Abstract
We have documented that exposure of rhesus monkeys to house dust mite aeroallergen during postnatal development resulted in significant recruitment of eosinophils into the airway mucosa (Clin Exp Allergy 33:1686–1694, 2003). Because eosinophils were not uniformly distributed throughout the five conducting airway generations examined, we speculated that trafficking within anatomic microenvironments of the lung is mediated by differential chemokine expression. To address this question, we used quantitative real-time RT-PCR to evaluate the related eosinophilic chemokines, eotaxin (CCL11), eotaxin-2 (CCL24), and eotaxin-3 (CCL26) within isolated airways of infant monkey lung. Overall, chemokine mRNA expression levels in house dust mite–exposed airways were as follows: eotaxin-3 > eotaxin > eotaxin-2. Immunofluorescence staining for eotaxin-3 and CC chemokine receptor 3 (CCR3) showed positive cells within epithelium and peripherally located nerve fiber bundles of the airway wall. Epithelial volume of eotaxin-3 within the trachea correlated with epithelial volume of major basic protein. CCR3+ and MHC Class II+ dendritic cells, but not eosinophils or mast cells, co-localized within eotaxin-3+ nerve fiber bundles. We conclude that localized expression of eotaxin-3 plays an important role in the recruitment of diverse CCR3+ cell populations to different anatomic microenvironments within the infant airway in response to chronic allergen exposure.
Keywords: lung, development, chemokine, eosinophil
Because eosinophilic infiltration is a cardinal feature of asthma, it has long been speculated that the eosinophil is a key effector cell in the pathogenesis of this disease in humans (1, 2). Eosinophils are a potent cellular source of many cytokines and mediators of allergy, including eosinophilic cationic protein, major basic protein, eosinophilic-derived neurotoxin, interleukin (IL)-2, IL-3, IL-4, IL-5, granulocyte macrophage colony-stimulating factor, IL-6, IL-10, interferon-γ, transforming growth factor (TGF)-α, TGF-β, leukotrienes, and prostaglandins (reviewed in Ref. 3). Despite the characteristic association of eosinophils with the allergic asthma phenotype, their role in directly mediating the development of reactive airways remains controversial. In animal models, direct administration of major basic protein can directly induce airway hyperresponsiveness, and inhibition of IL-5 (a cytokine essential for eosinophil differentiation) can inhibit airway hyperresponsiveness (4–7). However, recent clinical trials using a humanized monoclonal antibody against IL-5 (mepolizumab) showed no attenuation of airway hyperresponsiveness, despite significant reductions in circulating and lavage eosinophils; this outcome may, in part, be explained by the inability of IL-5 depletion methods to completely eliminate tissue eosinophils and major basic protein (8, 9). Treatment of individuals with asthma with anti–IL-5 therapy does significantly attenuate the deposition of extracellular matrix components within the reticular basement membrane and expression of TGF-β by airway eosinophils, suggesting that a major contribution of this inflammatory cell type on lung function decrement is mediation of airways remodeling (10).
Defining the effector function of airway eosinophils is further complicated in younger populations, where distinguishing between children with transient wheezing versus children with persistent wheezing (and the propensity to develop asthma) can be difficult to predict at infancy. Serum and urine eosinophil-specific proteins have been used as noninvasive measures of eosinophilia for very young children, but their value in predicting or diagnosing asthma has been limited; this may be due in part to sensitivity and reproducibility of assays (11). It has been reported that human infants with a familial history of atopy exhibit elevated levels of eosinophil cationic protein and eosinophil protein X in nasal lavages as early as 4 wk of age (12), yet levels of nasal lavage eosinophil cationic protein do not associate with respiratory symptoms at 1 yr of age (13). Direct sampling of the airways is likely to be more informative with regard to cellular profiles and effector functions, yet endoscopic assessment of normal healthy young children is often considered unethical. As such, experimental study groups consisting of infants and young children have been problematic and difficult to interpret due to the lack of appropriate controls.
Eosinophil trafficking within the lung is mediated by a complex multistep process, in which signaling via locally expressed adhesion molecules and chemokines result in the extravasation of inflammatory/immune cells into the airway mucosa and lumen. Eotaxin (CCL11) was first identified as a potent eosinophilic chemoattractant in guinea pig airways; eotaxin-2 (CCL24) and eotaxin-3 (CCL26) were later identified as functionally analogous chemokines (reviewed in Ref. 14). Although eotaxin, eotaxin-2, and eotaxin-3 do not exhibit a high degree of sequence homology (34–38%), all three chemokines signal through CC chemokine receptor 3 (CCR3), a seven-transmembrane G protein–linked receptor (15, 16). CCR3 is highly expressed by eosinophils, as well as other cell types involved in allergic inflammation. In addition to the eotaxin family, CCR3 functions as a receptor for other eosinophilic chemokines, including MEC (CCL28), RANTES (CCL5), MCP-3 (CCL7), and MCP-4 (CCL13). Eosinophils express other chemokine receptors, but CCR3 has been an attractive target for antagonistic therapy because most eosinophilic chemokines signal through this receptor.
We have previously shown that recurrent exposure of newborn rhesus monkeys to house dust mite aeroallergen (HDM) can result in the development of pulmonary and systemic immune responses at 2 mo of age (17). In addition to effects on the maturation and activation of T helper lymphocytes, significant recruitment of eosinophils into the airway mucosa was observed in response to HDM aerosols. Because the distribution of eosinophils throughout five different airway generations and compartments was not uniform in this study, we hypothesized that airway specific expression of eosinophilic chemokines contribute to spatial recruitment of eosinophils within the lung. Identification of anatomic microenvironments where specific leukocyte subpopulations preferentially traffic is of particular relevance to the development of novel therapeutic approaches that efficiently target the most affected sites within the lung during an inflammatory response. In the current investigation, we have assessed the expression of eotaxin, eotaxin-2, eotaxin-3, and CCR3 at different airway generations in the house dust mite aeroallergen–exposed infant lung. Our findings support a prominent role for eotaxin-3 in the modulation of airways inflammation in response to house dust mite exposure, by microenvironment-specific recruitment of CCR3+ eosinophils and dendritic cells into the infant lung following inhalation of aeroallergen.
