Abstract
We have evaluated five bioconjugation chemistries for immobilizing DNA onto silicon substrates for microfluidic biosensing applications. Conjugation by organosilanes is compared with linkage by carbonyldiimidazole (CDI) activation of silanol groups and utilization of dendrimers. Chemistries were compared in terms of immobilization and hybridization density, stability under microfluidic flow-induced shear stress, and stability after extended storage in aqueous solutions. Conjugation by dendrimer tether provided the greatest hybridization efficiency; however, conjugation by aminosilane treated with glutaraldehyde yielded the greatest immobilization and hybridization densities, as well as enhanced stability to both shear stress and extended storage in an aqueous environment. Direct linkage by CDI activation provided sufficient immobilization and hybridization density and represents a novel DNA bioconjugation strategy. Although these chemistries were evaluated for use in microfluidic biosensors, the results provide meaningful insight to a number of nanobiotechnology applications for which microfluidic devices require surface biofunctionalization, for example vascular prostheses and implanted devices.
Keywords: Microfluidics/microfabrication, Biosensors, DNA immobilization, Surface functionalization, Bioconjugation, Silane stability
Introduction
The immobilization of nucleic acids onto glass and silicon substrates has relevance in a number of nanobiotechnology applications, including the detection of single nucleotide polymorphism [1, 2], disease surveillance for identification of emerging strains [3], and biosensor development [4–6]. In choosing a surface immobilization chemistry for these applications, there are a number of factors which must be considered. Of primary importance are probe density and uniformity; however, specific applications may have additional requirements. Biosensors that are intended to be reused multiple times and implanted biomedical devices must possess long-term stability in aqueous environments. Initial functionalization typically involves deposition of an organosilane monolayer; however, conjugation by a silane linkage may not provide the necessary long-term stability for these applications [7, 8]. In the case of many optical biosensors [9–11], it is important to limit the distance between the sensor surface and the immobilized probe in order to contain the hybridization event within the region of highest optical intensity. There are a few recent reports on conjugation of various ligands [12] and proteins [13] to silicon supports by 1,1-carbonyldiimidazole (CDI), indicating that it may be possible to directly link DNA capture probes to silicon substrates. Bypassing the need for a silane monolayer would bring the hybridization event closer to the substrate, a potential benefit to label-free detections.
Nucleic acid probe density is often reported in terms of the amount of immobilized capture probe; however, it has been reported that beyond an optimal probe density, steric hindrance can prevent hybridization efficiency [14, 15]. Therefore, while immobilized probe density is an important parameter to consider when selecting surface functionalization chemistries, it is important to also consider hybridization efficiency, which can be defined as the ratio of hybridized target nucleic acids to immobilized capture probes. Branched or dendritic tethers can increase immobilization density by providing more reactive sites per unit area for ligand immobilization. Tether molecules are also reported to improve hybridization efficiency in microarrays by reducing steric effects of the surface immobilized capture probe and bringing the hybridization event into a solution-like state [16–18]. Dendrimers in particular have been reported to improve binding capacity, homogeneity, stability, and hybridization efficiency in microarray applications [19–22].
Although there are many reports evaluating surface functionalization techniques for immobilizing nucleic acids on silica for microarray applications, there are few published data on the use of these techniques in microfluidic applications. Incorporating microfluidics in bioanalysis can reduce the necessary sample size, significantly reduce analysis time, and allow the potential for automated, miniaturized lab-on-a-chip biodevices [23–30]. However, the tight confinement of the flow layer in pressure-driven or electrokinetic microfluidic devices can exert high surface shear stresses, which may in turn affect the stability of immobilized biomolecules [1]. The stability of surface functionalization chemistries and immobilized biomolecules to shear stresses exerted in microfluidic flow regimes has not been thoroughly investigated. It was therefore our goal to evaluate several DNA immobilization chemistries in order to compare immobilized capture probe density and hybridization efficiency in a microfluidic system. The bioconjugation chemistries evaluated herein are designed to perform on oxidized silica surfaces such as those presented on glass slides as well as the native oxide present on fabricated silicon devices. Each bioconjugation chemistry was evaluated for its stability under microfluidic flow and for long-term stability in an aqueous buffered environment. We compared two frequently used silicon surface functionalization techniques, glycidoxypropyltrimethoxysilane (GOPS) and aminopropyltrimethoxysilane (APTMS), as well as emerging techniques including direct linkage by CDI activation and linkage by a dendrimer tether. As a model system, we detected a synthetic oligonucleotide corresponding to human influenza A virus, subtype H3N2.
Materials
Superclean microarray slides were purchased from ArrayIt, TeleChem International (Sunnyvale, CA, USA). For contact angle and Fourier transform infrared (FTIR) measurements, chemistries were performed on P-type silicon wafers (WaferWorks, Helitek Company, Fremont, CA, USA). Glutaraldehyde (50%) and cyanoborohydride coupling buffer (3 g/L sodium cyanoborohydride) were purchased from Sigma Aldrich (St. Louis, MO, USA). Polydimethyl-siloxane (PDMS, Sylgard 184) components were purchased from Dow Corning (Midland, MI, USA). 3-Glycidoxypropyltrimethoxysilane (GOPS, 97%) was purchased from Acros Organics (Morris Plains, NJ, USA). 1,1′-Carbonyldiimidazole (CDI) was purchased from Nova Biochem (Darmstadt, Germany). Generation 4.5 carboxylic-acid-terminated PAMAM dendrimers and Generation 5 amine-terminated PAMAM dendrimers were purchased from Dendritech (Midland, MI, USA). 3-Aminopropyltrimethoxysilane (APTMS), 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide (EDC), ficoll, form-amide, N-hydroxysuccinimide (NHS), triethylamine (TEA), 2-(N-morpholino)ethanesulfonic acid (MES), sodium dodecyl sulfate (SDS), Triton X-100, 20× sodium saline citrate (SSC) buffer concentrate [20× SSC is 3 M NaCl, 0.3 M sodium citrate], and 10× phosphate-buffered saline (PBS) concentrate [1× PBS is 137 mM NaCl, 2.7 mM KCl, 10 mM phosphate buffer] were purchased from VWR (West Chester, PA, USA). All other reagents were laboratory grade or better and were used as received.
DNA probes corresponding to serotype H3N2 of influenza A virus were synthesized by Operon Biotechnologies (Huntsville, AL, USA) and purified by HPLC. Capture probes were 5′ modified with an amine functionality and 3′ modified with tetramethylrhodamine (TAMRA) fluorescent dye. Target probes were 5′ modified with fluorescein (FL). Sequences used for capture and target probes are listed in Table 1. All probes were diluted to 100μM in 0.1 M phosphate buffer, pH8.5, containing 1 mM ethylenediamine tetraacetic acid (EDTA) and 0.01 wt.% sodium azide and stored at −20°C until use. Hybridization buffer was prepared from 60% formamide, 6× SSC, 0.8% ficoll, and 0.01% Triton X-100. The sequential rinses used after hybridization were prepared as follows: rinse 1 [0.1% SDS, 2× SSC], rinse 2 [0.1% SDS, 0.2× SSC], and rinse 3 [0.1× SSC].
Table 1.
