.5% β-mercaptoethanol, and 0.002% bromophenol blue. Samples containing 50 μg of total protein per lane were loaded in duplicate and separated by SDS-polyacrylamide gel electrophoresis (PAGE) for 60 min at 150 V using a 7.5% gel for FoxO proteins and 15% gel for myogenin and MuRF1.
Following SDS-PAGE, proteins were transferred to polyvinylidene diflouride membranes (BioRad, Hercules, California) at 50 V for 60 min. Once transferred, membranes were placed in blocking buffer [5% nonfat dry milk (NFDM) in Tris-buffered saline and 0.1% Tween-20 (TBST)] for 1 h. Blots were then serially washed two times in distilled water, two more times in TBST and incubated with primary antibody in 5% NFDM in TBST overnight at 4°C with constant agitation. Blots were washed in TBST twice and incubated with secondary antibody for 1 h in 5% NFDM in TBST at room temperature with constant agitation. Blots were washed for 15 min followed by three additional washes lasting 5 minutes with TBST. Blots were then incubated for 5 min with enhanced chemiluminescence (ECL) reagent (ECL plus Western Blotting Detection System; Amersham Biosciences, Piscataway, New Jersey) to detect horseradish peroxidase activity. Optical density measurements were obtained by a ChemiDoc XRS imaging system (BioRad, Hercules, California). Once the appropriate image was captured, densitometric analysis was performed using Quantity One 1-D analysis software (version 4.5.2; BioRad, Hercules, California). Complete transfer and equal protein loading was determined using Coomassie blue and Ponceau S staining.
Data were expressed as raw value of the band minus a representative background sample from the membrane, divided by an internal loading control (50 μg/lane), which was loaded on every gel to ensure comparability across membranes. After the raw data were normalized to an internal control, phospho-FoxO1 and 4 were made relative to their respective total proteins, whereas FoxO3 was expressed as phosphorylation (arbitrary units).
Antibodies
Myogenin (M-225; 1:500) and MuRF1 (H-145; 1:1000) were purchased from Santa Cruz Biotechnology (Santa Cruz, California), and phospho-FoxO [1:1000; FoxO1 (Thr24), 3 (Thr32), and 4 (Thr28)] and total FoxO (detects FoxO1 and 4; 1:1000) were purchased from Cell Signaling (Beverly, Massachusetts). Anti-rabbit IgG horseradish peroxidase- conjugated secondary antibody was purchased from Amersham Bioscience (1:2000).
Statistical Analysis
All values are expressed as mean ± SE. Comparisons between control and paraplegic rats were performed with an independent t-test. Significance was set at P < 0.05.
RESULTS
Rat Body Weight and Muscle Characteristics
At the time of killing, paraplegic rat body weight tended to be lower than that of control (619.7 ± 8.7 vs. 567 ± 22.9 g; P = 0.06), and the mean soleus absolute weight was lower in paraplegic (154.0 ± 10.4 g) than control (224.0 ± 25.1 g; P = 0.03) rats. However, after normalizing the soleus to body weight, the relative soleus weight was not different between control and paraplegic rats (0.36 ± 0.04 vs. 0.27 ± 0.02 mg/g, respectively). There were no differences in total RNA (111.6 ± 17.2 vs. 121.0 ± 13.5 μg/soleus) or protein (33.3 ± 3.1 vs. 26.9 ± 2.2 mg/soleus) concentrations, when normalized to the relative soleus weight, between control and paraplegic rats. Total RNA/protein, an index of the capacity of the muscle to translate protein from RNA, was reduced in paraplegic rats (6.1 ± 0.4 μg/mg) as compared with control rats (8.2 ± 0.7 μg/mg; P = 0.02).
mRNA and Protein Expression
Associated with Muscle Protein Synthesis
IGF-1 (1.06 ± 0.14 vs. 0.71 ± 0.27 fold change), MGF (1.07 ± 0.17 vs. 1.51 ± 0.40 fold change), and mTOR mRNA (1.10 ± 0.22 vs. 0.87 ± 0.19 fold change) were unchanged between control and paraplegic rats, respectively. However, when compared with control rats, Akt mRNA expression decreased by 45% (1.06 ± 0.17 vs. 0.58 ± 0.09 fold change; P = 0.04) and S6K1 mRNA expression decreased by 31% (1.01 ± 0.07 vs. 0.70 ± 0.12 fold change; P = 0.02) in the paraplegic rats. Furthermore, myogenin mRNA expression decreased by 83% in the paraplegic rats (1.04 ± 0.13 vs. 0.28 ± 0.08 fold change; P = 0.0001), whereas myogenin protein tended to decrease (0.75 ± 0.13 vs. 0.47 ± 0.04 arbitrary units), but was not significant (P = 0.07) between groups.