MATERIALS AND METHODS
Experimental Design
Rhesus macaque (Macaca mulatta) monkeys used for this study were described previously (17). Male monkeys were housed in 4.2 m3 capacity exposure chambers within the California Regional Primate Research Center, starting at 1–2 d of age. Chambers were ventilated at a rate of 30 changes per hour with filtered air. Each exposure chamber housed three animals throughout the study. Animals were selected for the study based on physical examination and negative intradermal skin test reactivity to HDM. Intradermal skin testing was performed as previously described (18). Body weight between control and HDM-exposed animals at 2 d of age was comparable (control: mean = 0.508 ± 0.027 kg; HDM: mean = 0.510 ± 0.018 kg). Both control and HDM animal groups maintained comparable weight gain throughout the 9-wk study period (17).
One-week-old animals (n = 6) were exposed to HDM aerosols for 2 h per day, 3 d per week (Monday, Wednesday, Friday), for a total of 8 wk. Aerosols were generated with an extract of Dermatophagoides farinae (Greer Laboratories, Lenoir, NC) diluted in phosphate-buffered saline (PBS) and nebulized with a high-flow rate nebulizer as previously described (18). Protein concentration of the HDM aerosols consisted of 506 ± 38 μg/m3/d (n = 6), a concentration comparable to that previously used to induce symptoms of allergic asthma in adult rhesus monkeys (18). Control animals (n = 5) remained in filtered air for the duration of the study period. All animals were vaccinated with diptheria, tetanus, and acellular pertussis (DTaP; Darby Drug Co., Westbury, NY) at 5 wk of age. Because human infants in the United States are routinely inoculated with DtaP vaccine at 2 mo of age, animals in this study were immunized to obtain a comparable stimulation of systemic immunity. Animals were necropsied at 9 wk of age.
RNA Isolation and Quantitative Real-Time RT-PCR
Immediately after necropsy, specific airway generations were microdissected from the unfixed right caudal lobe of each monkey as previously described (19). Each airway sample was snap-frozen in liquid nitrogen, homogenized, and extracted for RNA using TRIzol reagent (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Reverse transcription was performed with random hexamer primers and MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA). Oligonucleotide primers for eotaxin-1, -2, -3 were designed using Primer Express software (Applied Biosystems), using human sequences obtained through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) (Table 1). Using SYBR green buffer (Applied Biosystems), real-time PCR analysis was performed on an Applied Biosystems PRISM 5700 Sequence Detection System, using default settings for amplification. PCR amplification with commercially available primers for 18S (Applied Biosystems) was performed for each sample to control for sample loading; sample values were normalized with 18S values according to manufacturer's instructions. There were no significant differences in 18S values for all airway generations sampled (data not shown). Analysis of relative mRNA quantitation was analyzed using the comparative Ct Method (User Bulletin #2, ABI PRISM Sequence Detection System; Applied Biosystems).
TABLE 1.
Oligonucleotide sequences used for quantitative real-time rt-pcr analysis
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| Eotaxin | CAAGACCAAACTGGCCAAGG | GAATCCTGCACCCACTTCTTCT |
| Eotaxin-2 | TTGGCGTCCAGGTTCTTCAT | TGTGGCGACCCCAAGC |
| Eotaxin-3 | ACCTGCTGCTTCCAATACAGC | CATAGCTTCGCACCCAGGTC |
Immunofluorescence Staining for Eotaxin-3, CCR3, and PGP 9.5
At necropsy, the left caudal lobe from each animal was inflated with a 50% vol/vol mixture of OCT freezing compound (Sakura Finetek, Torrance, CA) and PBS, then sliced perpendicular to the long axis of the main intrapulmonary conducting airway. Each left caudal lobe slice was numbered in sequence before freezing in OCT molds. Tissue blocks (slices) were cut in ∼ 7–8 mm thicknesses, the entire lung lobe consisted of 9–10 blocks. The trachea was also embedded for cryosectioning. Five-micrometer frozen sections from alternately numbered OCT blocks were used for immunofluorescence staining.
For eotaxin-3 protein detection, sections were stained with biotinylated goat anti-human eotaxin-3 (R&D Systems, Minneapolis, MN), followed by streptavidin ALEXA 488 (Molecular Probes, Eugene, OR). For CCR3 protein detection, sections were stained with mouse anti-rhesus CCR3 (clone 5B9) (20), followed by goat anti-mouse ALEXA 488 (Molecular Probes). Polyclonal rabbit anti-human PGP 9.5 (Biogenesis Ltd., Poole, UK) followed by goat anti-rabbit ALEXA 568 (Molecular Probes) was used for detection of neuronal cell bodies and axons (21). We have previously reported that an antibody directed against human major basic protein can detect the eosinophil phenotype in both adult and infant rhesus monkeys (17, 18). Eosinophils were detected by mouse anti-human major basic protein antibody (clone BMK-13; BIODESIGN International, Saco, ME). T lymphocytes, mast cells, and MHC class II were detected by mouse anti-rhesus CD3 (BIOSOURCE, Camarillo, CA), goat anti-CD117/c-kit (Santa Cruz Biotechnology, Santa Cruz, CA), and mouse anti-human HLA-DR (Dako, Carpenteria, CA), respectively. Purified MOPC 21 mouse IgG1 (ATCC, Manassas, VA) and goat IgG (Sigma-Aldrich, St. Louis, MO) were used as immunoglobulin controls for immunofluorescence staining. Mouse monoclonal antibodies and secondary detection reagents were used at 1 μg/ml, the rabbit anti-human PGP 9.5 was diluted to 1:1,000.