Sequences of DNA probes
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| Capture probe | [Amine]CAT GAT TGA AGC CGA GTC CTC[TAMRA] |
| Target probe | [FL]GAG GAC TCG GCT TCA ATC ATG |
Methods
Cleaning
To obtain optimal fluorescence signal, glass slides were used as substrates to quantify fluorescently tagged capture and target probes. To avoid the high background infrared absorption associated with glass substrates and thus enhance sensitivity, silicon wafers were used for grazing angle attenuated total reflectance FTIR (ATR-FTIR) analysis of the silane monolayers. The chemistries outlined below perform equally well on glass slides and the native oxide present on silicon wafers. Glass slides and silicon wafers were cleaned in piranha solution (3:1 mixture of concentrated sulfuric acid and 30% hydrogen peroxide) at 70°C for 30 min, followed by rinsing in copious deionized water, in order to clean the surface and generate surface silanol (Si–OH) groups. Unless otherwise noted, surface-modified substrates were rinsed in deionized water, dried under 0.45μm filtered air, and stored in clean glass petri dishes until further functionalization or analysis.
Functionalization of oxidized silicon substrates
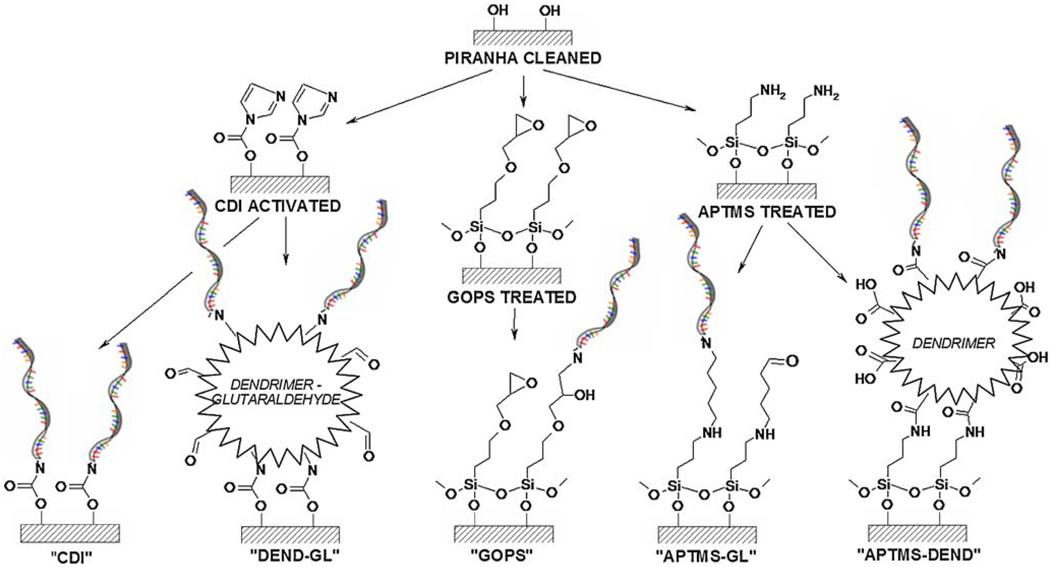
The reaction schemes for each surface functionalization evaluated in this study and the corresponding nomenclature are depicted in Fig. 1. The active groups present on the surface modified substrates were stable in dry storage for several days. After initial functionalization, subsequent analyses or functionalizations were performed within 1 to 2 days of initial functionalization to prevent surface contamination by adventitious carbon or other environmental sources. CDI—Piranha-cleaned glass or silicon substrates were shaken for 2 h at 37°C in 0.5 M CDI in dioxane, followed by rinsing in acetone and water. The CDI-functionalized surface possessed an imidazole carbamate functionality, which forms a stable carbamate linkage upon reaction with amine groups. DEND-GL—CDI-activated substrates were shaken for 5 h at room temperature in 0.15 M sodium carbonate buffer, pH9.6, containing 37.5μM amine-terminated dendrimers and 1 M NaCl. Following amino-dendrimer conjugation, substrates were shaken for 2 h at room temperature in 1× PBS, pH7.4, containing 10 mM glutaraldehyde and 10 mM sodium cyanoborohydride, followed by rinsing in deionized water and drying under filtered air. The DEND-GL-functionalized surface had aldehyde functionality, which can react with amine groups in a reductive amination reaction to form a secondary amine linkage. GOPS—Clean substrates were shaken in 2% GOPS in anhydrous toluene, containing 0.2% TEA, for 1 h at room temperature, after which they were rinsed in toluene to remove adsorbed silane, then isopropanol, methanol. GOPS-treated substrates were cured for 2 h at 80°C to cross-link the monolayer. The GOPS-functionalized surface possessed an epoxide functionality, which forms a secondary amine bond upon reaction with amine groups. APTMS—Clean substrates were shaken in 2% APTMS dissolved in 95% ethanol (5% water) for 10 min at room temperature, followed by rinsing in ethanol to remove adsorbed silane, and finally deionized water. As with GOPS treatment, APTMS-treated substrates were cured for 2 h at 80°C for cross-linking. APTMS-GL—Cured, APTMS-treated substrates were shaken for 2 h at room temperature in 1× PBS, pH7.4, containing 10 mM glutaraldehyde and 10 mM sodium cyanoborohydride, followed by rinsing in deionized water and drying under filtered air. The APTMS-GL-functionalized surface had aldehyde functionality, which can react with amine groups in a reductive amination reaction to form a secondary amine linkage. APTMS-DEND—Cured, APTMS-treated substrates were shaken for 2 h at room temperature in 50 mM MES buffer, pH6.8, containing 37.5μM carboxylic-acid-terminated dendrimers, 50 mM EDC, 5 mM NHS, and 0.5 M NaCl, followed by rinsing in deionized water and drying under filtered air. The APTMS-DEND-functionalized surface possessed a carboxylic acid active group, which can be activated by EDC/NHS to form an active intermediate which will form an amide bond upon reaction with amine groups.
Fig. 1.
Schematic of bioconjugation chemistries
Immobilization of DNA onto functionalized substrates
Although pH and use of specific bioconjugation reagents vary among the different immobilization chemistries, factors such as DNA concentration, salt concentration, ionic strength of buffer, reaction time, and spot size were kept constant in order to allow direct comparison of immobilization density between the various methods. Because the different functionalization procedures yielded different water contact angle (see “Results and discussion” below) spot size was maintained through the use of a PDMS mask (1.91-mm holes punched in a sheet of PDMS). A schematic of the use of PDMS to maintain DNA spot size is provided in the Electronic supplementary material. For each conjugation, 2.5μl of DNA conjugation solution (detailed below) was spotted onto the substrate, confined within a PDMS well, and allowed to react at room temperature in the dark. To prevent drying during conjugation, substrates were stored over a moist filter round in a petri dish sealed with parafilm. After 7 h, DNA conjugation solutions were withdrawn from the PDMS wells, and the wells were rinsed twice with deionized water before being shaken for 5 min in rinse 1, rinse 2, rinse 3, and finally 1× PBS to remove non-covalently bound DNA. CDI—DNA capture probes were diluted to 10μM in 0.15 M sodium carbonate buffer, pH9.6, containing 1.0 M NaCl. DEND-GL— DNA capture probes were diluted to 10μM in 0.15 M sodium carbonate buffer, pH9.6, containing 10 mM sodium cyanoborohydride and 1.0 M NaCl. GOPS—DNA capture probes were diluted to 10μM in 0.15 M sodium carbonate buffer, pH9.6, containing 1.0 M NaCl. APTMS-GL—DNA capture probes were diluted to 10μM in 0.15 M sodium carbonate buffer, pH9.6, containing 10 mM sodium cyanoborohydride and 1.0 M NaCl. APTMS-DEND—Surface carboxylic acids were first activated in 50 mM MES, pH6.0, containing 50 mM EDC, 5 mM NHS, and 0.5 M NaCl for 15 min at room temperature. Then, DNA capture probes were diluted to 10μM in 0.15 M phosphate buffer, pH7.4, containing 1.0 M NaCl.