Associated with Muscle Protein Breakdown
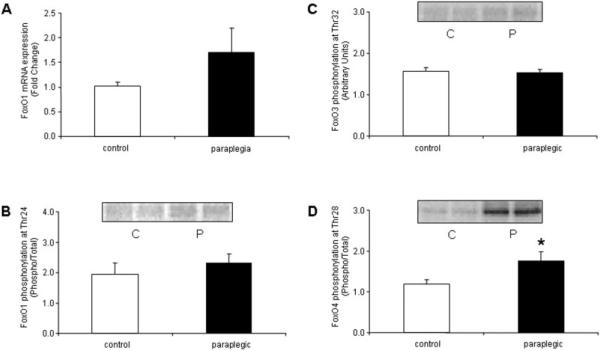
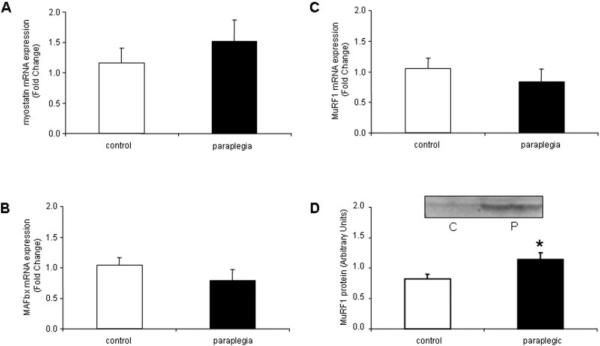
FoxO1 (Fig. 1A) mRNA and phospho-FoxO1 (Fig. 1B) and 3 (Fig. 1C) protein were not different between control and paraplegic rats. However, compared with controls, phospho-FoxO4 increased by 34% (Fig. 1D; P = 0.04) in the paraplegic rats. Myostatin (Fig. 2A), MAFbx (Fig. 2B), and MuRF1 (Fig. 2C) mRNA were not different between control and paraplegic rats. On the other hand, MuRF1 protein increased by 28% (Fig. 2D; P = 0.04) in the paraplegic rats.
FIGURE 1.
Soleus muscle. (A) FoxO1 mRNA expression and phospho- (B) FoxO1, (C) FoxO3, and (D) FoxO4 protein expression from six control and six paraplegic rats. FoxO1 mRNA expression is labeled as fold change, FoxO1 and 4 protein are labeled as phospho/total, and FoxO3 protein is labeled in arbitrary units. All data are expressed as mean ± SE. (B-D) Representative blot images for respective phospho-FoxO protein. Protein images are loaded in duplicate for control (C) and paraplegic (P) rats. *Significantly different from controls (P < 0.05).
FIGURE 2.
Soleus muscle (A) myostatin and (B) MAFbx mRNA expression, and MuRF1 (C) mRNA and (D) protein expression from six control and six paraplegic rats. Data are labeled as either fold change or arbitrary units and expressed as mean ± SE. All data expressed as mean ± SE. (D) Representative blot image for MuRF1 protein. Protein images are loaded in duplicate for control (C) and paraplegic (P) rats. *Significantly different from controls (P < 0.05).
DISCUSSION
We found that a majority of the mRNAs associated with skeletal muscle protein synthesis were reduced following chronic paraplegia in comparison to age-matched controls, whereas there were no detectable changes in the mRNAs associated with muscle protein breakdown. However, MuRF1, a key protein linked to muscle protein breakdown, was elevated in paraplegic rats, indicating that the mRNAs associated with proteolytic pathways do not reflect their protein expression and are likely controlled post-transcriptionally. Our results suggest that some mRNAs and proteins involved in the regulation of both muscle protein synthesis and breakdown continue to play a role in promoting muscle loss during chronic paraplegia.