Quantitation of Eotaxin-3+ and CCR3+ Cells
Images of eotaxin-3 or CCR3 immunostained cryosections were visualized on an Olympus (Ballerup, Denmark) PROVIS fluorescence microscope at ×600, using the appropriate excitation and emission filters for the cellular labeled fluorochromes previously listed. Images were captured using a Zeiss camera (Zeiss, Oberkochen, Germany) at a resolution of 150 pixels/inch in a 1,300 × 1,130 pixel image for each field of a selected airway, using stratified sampling with a random start for each block. At least 10 stratified random fields were sampled around one airway per section. Each field contained a region of epithelium and interstitium internal to the cartilaginous ring (if present). In addition to the trachea, four cryosections from the left caudal lobe were sampled per animal, representing the most proximal portion (lobar bronchus) through the most distal portion of the intrapulmonary conducting airway (respiratory bronchiole). Captured images were imported into Stereology Toolbox software (version 1.1; Morphometrix, Davis, CA), volume density for epithelium, interstitium, and positive fluorescence was determined by point counts. The volume density of eotaxin-3 or CCR3+ cells was calculated as follows:
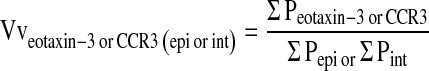 |
Using CCR3+ cells as an example, points of a 125-point grid which fell on CCR3 fluorescence-positive cells were counted (PCCR3), and those points which fell on either epithelium or interstitium were counted as the reference volume of epithelium (Pepi) or interstitium (Pint).
The surface of epithelial basal lamina per unit volume of epithelium (Svbl,epi) or interstitium (Svbl,int) was calculated on cross-sections of airways at ×100 using an Olympus BH-2 microscope with the CAST version 2.00.04 software (Olympus) as
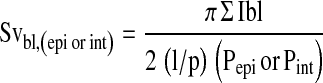 |
where l/p = length per test point on four lines oriented either horizontally or vertically in a counting frame, Ibl is the number of line intersections of the epithelial basal lamina and Pepi or Pint, the number of points that hit epithelium or interstitium, respectively. The volume of eotaxin-3 or CCR3+ cells within the epithelial or interstitial compartment per surface area of basement membrane (mm3/mm2) was then calculated as
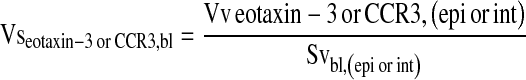 |
Data Analysis
Unless indicated, all data are reported as mean ± SE. Statistical analysis was conducted using GraphPad Prism and Instat statistical analysis software (Graphpad Software, San Diego, CA). Treatment groups and airway levels were compared using either one- or two-way ANOVA, where P < 0.05 was considered significant.
RESULTS
Expression of Eotaxin, Eotaxin-2, and Eotaxin-3 mRNA by Airway Generation
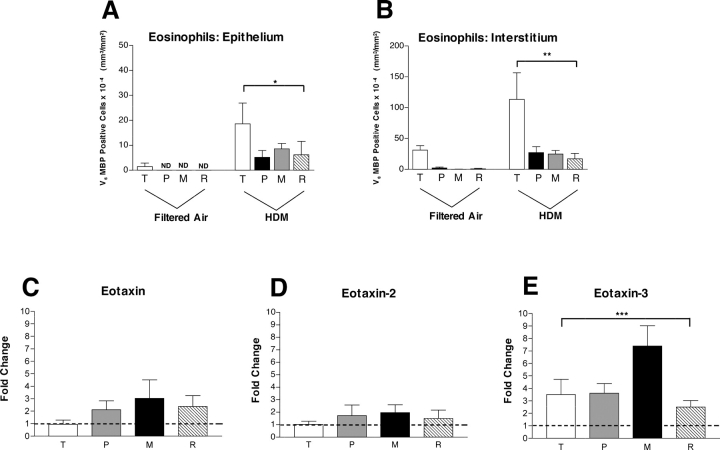
The effect of chronic aeroallergen exposure on tissue recruitment of eosinophils by airway generation in infant monkeys was determined by quantitative immunohistologic assessment of major basic protein volume within the trachea and left caudal lobe (17). Eosinophils (defined as cells that express major basic protein) were more abundant within the interstitial compartment as compared with the epithelium; however, the distribution profiles were similar: eosinophils accumulated at maximal volume within the trachea (Figures 1A and 1B). To determine if sites of eosinophil accumulation within the lung could be correlated with localized expression of one or more eosinophilic chemokines, we assessed airway samples collected from this study for expression of eotaxin, eotaxin-2, and eotaxin-3. In addition to the trachea, intrapulmonary conducting airway samples for mRNA analysis were freshly isolated from the right caudal lobe of each monkey immediately after necropsy. Conducting airways were microdissected from parenchyma; samples were collected from the most proximal portion of the intrapulmonary airway, a midlevel airway (a region containing the last generation of cartilaginous subsegmental bronchi), and respiratory bronchiole. To assess relative chemokine gene expression, each HDM-exposed monkey was compared with a pool of values from 4–5 filtered air age-matched control animals (with 2–3 replicates for each airway generation). HDM exposure had a minimal effect on mRNA expression levels for eotaxin and eotaxin-2 within the trachea (Figures 1C and 1D). Within intrapulmonary conducting airways, we observed a modest increase in eotaxin and eotaxin-2 mRNA levels in HDM-exposed infant monkeys as compared with controls; there were no significant effects by airway generation. The range of eotaxin expression levels within intrapulmonary airways was ∼ 2- to 3-fold over controls, whereas eotaxin-2 expression was lower (< 2). In contrast with eotaxin and eotaxin-2, eotaxin-3 mRNA expression within the trachea was elevated over controls (Figure 1E). Expression levels for eotaxin-3 mRNA was significantly affected by airway generation (P = 0.0285, by one-way ANOVA), peaking at ∼ 7-fold over controls within midlevel intrapulmonary airways. Overall, mRNA expression levels for eotaxin-3 in HDM-exposed monkey airways (relative to filtered air controls) were significantly greater than eotaxin (P = 0.0073, by two-way ANOVA).