Microfluidic hybridization assays
After functionalization of the substrates in bulk and immobilization of the DNA probes by confinement in PDMS wells, microfluidic hybridization assays were performed. Hybridization of synthetic target probes was conducted at room temperature under microfluidic flow, in a PDMS channel (1 mm × 2 cm × 45μm) prepared by casting a 1:10 ratio of curing agent to elastomer base over a positive relief master [31]. A schematic of the microfluidic channel is provided in the Electronic supplementary material. All reagents were withdrawn through the channel at 2μl/min by a syringe pump. Hybridization buffer was drawn through the channel for 15 min to precondition the channel and substrate, after which target probes (diluted to 10μM in hybridization buffer) were drawn through the channel for 15-min hybridization. After emptying the channel of hybridization mixture, rinse 1, rinse 2, and finally rinse 3 were withdrawn through the channel for 5 min each. PDMS fluidics were removed, the surface was air-dried, and fluorescence images were captured as described below.
Surface analysis
Contact angle
Water contact angles of functionalized substrates (averages of n=4 determinations) were determined using a VCA Optima contact angle analyzer (AST Products, Billerica, MA, USA). Contact angle was determined at each step in the surface functionalization prior to DNA immobilization.
FTIR spectroscopy
FTIR spectroscopy was conducted on silane-treated substrates to ensure proper deposition of the APTMS and GOPS silane monolayers. FTIR spectroscopy was conducted on a Vertex 80v vacuum FTIR spectrometer using the VeeMAX™ II accessory equipped with a germanium ATR crystal and a zinc selenide polarizer set to parallel (p) polarization (Bruker Optik GmbH, Ettlingen, Germany). After taking a background spectra of piranha-cleaned silicon wafer, spectra of silicon wafers functionalized with APTMS or GOPS silane monolayers were collected using 256 scans with 4 cm−1 at 60° angle of incidence and were analyzed with KnowItAll® Informatics System 7.9 (Bio-Rad, Hercules, CA, USA).
Fluorescence microscopy
Fluorescence images were taken of immobilized capture probes and hybridized target probes using a SensiCam CCD camera (Cooke Corporation, Eugene, OR, USA) through a ×4 objective. Image acquisition parameters were kept consistent in order to allow comparisons of fluorescence intensity between samples (250 ms, 1320 white point, 1520 black point for TAMRA modified capture probes; 250 ms, 500 white point, 100 black point for fluorescein modified target probes). Fluorescence intensities were determined using ImageJ Image Analysis program (National Institutes of Health, USA) and were converted to picomole per square centimeter through use of a standard curve made from dilutions of capture and target probe DNA in deionized water, spotted onto glass slides, dried, and imaged as described above. Initial immobilization density (number of immobilized capture probes per unit area before microfluidic flow), final immobilization density (number of immobilized capture probes per unit area after microfluidic flow), and hybridization density (number of hybridized target probes per unit area) were normalized by subtracting fluorescence of regions of functionalized substrates with no immobilized probes. Hybridization efficiencies were calculated as the ratio of hybridization density with respect to final immobilization density (after microfluidic flow).
Results and discussion
Surface analysis
Contact angle
Contact angles provided rapid confirmation of successful surface modification at each stage of the surface functionalization depicted in Fig. 1 (Table 2). Piranha-cleaned substrates were fully wetting, indicating a clean surface with available silanol groups. CDI activation only slightly increased the contact angle as the imidazole carbamate functionalization contributes only slightly hydrophobic character. Immobilization of amino-terminated dendrimers onto CDI-activated substrates, along with the subsequent immobilization of glutaraldehyde, further raised the contact angle due to the ethylenediamine core of the dendrimer molecule and the alkane core of the glutaraldehyde. APTMS and subsequent glutaraldehyde treatments presented a somewhat hydrophobic character, as expected from the alkane linkages in both molecules. Immobilization of carboxy-terminated dendrimers onto APTMS-treated substrates regenerated the hydrophilic character of the substrate. It is interesting to note that the amine-and carboxylic-acid-terminated dendrimers present similarly wetting surfaces, despite the difference in hydrophilicity of the initial functionalization (amino-dendrimers were conjugated to fully wetting CDI-activated surfaces, and carboxy-dendrimers were conjugated to the hydrophobic APTMS-treated surfaces). It is likely that the relatively large size of the dendrimers (~5.4 nm diameter [32]) combined with the short time between placing the water droplet on the surface and taking the measurement (1–2 s) prevented the underlying surface chemistry from contributing to the measured wettability. Finally, treatment with GOPS generated the most hydrophobic of the surfaces, as expected since it possesses hydrophobic alkane core, similar to APTMS, but without the ionizable terminal group.
Table 2.