Many studies over the past decade have explored the mechanisms that occur following an acute loss of muscle mass. We previously evaluated the signaling mechanisms associated with muscle protein synthesis using a chronic paraplegia rat model and found that Akt, mTOR, and S6K1 total protein and S6K1 activity were reduced.15 We have now examined mRNAs associated with muscle protein synthesis and mRNAs and proteins associated with muscle protein breakdown, particularly FoxO, MAFbx, and MuRF1. There are no published studies available that have evaluated genes associated with protein synthesis and breakdown beyond 4 weeks of rat paraplegia.33 Individuals with spinal cord injury have a nearly equal life expectancy to that of ambulatory persons, thus this information could provide insight into the mechanisms associated with chronic spinal cord injury, and it may target interventions that could minimize muscle loss and restore muscle growth.
mRNA and Protein Expression Associated with Muscle Protein Synthesis
We have demonstrated that Akt1 and S6K1 mRNA levels were significantly lower compared with the sham-control rats, whereas IGF-1, MGF, and mTOR were unchanged. The data complement and are consistent with the reduction in Akt and S6K1 protein and S6K1 activity found previously.15 Further, paraplegic rats had a reduced RNA/ protein ratio compared with control rats. This accords with the decreased gene and protein expression as currently and previously reported,15 thus substantiating the deterioration of skeletal muscle due to downregulation of anabolic pathways associated with muscle protein synthesis.
Myogenin mRNA was significantly downregulated, and myogenin protein tended to be lower in paraplegic rats compared with controls. This could suggest a mechanism to reduce muscle growth by decreasing the number of differentiating cells or decreasing the number of genes activated transcriptionally by myogenin. However, contrary to our results, studies have shown that myogenin mRNA and protein are upregulated initially (within days and weeks) and return to basal levels or remain higher after several months of denervation.2,10,20,24,25 We believe these inconsistencies are possibly a result of differences in muscle type composition. We evaluated the soleus muscle, a slow-twitch muscle, whereas others have studied fast-twitch muscles. For instance, Alway et al. demonstrated that myogenin mRNA was upregulated in plantaris (a fast-twitch muscle) after 21 days of hindlimb suspension, but unchanged in the soleus muscle.4 A majority of studies have indicated that slow-twitch muscle tends to be more susceptible to acute muscle loss than fast-twitch muscle.40 Possibly this is because slow-twitch muscles such as the soleus participate continuously in postural and locomotive activities, whereas fast-twitch muscles are generally recruited as force requirements increase. Finally, myogenin resides in greater abundance in slow-twitch muscle,19,40 implying that transcription factors such as myogenin may be more sensitive to muscle loss and less likely to contribute to the maintenance of muscle mass.
mRNA and Protein Expression Associated with Muscle Protein Breakdown
The IGF-1/Akt pathway appears to be a major regulator of FoxO activity.36,37 Therefore, a primary goal of this study was to further clarify the role of IGF-1, Akt, and FoxO following chronic paraplegia in rats. Akt is known to translocate to the nucleus,5,30 phosphorylate all three FoxO proteins (FoxO1, 3, and 4) on residues Thr24, Thr32, and Thr28, respectively, and thereby reduce the ability of FoxO to transcriptionally regulate mRNAs such as the ubiquitin ligases MAFbx and MuRF1.6,9,38 As previously mentioned, Akt mRNA and protein15 were reduced following rat paraplegia. Surprisingly, we noted an increase in FoxO4 protein phosphorylation (Fig. 1D), whereas FoxO1 mRNA (Fig. 1A) and phospho-FoxO1 (Fig. 1B) and FoxO3 (Fig. 1C) were unchanged following chronic paraplegia. These findings suggest that skeletal muscle protein breakdown occurs independently of FoxO hypophosphorylation in chronic paraplegia and that FoxO4 protein is targeted independently of the other FoxO proteins by an unknown upstream kinase other than Akt. The function of FoxO4 remains elusive, but apparently it does not participate in skeletal muscle breakdown following 10 weeks of chronic paraplegia. Together, these data suggest that Akt mRNA expression is reduced independent of a decreased FoxO phosphorylation, indicating a dysregulation between Akt and FoxO. However, future work to determine the importance of FoxO4 phosphorylation in skeletal muscle breakdown is warranted.
We chose to examine MAFbx and MuRF1, two ubiquitin ligases that are associated with the stimulation of skeletal muscle protein breakdown.7,26 Neither MAFbx nor MuRF1 mRNA were differentially expressed in paraplegic rats compared with controls (Fig. 2B and C, respectively). However, MuRF1 protein expression was elevated (Fig. 2D). We are unaware of direct regulators of MuRF1, although activated FoxO (at least 1 and 3) appears to be involved.37 Because FoxO1 and 3 were unchanged after chronic paraplegia, this indicates that another upstream factor may have upregulated MuRF1 protein expression. Alternately, because MuRF1 mRNA expression was unchanged (Fig. 2C), possibly the stability of the protein was enhanced. However, the current literature has clarified that FoxO1 and FoxO3 are direct activators of MAFbx35,36 and therefore may explain why we did not identify any changes in MAFbx mRNA (Fig. 2B). These results indicate that an increase in MuRF1 protein expression, but not MAFbx mRNA, is an important step in the breakdown of specific skeletal muscle proteins following chronic paraplegia.