Figure 1.
Effect of house dust mite exposure on gene expression levels for eotaxin, eotaxin-2, and eotaxin-3 within four different airway generations: comparison with distribution of eosinophils. (A and B) Volume of major basic protein (eosinophils) within four different airway generations of infant monkeys (filtered air, n = 5; HDM, n = 6). Columns represent the volume of major basic protein–positive cells within either epithelial (A) and interstitial (B) compartments sampled from trachea, proximal airway, midlevel airway, and respiratory bronchiole, as previously reported (17). T, trachea; P, proximal airway; M, midlevel airway; R, respiratory bronchiole. All data are represented as mean ± SEM and analyzed by two-way ANOVA. *P = 0.0012 as compared with filtered air control animals, **P = 0.0017 as compared with filtered air control animals. (C–E) Columns represent the average relative fold change of eotaxin (C), eotaxin-2 (D), and eotaxin-3 (E) mRNA for indicated genes by RT-PCR analysis using isolated airway generations from HDM-exposed infant monkeys (n = 5–6) after normalization with 18S mRNA levels, relative to filtered air control infant monkeys (n = 4–5). RT-PCR analysis for each monkey and airway generation was repeated over 2–3 experiments; control monkeys represent a pool of 12–15 separate replications by RT-PCR. The dotted line represents the point at which expression of experimental versus control is equivalent (no change in expression). *** P = 0.0285 by one-way ANOVA.
Distribution of Eotaxin-3 by Airway Generation
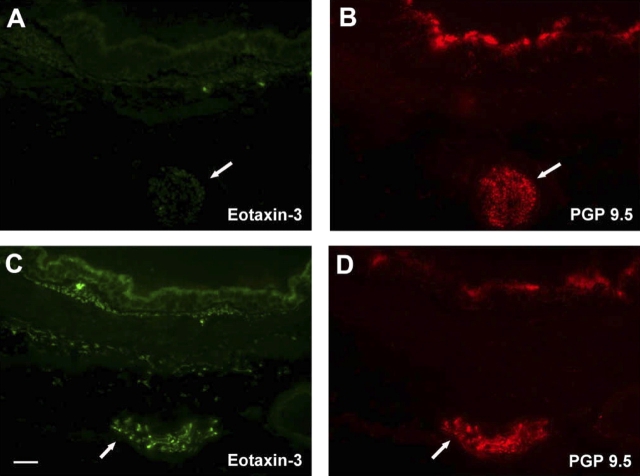
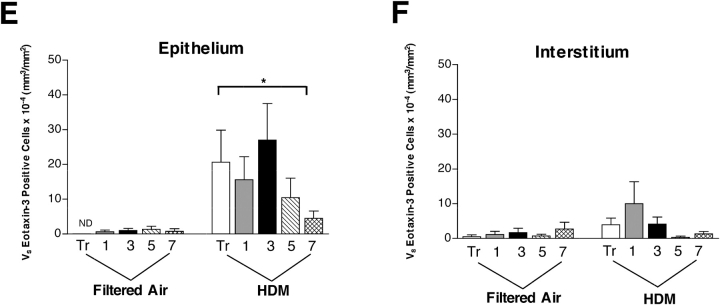
Because eotaxin-3 was the most prominently expressed eotaxin chemokine family member in response to HDM exposure, we subsequently evaluated infant monkey airways for cellular sources of eotaxin-3 protein. Immunostaining of airways from HDM-exposed monkeys with an antibody directed against eotaxin-3 showed strong immunoreactivity within the epithelium; comparatively, filtered air control animals had few positive cells within the epithelial compartment (Figure 2A). Quantitative assessment of five different airway generations confirmed a significant increase in volume of eotaxin-3–positive epithelium within HDM-exposed animals, relative to filtered air controls (P = 0.0003) (Figure 2E). In addition to the epithelium, occasional mononuclear cells within the interstitial compartment were positively stained for eotaxin-3. Quantitative assessment of the interstitial region below the epithelium (internal to the cartilaginous ring) reflected the small numbers of eotaxin-3–immunoreactive cells within this compartment; although there was a modest elevation in volume within proximal conducting airways of HDM-exposed animals, the increase was not statistically significant as compared with filtered air controls (P = 0.1199) (Figure 2F). We did not observe a significant airway generation/block dependent effect of eotaxin-3 protein staining within epithelial or interstitial compartments (epithelium, P = 0.4105; interstitium, P = 0.3912); however, the volume of eotaxin-3 within tracheal epithelium did significantly correlate with volume of major basic protein at this site (P = 0.012 by multiple regression analysis). Because the volume of eotaxin-3–positive cells within the epithelial and interstitial compartments did not parallel the mRNA expression profile for different airway generations, we further examined peripheral sites external to the cartilaginous ring of the midlevel bronchus and identified large eotaxin-3–immunoreactive nerve fiber bundles; these structure were confirmed to be PGP 9.5–immunopositive, a marker for cells of neuronal origin (Figure 2D). Filtered air control animals did not exhibit eotaxin-3/PGP 9.5–double positive staining (Figure 2B). Eotaxin-3/PGP 9.5–double immunopositive nerve fiber bundles were found only within blocks 3 and 5; these airway generations correspond to the midlevel airway sampled for RNA analysis (Figures 1C–1E).
Figure 2.