Contact angle of control and functionalized substrates
| Treatment | (Abbr.) | Contact anglea |
|---|---|---|
| Piranha-cleaned silicon | (PIRANHA) | 4.85±0.30 |
| CDI-activated silicon | (CDI) | 9.33±0.48 |
| Amino-dendrimer immobilized via CDI | (DEND) | 13.58±0.93 |
| Glutaraldehyde onto amino-dendrimer | (DEND-GL) | 22.75±1.56 |
| GOPS-treated | (GOPS) | 70.68±0.28 |
| APTMS-treated | (APTMS) | 43.78±2.16 |
| Glutaraldehyde onto APTMS | (APTMS-GL) | 58.45±0.86 |
| Carboxy-dendrimer onto APTMS | (APTMS-DEND) | 13.68±0.34 |
Values represent means of four determinations±standard error
FTIR spectroscopy
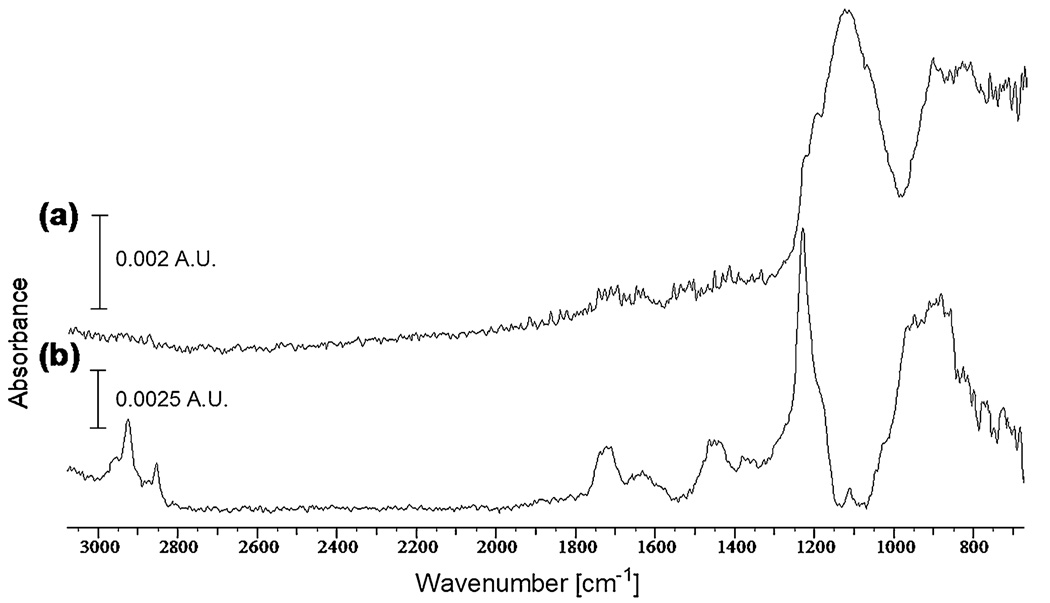
In order to obtain detailed information on the chemistry of the immobilized APTMS and GOPS monolayers and confirm the reaction chemistry as depicted in Fig. 1, ATRFTIR spectroscopy was performed (Fig. 2). Assignment of infrared bands was based on values reported in the literature [33–35] as well as those provided in the KnowItAll® spectral analysis software. The presence of Si–O–Si bands at 1,130-1,040 cm−1 in both spectra suggests successful cross-linking of the silane monolayer. In the GOPS spectra (Fig. 2a), the strong absorbance between 1,080 and 1,180 cm−1 is representative of the Si–C band, as well as the aliphatic ether band. The high intensity of this band, relative to the others, suggests the formation of a GOPS multilayer. The aromatic ether band at 1,220–1,260 cm−1 corresponds to the epoxy functionality of the silane. In the APTMS spectra (Fig. 2b), a strong Si–O stretching band at ~1,230 cm−1 suggests formation of the siloxane linkage. The sharp, but less intense, bands around 2,850 and 2,925 cm−1 correspond to the C–H stretching vibrations of the three carbon linkage within the APTMS silane. The presence of an absorbance band ~1,630 cm−1 represents the primary amine functionality of the silane.
Fig. 2.
FTIR spectra of silanetreated substrates (a) GOPS and (b) APTMS
Immobilization density
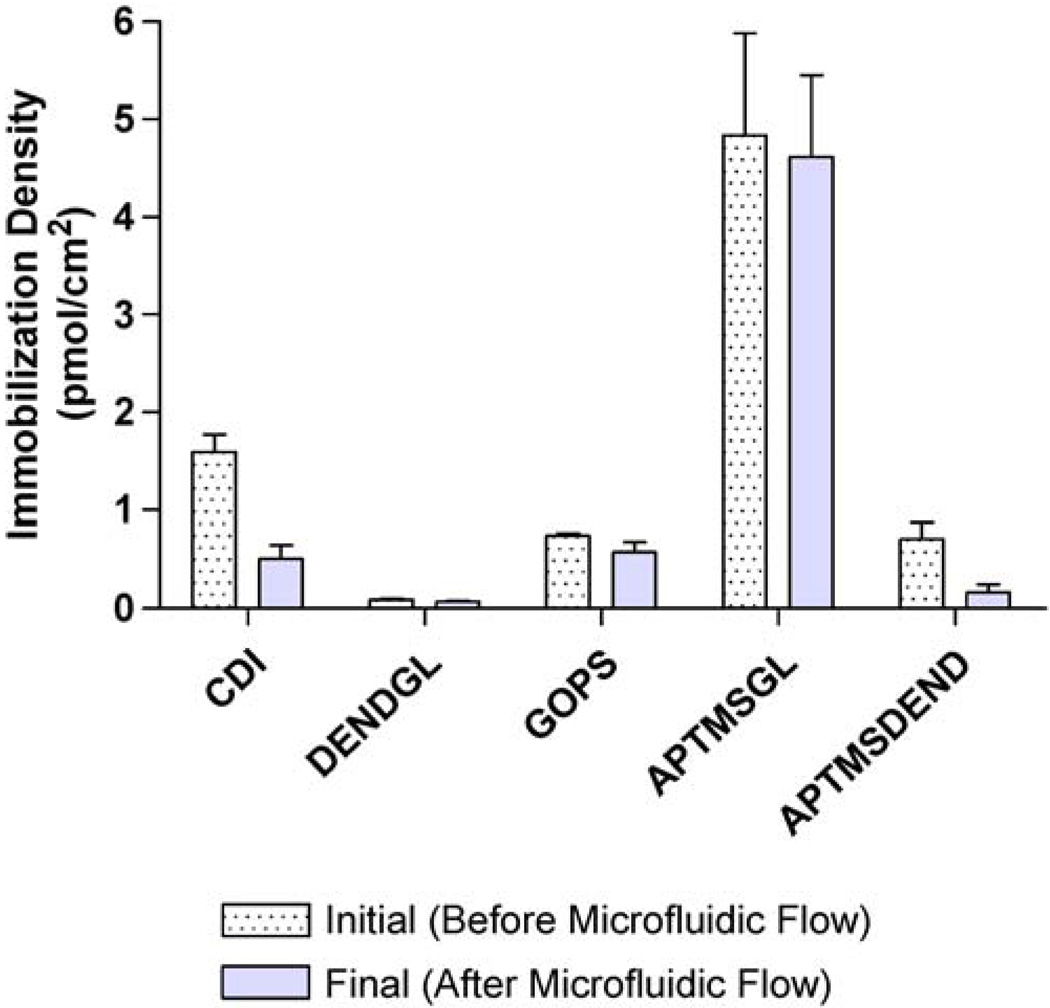
The density of immobilized capture probes in picomole per square centimeter was determined by comparison of fluorescence images to a standard curve prepared at the same exposure, gain, white, and black points (Fig. 3). Because each dendrimer molecule possesses 128 reactive functional groups, dendrimers provide the potential for increased surface functionalization per unit area in comparison to a silane monolayer. However, their charged nature can prevent high packing onto the silicon substrate and interfere with capture probe immobilization. Such repulsion may have contributed to the low capture probe immobilization density of both DEND-GL and APTMS-DENDfunctionalized substrates (0.070±0.01 and 0.169±0.08 pmol/cm2, respectively). Despite using a high saline concentration during dendrimer and DNA conjugation, it is possible that charge-charge repulsion of the dendrimer core tertiary amines prevented maximum packing density, thus reducing optimal capture probe immobilization densities. Utilization of cloud point conditions during dendrimer immobilization may improve packing density and resulting DNA capture probe immobilization yields [36, 37]. GOPSand CDI-functionalized substrates provided similar immobilization densities (0.577±0.10 and 0.509±0.14 pmol/cm2, respectively). Interestingly, the greatest immobilization density (4.617±0.83) was achieved by functionalization with APTMS-GL. It was expected that CDI, GOPS, and APTMS-GL would provide similar immobilization densities since none of these strategies utilized a branched, polyfunctional, or dendritic tether. However, glutaraldehyde has been reported to polymerize to a number of molecular structures in aqueous solution [38–40], which may have lead to immobilization of a branched, polyaldehyde tether rather than the native bifunctional molecule (Fig. 1). Such polymerization during the gluteraldehyde functionalization step would enable the observed increase in immobilization yield.