Myostatin is a negative regulator of muscle mass.21,28,29,39 Zhang et al. reported that myostatin mRNA peaks at 28 days of denervation in rat gastrocnemius muscle and remains elevated (albeit less) at 56 days.43 Contrary to our hypothesis, myostatin mRNA was unchanged in the soleus muscle of paraplegic rats (Fig. 2A). In support of our findings, Carlson et al. did not identify any changes in myostatin mRNA in the soleus muscle after 7 days of hindlimb unloading even though this muscle underwent the most muscle loss.11 Careful analysis of the current literature has identified an association between myostatin and the FoxO protein family members. Recent work has identified that FoxO1 can activate myostatin gene expression in C2C12 cell lines.3 Therefore, myostatin mRNA may not have increased in our paraplegia model because FoxO1 mRNA (Fig. 1A) and protein phosphorylation (Fig. 1B) were unchanged compared with sham-controls. It is possible that myostatin does not play a role in muscle protein breakdown of the soleus after chronic paraplegia.
We conclude that a decrease in the expression of specific genes and proteins associated with the regulation of protein synthesis indicate a likely reduced rate of muscle protein synthesis following chronic paraplegia. Furthermore, MuRF1 protein expression is increased suggesting that the rate muscle protein breakdown may remain elevated in long-term paraplegia. Therefore, clinical interventions designed to increase muscle protein synthesis such as electrical stimulation, assisted-treadmill walking, and/or nutritional supplementation with leucine-enriched essential amino acids may be helpful in preventing excessive muscle loss during long-term paraplegia.
Acknowledgments
This study was funded by grant R01 HL074122 from the National Heart, Lung, and Blood Institute of the National Institutes of Health, and by grant R01 AR049877 from the National Institute for Arthritis and Musculoskeletal and Skin Diseases.
Abbreviations
- β2M
beta2-microglobulin
- EDTA
ethylene-diamine tetraacetic acid
- EGTA
ethylene-glycol tetraacetic acid
- FoxO
forkhead transcription factor
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IGF-1
insulin-like growth factor-1
- MAFbx/atrogin-1
muscle atrophy F-box
- MGF
mechano-growth factor
- mTOR
mammalian target of rapamycin
- MuRF1
muscle ring finger 1
- NFDM
nonfat dry milk
- PCR
polymerase chain reaction
- PKB/Akt
protein kinase B
- PMSF
phenylmethylsulfonylfluoride
- S6K1
ribosomal S6 kinase 1
- SDS-PAGE
sodium dodecylsulfate-polyacrylamide gel electrophoresis
- TBST
Tris-buffered saline and 0.1% Tween-20
REFERENCES
- 1.Adams GR, Cheng DC, Haddad F, Baldwin KM. Skeletal muscle hypertrophy in response to isometric, lengthening, and shortening training bouts of equivalent duration. J Appl Physiol. 2004;96:1613–1618. doi: 10.1152/japplphysiol.01162.2003. [DOI] [PubMed] [Google Scholar]
- 2.Adams L, Carlson BM, Henderson L, Goldman D. Adaptation of nicotinic acetylcholine receptor, myogenin, and MRF4 gene expression to long-term muscle denervation. J Cell Biol. 1995;131:1341–1349. doi: 10.1083/jcb.131.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol. 2007;292:C188–199. doi: 10.1152/ajpcell.00542.2005. [DOI] [PubMed] [Google Scholar]
- 4.Alway SE, Lowe DA, Chen KD. The effects of age and hindlimb supension on the levels of expression of the myogenic regulatory factors MyoD and myogenin in rat fast and slow skeletal muscles. Exp Physiol. 2001;86:509–517. doi: 10.1113/eph8602235. [DOI] [PubMed] [Google Scholar]
- 5.Andjelkovic M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, et al. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 6.Biggs WH, III, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 8.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 9.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 10.Carlson BM, Borisov AB, Dedkov EI, Khalyfa A, Kostrominova TY, Macpherson PC, et al. Effects of long-term denervation on skeletal muscle in old rats. J Gerontol A Biol Sci Med Sci. 2002;57:B366–374. doi: 10.1093/gerona/57.10.b366. [DOI] [PubMed] [Google Scholar]
- 11.Carlson CJ, Booth FW, Gordon SE. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am J Physiol. 1999;277:R601–606. doi: 10.1152/ajpregu.1999.277.2.r601. [DOI] [PubMed] [Google Scholar]