Effect of HDM exposure on eotaxin-3 protein expression within different airway generations and compartments. (A–D) Immunofluorescence staining of cryosections obtained from midlevel airways, a representative filtered air control infant monkey (A and B) is compared with a representative HDM-exposed infant monkey (C and D). Cryosections were double labeled for eotaxin-3 (green, A and C) and PGP 9.5 (red staining, B and D). Arrows point to large nerve fiber bundles peripherally located within the airway wall. Scale bar = 50 μm. (E and F) Abundance and distribution of eotaxin-3–immunoreactive cells within five different airway generations of infant monkeys (filtered air, n = 5; HDM, n = 6). Columns represent the volume of eotaxin-3–positive cells within epithelial (E) and interstitial (F) compartments sampled from cryopreserved blocks of the left caudal lobe. Blocks Tr 1, 3, 5, and 7 correspond to the trachea (Tr) and regions from the most proximal (1) to distal (7) airways of the lobe. All data are represented as mean ± SEM. *P = 0.0003 by two-way ANOVA as compared with filtered air control animals.
Distribution of CCR3 by Airway Generation
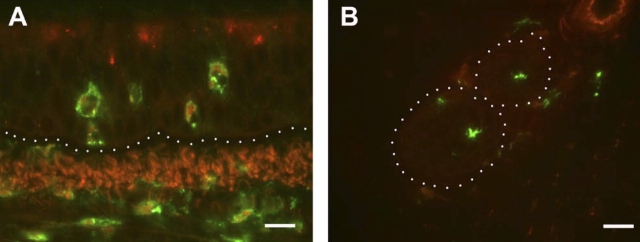
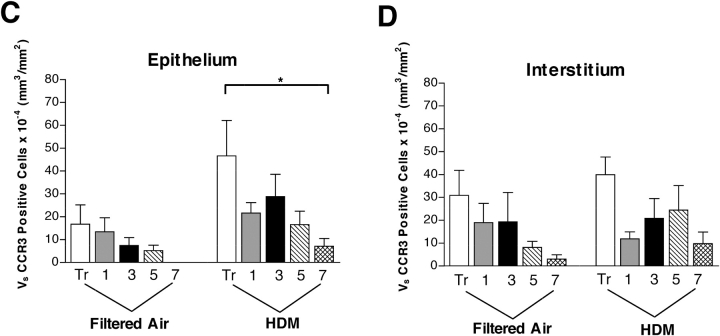
Eotaxin-3 is only known to have agonist interactions with CCR3 (22). In addition to eosinophils, CCR3 is expressed by basophils, Th2 cells, mast cells, and dendritic cells (23–26). To determine if other additional cell types besides eosinophils may respond to eotaxin-3 in the infant lung, we evaluated airways using a rhesus-specific monoclonal antibody against CCR3. Immunofluorescence staining showed an abundance of CCR3+ cells within the epithelial compartment as well as in the interstitial compartment (Figure 3A); many of the CCR3+ cells exhibited an eosinophil phenotype (bilobed nucleus, granulated cytoplasm). CCR3+ cells consisted of both eosinophils and mast cells (data not shown). Within peripherally located nerve fiber bundles, we also observed CCR3+ cells with a dendritic morphologic phenotype. In parallel with eotaxin-3, quantitative assessment of five airway generations showed an increase in CCR3+ cells within the epithelial compartment as compared with control animals, and a significant airway dependent effect (P = 0.0118) (Figure 3C). The volume of CCR3+ cells within the interstitium were not significantly affected by HDM exposure, although there was an airway-dependent effect (P = 0.012) (Figure 3D).
Figure 3.
Effect of HDM exposure on CCR3 protein expression within different airway generations and compartments. (A and B) Immunofluorescence staining for CCR3; a midlevel airway from a representative HDM-exposed infant monkey is shown. The dotted line in A defines the epithelial compartment. The dotted line in B surrounds two small nerve fiber bundles peripherally located within the airway wall. Scale bar = 20 μm. (C and D) Abundance and distribution of CCR3-immunoreactive cells within five different airway generations of infant monkeys (filtered air, n = 5; HDM, n = 6). Columns represent the volume of CCR3+ cells within epithelial (C) and interstitial (D) compartments sampled from cryopreserved blocks of the left caudal lobe. Blocks Tr 1, 3, 5, and 7 correspond to the trachea (Tr) and regions from the most proximal (1) to distal (7) airways of the lobe. All data are represented as mean ± SEM. *P = 0.0025 by two-way ANOVA as compared with filtered control animals.
Phenotype of CCR3+ Cells within Airway Nerve Bundles
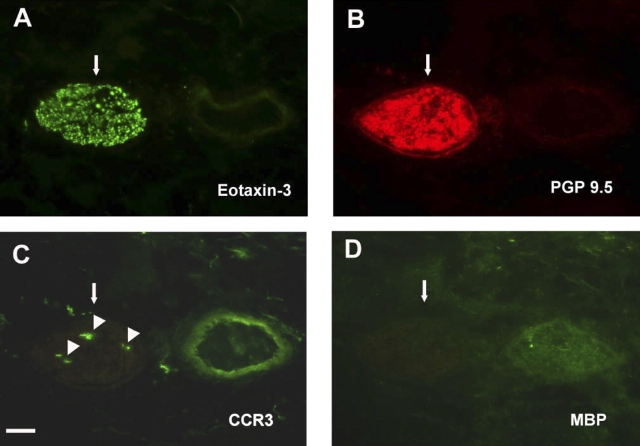
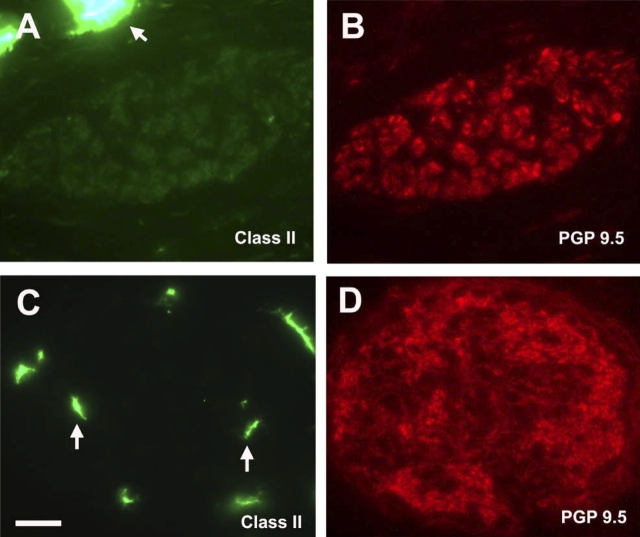
The immunophenotype of CCR3+ cells within airway nerves was further defined by cell surface markers. As shown in Figure 4, CCR3+ cells co-localized within eotaxin-3/PGP 9.5–double positive nerves did not express major basic protein, indicating that eosinophils do not accumulate in or around peripherally located nerve fiber bundles. To determine if the CCR3+ cells were mast cells, we used CD117/c-kit as a cell surface marker, because it is highly expressed on mast cells throughout their life cycle, from circulating progenitor cell to differentiated tissue cells. Immunostaining of airway nerve bundles showed expression of CD117/c-kit within individual axons, consistent with findings from other laboratories, suggesting a role for this receptor tyrosine kinase in neural growth (Figure 5A) (27, 28). However, CCR3+ cells within nerve bundles did not co-express CD117/c-kit (Figure 5E). In addition, no immunostaining was detected for CD3 within nerve bundles, indicating that the CCR3+ cells were not T lymphocytes (data not shown). Because the CCR3+ cells associated with airway nerves exhibited a dendritic cell morphologic phenotype, we immunostained midlevel airways for MHC class II expression (Figure 6). In filtered air control monkeys, Class II–positive cells were observed throughout the interstitium but not within airway nerves. In contrast, Class II–positive cells were co-localized with airway nerves in HDM-exposed monkeys.