Fig. 3.
Immobilization density of capture probes before (dotted) and after (solid) microfluidic flow. Values are means of at least four determinations with error bars representing standard error
Hybridization density
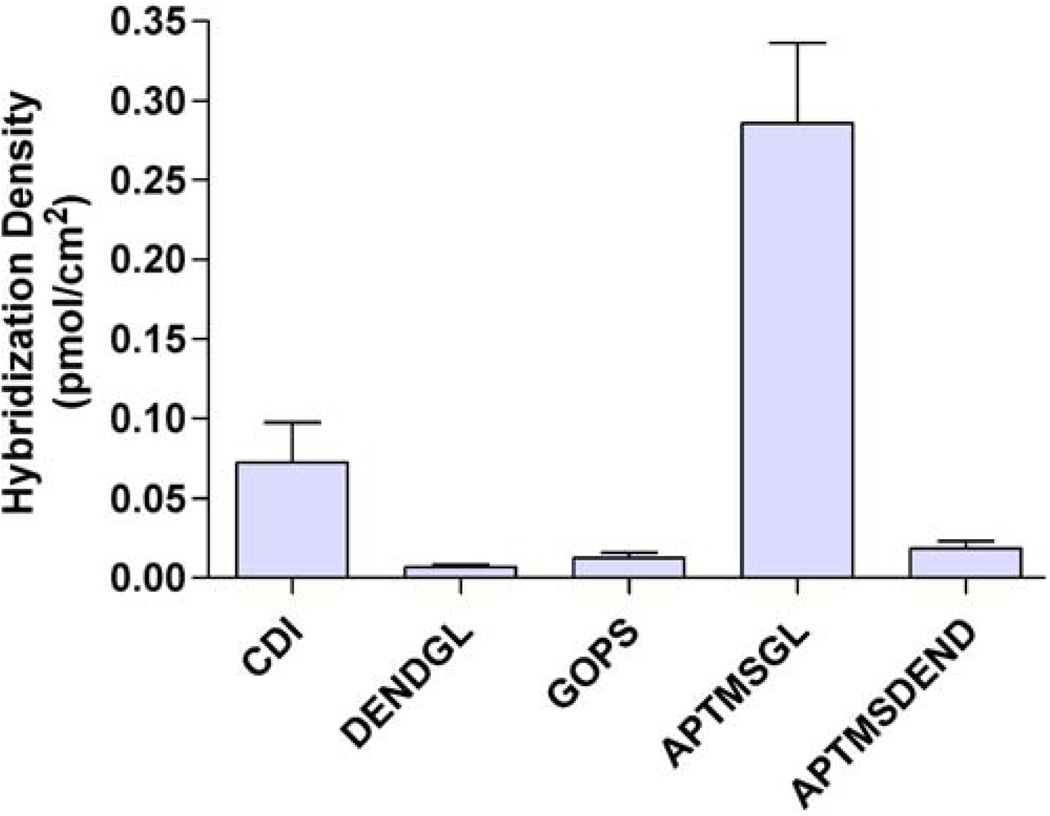
Each DNA conjugation chemistry was evaluated for its ability to hybridize synthetic oligonucleotide target probes in a microfluidic system under the same hybridization conditions. After channel preconditioning in hybridization buffer, 10μM capture probe was allowed to hybridize to the immobilized capture probes for 15 min at 2μl/min, followed by rinsing. Fluorescent images were captured and compared to a standard curve to determine the number of hybridized DNA probes per unit area, as shown in Fig. 4. As expected from the high density of immobilized capture probes, APTMS-GL-functionalized surfaces had the maximum hybridization density, hybridizing 0.286± 0.05 pmol/cm2 of target probe, while DEND-GL-and APTMS-DEND-functionalized surfaces had low hybridization densities (0.007±0.002 and 0.019±0.05 pmol/cm2 of hybridized target probe, respectively). Interestingly, although CDI and GOPS surfaces provided similar densities of immobilized capture probe, CDI-activated surfaces allowed much greater hybridization densities than GOPSfunctionalized surfaces (0.072±0.02 versus 0.013± 0.003 pmol/cm2). There are a few published reports describing directly linking proteins and ligands to inorganic substrates by CDI activation [12, 41]; however, to the best of our knowledge, this is the first published report immobilizing DNA directly to a silicon substrate via CDI activation. These results suggest that such a bioconjugation chemistry provides a viable alternative to the more traditional organofunctional silane.
Fig. 4.
Hybridization density of target probes. Values are means of at least four determinations with error bars representing standard error
Variations in reaction conditions (time, concentration of reagents, etc.) make it difficult to draw comparisons between immobilization and hybridization densities observed in this work to other published reports; further, densities are often reported in terms of relative fluorescence units (RFU), which can only provide relative data. In an effort to quantify immobilization and hybridization densities, we have utilized a high-sensitivity, low-light camera to capture fluorescence images and compared mean RFU to a standard curve of synthetic DNA probe at varying concentrations at the same camera settings. The immobilization densities observed in this work are of the same order of magnitude as other reports, in which ssDNA has been immobilized in densities ranging from ~100 fmol/cm2 to ~20 pmol/cm2 [4, 5, 42, 43]. By directly observing the fluorescently tagged DNA immobilized on or hybridized to the substrate rather than performing a wash-off or cleavage assay, losses due to experimental error and photobleaching can be avoided.