- 12.Collins HL, Dicarlo SE. Acute exercise reduces the response to colon distension in T(5) spinal rats. Am J Physiol Heart Circ Physiol. 2002;282:H1566–1570. doi: 10.1152/ajpheart.00733.2001. [DOI] [PubMed] [Google Scholar]
- 13.Collins HL, DiCarlo SE. TENS attenuates response to colon distension in paraplegic and quadriplegic rats. Am J Physiol Heart Circ Physiol. 2002;283:H1734–1739. doi: 10.1152/ajpheart.00253.2002. [DOI] [PubMed] [Google Scholar]
- 14.Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 15.Dreyer HC, Glynn EL, Lujan HL, Fry CS, Dicarlo SE, Rasmussen BB. Chronic paraplegia-induced muscle atrophy downregulates the mTOR/S6K1 signaling pathway. J Appl Physiol. 2008;104:27–33. doi: 10.1152/japplphysiol.00736.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 17.Haddad F, Roy RR, Zhong H, Edgerton VR, Baldwin KM. Atrophy responses to muscle inactivity. I. Cellular markers of protein deficits. J Appl Physiol. 2003;95:781–790. doi: 10.1152/japplphysiol.00317.2003. [DOI] [PubMed] [Google Scholar]
- 18.Haddad F, Roy RR, Zhong H, Edgerton VR, Baldwin KM. Atrophy responses to muscle inactivity. II. Molecular markers of protein deficits. J Appl Physiol. 2003;95:791–802. doi: 10.1152/japplphysiol.01113.2002. [DOI] [PubMed] [Google Scholar]
- 19.Hughes SM, Taylor JM, Tapscott SJ, Gurley CM, Carter WJ, Peterson CA. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- 20.Hyatt JP, Roy RR, Baldwin KM, Edgerton VR. Nerve activityindependent regulation of skeletal muscle atrophy: role of MyoD and myogenin in satellite cells and myonuclei. Am J Physiol Cell Physiol. 2003;285:C1161–1173. doi: 10.1152/ajpcell.00128.2003. [DOI] [PubMed] [Google Scholar]
- 21.Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 22.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 23.Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 24.Kostrominova TY, Dow DE, Dennis RG, Miller RA, Faulkner JA. Comparison of gene expression of 2-mo denervated, 2-mo stimulated-denervated, and control rat skeletal muscles. Physiol Genomics. 2005;22:227–243. doi: 10.1152/physiolgenomics.00210.2004. [DOI] [PubMed] [Google Scholar]
- 25.Kostrominova TY, Macpherson PC, Carlson BM, Goldman D. Regulation of myogenin protein expression in denervated muscles from young and old rats. Am J Physiol Regul Integr Comp Physiol. 2000;279:R179–188. doi: 10.1152/ajpregu.2000.279.1.R179. [DOI] [PubMed] [Google Scholar]
- 26.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-/-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 29.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meier R, Alessi DR, Cron P, Andjelkovic M, Hemmings BA. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bbeta. J Biol Chem. 1997;272:30491–30497. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- 31.Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999;344:427–431. [PMC free article] [PubMed] [Google Scholar]
- 32.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 33.Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 34.Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab. 2004;287:E591–601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- 35.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 38.Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, et al. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc Natl Acad Sci USA. 1999;96:11836–11841. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor WE, Bhasin S, Artaza J, Byhower F, Azam M, Willard DH, Jr, et al. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am J Physiol Endocrinol Metab. 2001;280:E221–228. doi: 10.1152/ajpendo.2001.280.2.E221. [DOI] [PubMed] [Google Scholar]
- 40.Thomason DB, Booth FW. Atrophy of the soleus muscle by hindlimb unweighting. J Appl Physiol. 1990;68:1–12. doi: 10.1152/jappl.1990.68.1.1. [DOI] [PubMed] [Google Scholar]
- 41.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:0034.0031–0034.0012. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Euler M, Akesson E, Samuelsson EB, Seiger A, Sundstrom E. Motor performance score: a new algorithm for accurate behavioral testing of spinal cord injury in rats. Exp Neurol. 1996;137:242–254. doi: 10.1006/exnr.1996.0023. [DOI] [PubMed] [Google Scholar]
- 43.Zhang D, Liu M, Ding F, Gu X. Expression of myostatin RNA transcript and protein in gastrocnemius muscle of rats after sciatic nerve resection. J Muscle Res Cell Motil. 2006;27:37–44. doi: 10.1007/s10974-005-9050-5. [DOI] [PubMed] [Google Scholar]