Figure 4.
Immunofluorescence staining for CCR3+ cells and eosinophils within airway nerves of HDM-exposed infant monkeys. Adjacent cryosections from a midlevel airway of a representative HDM-exposed infant monkey were double immunostained for eotaxin-3 (A) and PGP 9.5 (B) or individually stained for CCR3 (C) or major basic protein (D). Large arrows point to the nerve fiber bundle in corresponding sections, small arrowheads point to CCR3 positive cells. Scale bar = 20 μm.
Figure 5.
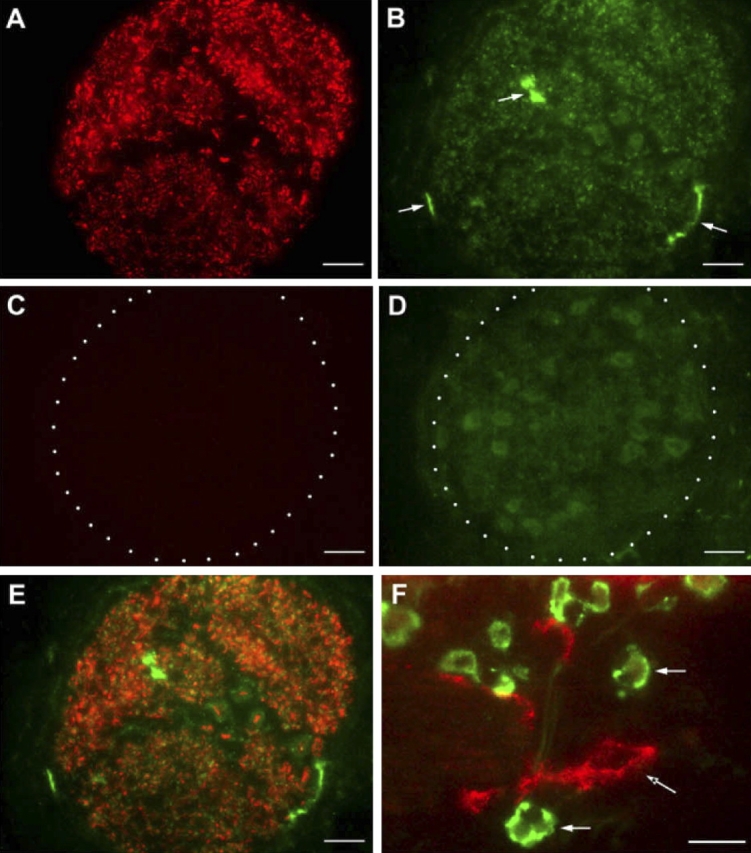
Immunofluorescence staining of CCR3+ cells and CD117/c-kit expression within airway nerves of HDM-exposed infant monkeys. Cryosections from a midlevel airway of a representative HDM-exposed infant monkey were double immunostained for CD117/c-kit and CCR3. Both CD117/c-kit (A) and CCR3 (B) expression were detected in a representative airway nerve bundle; arrowheads in B point to three CCR3+ cells. Isotype control immunostaining for CD117/c-kit (C) and CCR3 (D) antibodies are shown. Overlay of CD117/c-kit and CCR3 staining (E) shows no overlap in expression. As a positive control, CCR3+ eosinophils (white arrows) and a CD117/c-kit–positive mast cell (arrow with asterisk) are present within the interstitial region of the same cryosection (F). Scale bar = 20 μm.
Figure 6.
Immunofluorescence staining of MHC Class II–positive cells within airway nerves of HDM-exposed infant monkeys. Cryosections from a midlevel airway of a representative filtered air control (A and B) or HDM-exposed (C and D) monkey were double immunostained for MHC Class II (A and C) and PGP 9.5 (B and D). Arrows point to MHC Class II–positive cells in the periphery of nerve bundles (A) or within nerve bundles (C). Scale bar = 20 μm.