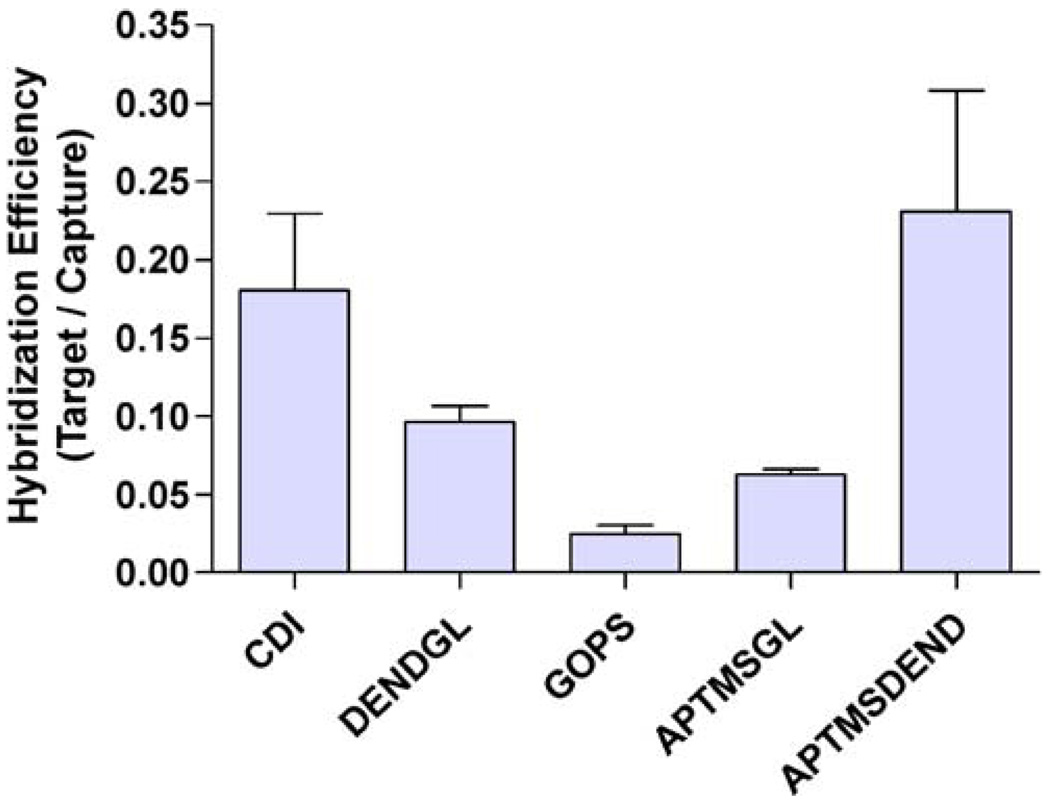
Hybridization efficiency
As described in the “Introduction” section, while density of hybridized target probe is an important factor to consider in selecting a bioconjugation chemistry, hybridization efficiency is also an important parameter in biosensor design. Hybridization efficiencies were determined by taking the ratio of hybridized target probes to immobilized capture probes, the results of which are shown in Fig. 5. In order of highest hybridization efficiency to lowest, surface functionalization chemistries ranked APTMS-DEND (highest efficiency)> CDI>DEND-GL>APTMS-GL>GOPS (lowest efficiency). It is not surprising that the chemistries that incorporated dendrimers provided the greatest hybridization efficiencies not only because of their proven ability to reduce steric hindrance by bringing the hybridization event into a solution-like state [16, 17] but also because they had the lowest densities of immobilized capture probe, therefore further reducing steric hindrance. APTMS-GL surfaces provided greater hybridization efficiency than GOPS, again suggesting the formation of branched glutaraldehyde polymers. It has been reported that functionalization via a polymer brush yields greater hybridization efficiency than functionalization by a self-assembled monolayer of the same functionality [18], further supporting this conclusion. Interestingly, DNA immobilized by CDI activation had the second highest hybridization efficiency. While the mechanism for such high hybridization efficiency is unclear, this result again points to the utility of CDI as a novel DNA conjugation strategy for microfluidic biosensors.
Fig. 5.
Hybridization efficiency. Values are means of at least four determinations with error bars representing standard error
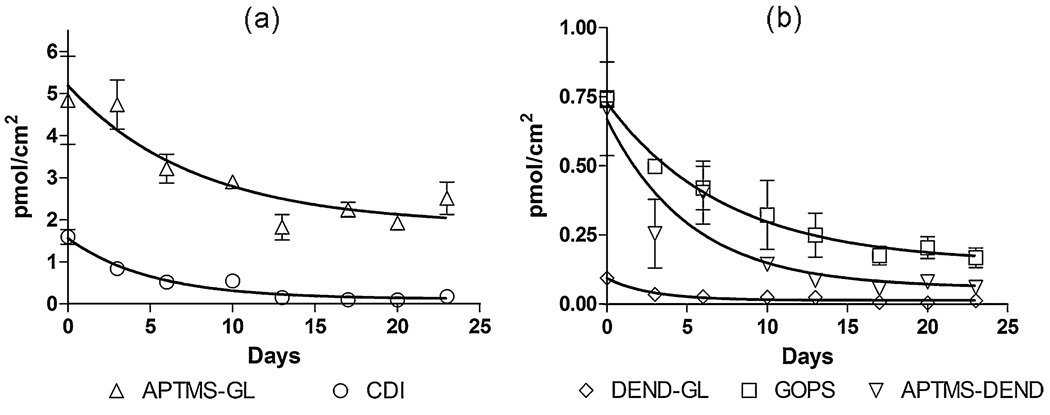
Probe stability in aqueous environments
The stability of DNA probes immobilized onto silicon substrates is critical to the design of robust biosensors that are intended for repeated use or continuous monitoring. In addition to nucleic acid sensing applications, stability of biomolecules (e.g., proteins, peptides, growth factors) onto inorganic substrates is of great importance to the biomedical field in development of bioactive implanted devices, which require extended stability in vivo. Unfortunately, it has been reported that silane monolayers do not have longterm stability under extended aqueous storage [7, 8]. We have monitored the stability of DNA capture probes immobilized onto glass substrates by the conjugation techniques reported above (Fig. 6). Functionalized slides were stored for 1 month at room temperature in 1× PBS and were monitored twice weekly. To prevent photobleaching, each substrate was evaluated only two times during the course of the study, and substrates were stored in a dark environment. Density of immobilized capture probes over time was fitted to an exponential decay model to compare rates of probe loss (Table 3). The equation for the exponential decay model used to assess stability of the immobilized probes is presented in Eq. 1, in which y is the amount of immobilized capture probe (in picomole per square centimeter), K is the decay rate constant, t is time (in days), B is the plateau value, and the sum of A and B is the initial amount of immobilized capture probe (in picomole per square centimeter). Relative stabilities of the evaluated silicon functionalization chemistries were determined by comparison of decay rate constants and were ranked as follows in order of decreasing stability: APTMS-GL (highest stability)>GOPS>APTMS-DEND>CDI>DEND-GL (lowest stability).
| (1) |
Fig. 6.
Stability of immobilized capture probes. a APTMS-GL and CDI functionalizations, b DEND-GL, GOPS, and APTMS-DEND functionalizations. Values are means of at least three determinations with error bars representing standard error
Table 3.
Stability of immobilized capture probes
| Technique | Constant (A, pmol/cm2) | Decay rate constant (K, s−1) | Plateau (B, pmol/cm2) |
|---|---|---|---|
| CDI | 1.448 | 0.1983 | 0.115 |
| DEND-GL | 0.080 | 0.3568 | 0.013 |
| GOPS | 0.580 | 0.1359 | 0.148 |
| APTMS-GL | 3.315 | 0.1288 | 1.877 |
| APTMS-DEND | 0.610 | 0.1909 | 0.058 |
Although APTMS monolayers have been reported to have low stability in aqueous environments [7, 8], use of a glutaraldehyde tether may have improved the APTMS stability by cross-linking the surface and by converting the reactive nucleophilic amines to secondary amine linkages, contributing to the relative stability of the APTMS-GL surface. GOPS functionalization had the second greatest stability, after APTMS-GL. As with APTMS-GL, the functionalized substrate is uncharged, and the surface is therefore less subject to microenvironmental pH extremes. As noted in the “Surface analysis” section (above), there may have been multilayer formation during GOPS deposition; therefore, the observed loss in stability may be a result of delamination of the multilayer. A major factor in the instability of immobilized probes for APTMS-DEND, DEND-GL, and CDI bioconjugations is likely the presence of surface microenvironmental effects. The numerous tertiary amines that make up the PAMAM dendrimer core are reported to be charged at neutral pH [37, 44, 45]. Similarly, piranha-cleaned glass used in the CDI linkage presents a charged surface, resulting in local pH extremes and increased rates of hydrolytic cleavage. Because conjugation with CDI involves a direct linkage of DNA to the glass substrate, its stability is highly dependent on nanoscale surface impurities. Although substrates were aggressively cleaned and were handled aseptically during treatment, deposition of an adventitious carbon layer may have also contributed to the poor stability.