DISCUSSION
Currently, there is very limited understanding of pulmonary immune responses to environmental challenges during infancy, particularly in primate species. To address this deficiency, we have previously investigated pulmonary and systemic immune responses to chronic aeroallergen exposure, using infant rhesus monkeys as model of postnatal development in children (17). One hallmark feature of allergic airways disease is eosinophilia; recurrent exposure of newborn monkeys to HDM aeroallergen resulted in significant recruitment of eosinophils into the airway mucosa at 2 mo of age (Figures 1A and 1B). Further, tissue recruitment of eosinophils throughout the HDM-exposed infant lung is significantly affected by airway generation, with the trachea as a preferential site for accumulation. To investigate the cellular mechanisms for eosinophil trafficking in the infant lung, the goal of this study was to identify the key chemokine mediators for eosinophil recruitment into the airway mucosa in response to HDM exposure. We initially focused our analysis on the three members of the eotaxin chemokine family; this group of structurally and functionally related peptides has been reported by a number of laboratories to be prominently involved in allergic mechanisms (reviewed in Ref. 29).
The functional role for the expression of multiple eosinophilic chemokines in allergy and asthma has been speculated. Elevated mRNA and protein expression of eotaxin has been well documented in subjects with asthma relative to control subjects (30, 31). In both human asthma and animal models of allergic airways disease, eotaxin levels in the lung increase very early (4–7 h) after allergen challenge; this correlates well with the initial development of eosinophilia within the airway mucosa and bronchoalveolar lavage (32–35). However, blocking antibody treatment or targeted deletion mutants do not completely abolish eosinophil recruitment following allergen challenge, and later time points after allergen challenge (12–24 h) show a decline of eotaxin expression with persisting eosinophila (33–36). These findings suggest that eotaxin is most important for the initial recruitment of eosinophils to the lung after allergen challenge, but other chemotactic factors for eosinophils may play a role in later time points. Consistent with this notion, it has been recently reported that eotaxin and eotaxin-2 mRNA expression in bronchial biopsies of humans with asthma are constitutively elevated, with no further increase at 24 h after allergen challenge (37). In contrast, eotaxin-3 mRNA is significantly increased at 24 h after allergen challenge in individuals with asthma (but no difference at baseline), suggesting that this eosinophilic chemokine is important during the late phase response after allergen challenge.
More recently, analysis of single nucleotide polymorphisms (SNP) for all three members of the eotaxin gene family showed a significant association of serum IgE levels, peripheral blood eosinophilia, and asthma susceptibility with two different SNP within the eotaxin-3 gene (16, 38). Based on these findings, it can be postulated that each form of eotaxin has a specific functional role within the lung, relative to timing of allergen challenge and airway generation. By gene expression analysis, all three eotaxin gene family members were detected in HDM-exposed infant airways from this study at 24 h after allergen challenge; however, there were clear differences in magnitude of responses. In comparison with eotaxin and eotaxin-2, eotaxin-3 expression was most significantly affected by HDM exposure (Figure 1). Although we do not know if eotaxin and eotaxin-2 are important for the initial recruitment of eosinophils within 4–7 h after allergen challenge in very young monkeys, the gene expression profile of eotaxin family members at 24 h after allergen challenge is consistent with previously reported findings from human studies.
As evidenced by quantitative assessment of eotaxin-3 within the airways, the epithelium is a highly responsive compartment in the context of allergen exposure; only the epithelium was significantly affected to express this chemokine (Figure 2). In contrast, there were minimal differences in eotaxin-3 expression within the interstitial compartment of HDM-exposed infant airways. There was a significant correlation for eotaxin-3 expression in tracheal epithelium with volume of major basic protein, demonstrating a putative chemokine mechanism for recruitment of eosinophils from the vasculature and interstitial compartment. The chemoattractant mechanism for interstitial recruitment of eosinophils within the trachea remains unknown, but these data further support the notion that eosinophil recruitment into the lung is a complex multistep process that requires a succession (and upregulation) of chemokines/chemoattractants that drive extravasation from the vasculature, emigration into the epithelium, and movement into the airway lumen. CCR3+ cells within the epithelium were also significantly affected by HDM exposure, but did not directly correlate with eosinophil volume. This may be explained by the diverse population of cells that express CCR3, and our finding of resident CCR3+ cells constitutively present in control animals (both epithelium and interstitium).
As measured by our analysis, the expression profile of eotaxin-3 protein was inconsistent with mRNA distribution. It is possible that the differences between mRNA and protein expression levels are reflective of the relative contribution of epithelial volume and interstitial volume between the trachea and intrapulmonary airways. This discrepancy may also be explained by the presence of peripherally located nerve fiber bundles within the midlevel bronchus that express eotaxin-3, which were excluded from morphometric analysis in this study. The observation of eosinophilic chemokine expression by airway nerves would suggest a physical and functional relationship with eosinophils their cytotoxic contents (i.e., major basic protein); this notion has been speculated by other investigators (39, 40). In this study, CCR3+ cells that were identified within eotaxin-3–expressing nerve fiber bundles did not express major basic protein or the mast cell marker, CD117/c-kit. The subsequent observation of MHC Class II–positive cells with an identical dendritic cell phenotype within nerve bundles supports an antigen-presenting function for CCR3+ cells in this compartment.
The interaction of antigen-presenting cells within airway nerves is not unprecedented; it has been reported that pulmonary dendritic cells are normally located in close proximity to unmyelinated nerve fibers of the rat lung (41). Regardless of maturational state, all dendritic cells contain an intracellular pool of CCR3 and can respond to eotaxin and eotaxin-2 (26). Dendritic cells can express neurokinin receptors, and peripheral neuropeptides can attract blood-derived dendritic cells (42). Of clear interest is the functional role for interactions of dendritic cells with nerves. The nerves fiber bundle may function as a conduit for local recruitment of dendritic cells of a particular phenotype into the airways. Alternatively, signaling through CCR3 by eotaxin-3 may have functional implications for dendritic cell populations. Interestingly, it has been recently reported that eotaxin-3 is a natural antagonist for CCR1, CCR2, and CCR5, all known to be expressed by dendritic cell populations; it is tempting to propose that the interaction of dendritic cells with airway nerves that express eotaxin-3 may be pivotal for the differentiation of antigen-presenting cells that promote a Th2 phenotype (43).