Stability under microfluidic flow
Because our interest is in the development of microfluidic biosensors, we further evaluated the stability of the immobilized capture probes after subjecting them to 45 min of microfluidic flow. The density of immobilized capture probe before and after exposure to microfluidic flow conditions are reported in Fig. 3. The percent of immobilized capture probe remaining after microfluidic flow was determined by comparing immobilization density before and after microfluidic flow. Shear forces occurring at the surface of the microfluidic biosensor can induce mechanical breakage of the bonds within the DNA probe [46], as well as between the immobilized DNA and the functionalized substrate. Such shear stress can also result in delamination of silane monolayers, further reducing microfluidic stability of immobilized biomolecules. Shear rate, σ, can be defined as the change in velocity in the direction of fluid flow, νx, with respect to channel height at the channel surface (y=0) (Eq. 2) [47]. To quantify the forces acting on the biofunctionalized surfaces during microfluidic flow, shear rate was calculated by applying boundary conditions of Poiseuille flow (pressure-driven flow in a slot), in which Q is the flow rate (2μL/min), w is the channel width (1 mm), and h is the channel height (45μm).
| (2) |
Shear rate was determined to be 98.8 s−1, which when applied to the various functionalized surfaces for 45 min resulted in the following ranking of relative stabilities APTMS-GL (highest stability)>GOPS>DEND-GL>CDI> APTMS-DEND (lowest stability). This level of shear rate can also affect hybridization density and therefore efficiency. These values can be improved by modifying flow rate, channel dimensions, and hybridization buffer components. After 45-min microfluidic flow, CDI-functionalized substrates retained 32.0% of immobilized capture probe, while DEND-GL-and GOPS-functionalized substrates retained 74.5% and 77.8% immobilized capture probe, respectively. APTMS-GL-functionalized substrates had the greatest stability to shear stress, retaining 95.4% of the immobilized capture probe, while APTMS-DEND-functionalized substrates had the poorest stability to shear stress, retaining only 23.9% of the immobilized capture probe.
In general, the observed stabilities of the various bioconjugation chemistries to microfluidic flow conditions is similar to those observed after extended storage in aqueous environments, with the exception of DEND-GL functionalization. It is possible that glutaraldehyde crosslinked the amino-terminated dendrimer, providing enhanced stability to shear stress, whereas the major factor in the reduced stability of DEND-GL surfaces over extended periods is the formation of locally high pH from the charged dendrimer core, which is unaffected by glutaraldehyde cross-linking.
Conclusions
Five DNA bioconjugation chemistries were evaluated for their use in silicon based microfluidic biosensors. A summary of the comparisons drawn is as follows:
Immobilization density: APTMS-GL>GOPS>CDI> APTMS-DEND>DEND-GL
Hybridization density: APTMS-GL>CDI>APTMSDEND> GOPS>DEND-GL
Hybridization efficiency: APTMS-DEND>CDI>DENDGL> APTMS-GL>GOPS
Stability to shear stress: APTMS-GL>GOPS>APTMSDEND> CDI>DEND-GL
Storage stability: APTMS-GL>GOPS>DEND-GL> CDI>APTMS-DEND
Although dendrimer tethers allowed for enhanced hybridization efficiency, as has been previously reported [16, 17], they did not increase density of immobilized capture probe, as was expected. Further, their poor stability to both microfluidic flow conditions and storage in buffered aqueous solution suggests that while they are advantageous in microarray applications, their suitability in microfluidic biosensors may be limited. Conjugation by APTMS-GL yielded the greatest immobilization and hybridization densities, as well as the greatest stability to both shear stress and extended storage in aqueous environment. Although it does not offer the stability of APTMS-GL-functionalized substrates, linkage of DNA capture probes to silicon substrates by CDI activation represents a promising bioconjugation technique in terms of density of immobilized probes and hybridization efficiency. CDI is a zero-length cross-linker, which activates carboxylic acids and hydroxyl groups to an intermediate that is reactive to nucleophiles, and considerably more stable than water soluble carbodiimide cross-linkers, thus allowing longer conjugation times in aqueous environments. Although it is widely used in peptide synthesis, biomolecule labeling, and immobilization of proteins onto organic and inorganic solid supports [41, 48–50], direct linkage of DNA to silicon substrates by CDI activation is an emerging bioconjugation technique. This chemistry could be particularly useful in the fields of label-free biosensing, in which highest device sensitivities are achieved when the biorecognition event occurs in close proximity to the sensing surface.
Important considerations in ensuring stability of silane monolayers includes proper cleaning of surface, formation of single monolayer, complete cross-linking of monolayer by curing after deposition, and minimizing hydrolysis of siloxane linkages by nucleophilic terminal groups. To that end, preventing the generation of GOPS multilayers through use of anhydrous conditions [51] may improve stability by avoiding delamination within the multilayer as was observed in this work. Further, glutaraldehyde provided improved stability both by further cross-linking the APTMS monolayer and by converting the nucleophilic primary amines to secondary amines, thus preventing nucleophilic attack on the siloxane linkage.
Although our focus was on isothermal microfluidic devices, future work evaluating thermal stability would provide useful additional information regarding suitability of these bioconjugation chemistries in thermocycling microfluidic applications such as polymerase chain reaction. Our primary interest regarding stability under shear stress is in the development of microfluidic biosensors; however, understanding the relative stability of these bioconjugation chemistries is important in a number of other nanobiotechnology applications, including development of vascular prostheses, drug delivery systems, and other implanted biomedical devices.
Supplementary Material
The online version of this article (doi:10.1007/s00216-009-2731-y) contains supplementary material, which is available to authorized users.
Acknowledgments
Support for this work was provided by the National Institutes of Health-National Institute of Biomedical Imaging and Bioengineering (NIH-NIBIB) under grant number R21EB007031. This work made use of STC shared experimental facilities supported by the National Science Foundation under Agreement No. ECS-9876771. The authors gratefully acknowledge Dr. Sam Nugen and Prof. Antje Baeumner for technical assistance in determining hybridization conditions and Sudeep Mandal for preparation of PDMS master.
Contributor Information
Julie M. Goddard, Email: jmg26@cornell.edu.
David Erickson, Email: de54@cornell.edu.
References
- 1.Erickson D, Liu XZ, Venditti R, et al. Anal Chem. 2005;77:4000–4007. doi: 10.1021/ac050236r. [DOI] [PubMed] [Google Scholar]
- 2.Ng JKK, Liu WT. Anal and Bioanal Chem. 2006;386:427–434. doi: 10.1007/s00216-006-0552-9. [DOI] [PubMed] [Google Scholar]
- 3.Dawson E, Moore C, Smagala J, et al. Anal Chem. 2006;78:7610–7615. doi: 10.1021/ac061739f. [DOI] [PubMed] [Google Scholar]
- 4.Zammatteo N, Jeanmart L, Hamels S, et al. Anal Biochem. 2000;280:143–150. doi: 10.1006/abio.2000.4515. [DOI] [PubMed] [Google Scholar]
- 5.Wang YY, Prokein T, Hinz M, et al. Anal Biochem. 2005;344:216–223. doi: 10.1016/j.ab.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 6.Fritz J, Baller MK, Lang HP, et al. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 7.Wang AF, Tang HY, Cao T, et al. J Colloid Interface Sci. 2005;291:438–447. doi: 10.1016/j.jcis.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Szczepanski V, Vlassiouk I, Smirnov S. J Membr Sci. 2006;281:587–591. [Google Scholar]
- 9.Anker JN, Hall WP, Lyandres O, et al. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 10.Vollmer F, Arnold S. Nat Methods. 2008;5:591–596. doi: 10.1038/nmeth.1221. [DOI] [PubMed] [Google Scholar]
- 11.Mandal S, Erickson D. Opt Express. 2008;16:1623–1631. doi: 10.1364/oe.16.001623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aretaki IN, Koulouridakis P, Kallithrakas-Kontos N. Anal Chim Acta. 2006;562:252–257. [Google Scholar]
- 13.Pathak S, Singh AK, McElhanon JR, et al. Langmuir. 2004;20:6075–6079. doi: 10.1021/la036271f. [DOI] [PubMed] [Google Scholar]
- 14.Keighley SD, Li P, Estrela P, et al. Biosens Bioelectron. 2008;23:1291–1297. doi: 10.1016/j.bios.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Lee CY, Nguyen PCT, Grainger DW, et al. Anal Chem. 2007;79:4390–4400. doi: 10.1021/ac0703395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southern E, Mir K, Shchepinov M. Nat Genet. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- 17.Shchepinov MS, Case-Green SC, Southern EM. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirri G, Chiari M, Damin F, et al. Anal Chem. 2006;78:3118–3124. doi: 10.1021/ac0521091. [DOI] [PubMed] [Google Scholar]
- 19.Le Berre V, Trevisiol E, Dagkessamanskaia A, et al. Nucleic Acids Res. 2003;31:e88. doi: 10.1093/nar/gng088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatnagar P, Mark SS, Kim I, et al. Adv Mater. 2006;18:315–319. [Google Scholar]
- 21.Benters R, Niemeyer CM, Drutschmann D, et al. Nucleic Acids Res. 2002;30:e10. doi: 10.1093/nar/30.2.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caminade AM, Padie C, Laurent R, et al. Sensors. 2006;6:901–914. [Google Scholar]
- 23.Cote GL, Lec RM, Pishko MV. IEEE Sens J. 2003;3:251–266. [Google Scholar]
- 24.Erickson D, Li DQ, Krull UJ. Anal Biochem. 2003;317:186–200. doi: 10.1016/s0003-2697(03)00090-3. [DOI] [PubMed] [Google Scholar]
- 25.Becker H, Locascio LE. Talanta. 2002;56:267–287. doi: 10.1016/s0039-9140(01)00594-x. [DOI] [PubMed] [Google Scholar]
- 26.Bange A, Halsall HB, Heineman WR. Biosens Bioelectron. 2005;20:2488–2503. doi: 10.1016/j.bios.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Ros A. Anal Bioanal Chem. 2008;390:799–800. doi: 10.1007/s00216-007-1758-1. [DOI] [PubMed] [Google Scholar]
- 28.Tegenfeldt JO, Prinz C, Cao H, et al. Anal Bioanal Chem. 2004;378:1678–1692. doi: 10.1007/s00216-004-2526-0. [DOI] [PubMed] [Google Scholar]
- 29.Wei CW, Cheng JY, Huang CT, et al. Nucleic Acids Res. 2005;33:e78. doi: 10.1093/nar/gni078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dittrich PS, Tachikawa K, Manz A. Anal Chem. 2006;78:3887–3907. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 31.Duffy DC, McDonald JC, Schueller OJA, et al. Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 32.PAMAM Dendrimers. Midland, MI: Dendritech; 2008. [Accessed 08 Dec 2008]. http://www.dendritech.com/pamam.html. [Google Scholar]
- 33.Coates J. In: Encyclopedia of analytical chemistry. Meyers RA, editor. Chichester: Wiley; 2000. [Google Scholar]
- 34.Kempfert KD. Spectroscopy. 2007;22:22–23. [Google Scholar]
- 35.Kurth DG, Bein T. Langmuir. 1995;11:578–584. [Google Scholar]
- 36.Parrott MC, Valliant JF, Adronov A. Langmuir. 2006;22:5251–5255. doi: 10.1021/la0529355. [DOI] [PubMed] [Google Scholar]
- 37.Haba Y, Harada A, Takagishi T, et al. J Amer Chem Soc. 2004;126:12760–12761. doi: 10.1021/ja047755g. [DOI] [PubMed] [Google Scholar]
- 38.Kawahara J, Ohmori T, Ohkubo T, et al. Anal Biochem. 1992;201:94–98. doi: 10.1016/0003-2697(92)90178-a. [DOI] [PubMed] [Google Scholar]
- 39.Whipple EB, Ruta M. J Org Chem. 1974;39:1666–1668. [Google Scholar]
- 40.Migneault I, Dartiguenave C, Bertrand MJ, et al. Bio-Techniques. 2004;37:790–802. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 41.Saiyed ZM, Sharma S, Godawat R, et al. J Biotechnol. 2007;131:240–244. doi: 10.1016/j.jbiotec.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Pal S, Kim MJ, Song JM. Lab Chip. 2008;8:1332–1341. doi: 10.1039/b804066d. [DOI] [PubMed] [Google Scholar]
- 43.Steel AB, Levicky RL, Herne TM, et al. Biophys J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu YH, Sun L, Crooks RA. Macromolecules. 2003;36:5725–5731. [Google Scholar]
- 45.Cakara D, Kleimann J, Borkovec M. Macromolecules. 2003;36:4201–4207. [Google Scholar]
- 46.Quail M. DNA: mechanical breakage. Encyclopedia of life sciences. Chichester: John Wiley & Sons, Ltd; 2005. [Google Scholar]
- 47.Panton RL. Incompressible flow. 2nd edn. . New York: Wiley; 1996. [Google Scholar]
- 48.Hermanson GT. Bioconjugate techniques. New York: Academic; 1996. [Google Scholar]
- 49.Lee HJ, Nedelkov D, Corn RM. Anal Chem. 2006;78:6504–6510. doi: 10.1021/ac060881d. [DOI] [PubMed] [Google Scholar]
- 50.Bencina M, Babic J, Podgornik A. J Chromatogr. A. 2007;1144:135–142. doi: 10.1016/j.chroma.2006.12.083. [DOI] [PubMed] [Google Scholar]
- 51.Wong AKY, Krull UJ. Anal Bioanal Chem. 2005;383:187–200. doi: 10.1007/s00216-005-3414-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online version of this article (doi:10.1007/s00216-009-2731-y) contains supplementary material, which is available to authorized users.