In conclusion, we have found that eotaxin-3 is a prominent eosinophilic chemokine in HDM-exposed infant monkeys. Eotaxin-3 appears to play an important role in the recruitment of eosinophils into airway epithelium in response to house dust mite. The role of eotaxin-3 expression by airway nerves is not known, but a functional correlate between signaling via CCR3 and neurokinin receptors may be speculated.
Acknowledgments
The authors acknowledge the expert technical assistance of Brian Tarkington, Sarah Davis, and the California National Primate Center nursery staff during the course of this study.
This study was supported by NIEHS ES00628 (C.G.P.), NCRR RR00169, and T32HL007013 (D.L.C.)
Conflict of Interest Statement: None of the authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Wardlaw AJ, Dunnette S, Gleich GJ, Collins JV, Kay AB. Eosinophils and mast cells in bronchoalveolar lavage in subjects with mild asthma: relationship to bronchial hyperreactivity. Am Rev Respir Dis 1988;137:62–69. [DOI] [PubMed] [Google Scholar]
- 2.Kay AB, Menzies-Gow A. Eosinophils and interleukin-5: the debate continues. Am J Respir Crit Care Med 2003;167:1586–1587. [DOI] [PubMed] [Google Scholar]
- 3.Wills-Karp M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu Rev Immunol 1999;17:255–281. [DOI] [PubMed] [Google Scholar]
- 4.Mauser PJ, Pitman AM, Fernandez X, Foran SK, Adams GK, Kreutner W, Egan RW, Chapman RW. Effects of an antibody to interleukin-5 in a monkey model of asthma. Am J Respir Crit Care Med 1995;152:467–472. [DOI] [PubMed] [Google Scholar]
- 5.Hamelmann E, Oshiba A, Schwarze J, Bradley K, Loader J, Larsen GL, Gelfand EW. Allergen-specific IgE and IL-5 are essential for the development of airway hyperresponsiveness. Am J Respir Cell Mol Biol 1997;16:674–682. [DOI] [PubMed] [Google Scholar]
- 6.Gundel RH, Letts LG, Gleich GJ. Human eosinophil major basic protein induces airway constriction and airway hyperresponsiveness in primates. J Clin Invest 1991;87:1470–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamelmann E, Cieslewicz G, Schwarze J, Ishizuka T, Joetham A, Heusser C, Gelfand EW. Anti-interleukin 5 but not anti-IgE prevents airway inflammation and airway hyperresponsiveness. Am J Respir Crit Care Med 1999;160:934–941. [DOI] [PubMed] [Google Scholar]
- 8.Leckie MJ, ten Brinke A, Khan J, Diamant Z, O'Connor BJ, Walls CM, Mathur AK, Cowley HC, Chung KF, Djukanovic R, et al. Effects of an interleukin-5 blocking monoclonal antibody on eosinophils, airway hyper-responsiveness, and the late asthmatic response. Lancet 2000;356:2144–2148. [DOI] [PubMed] [Google Scholar]
- 9.Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS. Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am J Respir Crit Care Med 2003;167:199–204. [DOI] [PubMed] [Google Scholar]
- 10.Flood-Page P, Menzies-Gow A, Phipps S, Ying S, Wangoo A, Ludwig MS, Barnes N, Robinson D, Kay AB. Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J Clin Invest 2003;112:1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolthers OD. Eosinophil granule proteins in the assessment of airway inflammation in pediatric bronchial asthma. Pediatr Allergy Immunol 2003;14:248–254. [DOI] [PubMed] [Google Scholar]
- 12.Halmerbauer G, Gartner C, Koller D, Schierl M, Kühr J, Forster J, Urbanek R, Frischer T. Eosinophil cationic protein and eosinophil protein X in the nasal lavage of children during the first 4 weeks of life: SPACE Collaborative Study Team. Allergy 2000;55:1121–1126. [DOI] [PubMed] [Google Scholar]
- 13.Wojnarowski C, Halmerbauer G, Mayatepek E, Gartner C, Frischer T, Forster J, Kuehr J. Urinary leukotriene E(4), eosinophil protein X, and nasal eosinophil cationic protein are not associated with respiratory symptoms in 1-year-old children. Allergy 2001;56:883–888. [DOI] [PubMed] [Google Scholar]
- 14.Bandeira-Melo C, Herbst A, Weller PF. Eotaxins: contributing to the diversity of eosinophil recruitment and activation. Am J Respir Cell Mol Biol 2001;24:653–657. [DOI] [PubMed] [Google Scholar]
- 15.Daugherty B, Siciliano S, DeMartino J, Malkowitz L, Sirotina A, Springer M. Cloning, expression, and characterization of the human eosinophil eotaxin receptor. J Exp Med 1996;183:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponath P, Qin S, Post T, Wang J, Wu L, Gerard N, Newman W, Gerard C, Mackay C. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med 1996;183:2437–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller LA, Plopper CG, Hyde DM, Gerriets JE, Pieczarka E, Tyler N, Gershwin LJ, Schelegle ES, Van Winkle LS. Immune and airway effects of house dust mite aeroallergen exposures during postnatal development of the infant rhesus monkey. Clin Exp Allergy 2003;33:1686–1694. [DOI] [PubMed] [Google Scholar]
- 18.Schelegle ES, Gershwin LJ, Miller LA, Fanucchi MV, Van Winkle LS, Gerriets JP, Walby WF, Omlor AM, Buckpitt AR, Tarkington BK, et al. Allergic asthma induced in rhesus monkeys by house dust mite (Dermatophadoides farinae). Am J Pathol 2001;158:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan X, Buckpitt AR, Plopper CG. Variation in antioxidant enzyme activities in anatomic subcompartments within rat and rhesus monkey lung. Toxicol Appl Pharmacol 1993;123:73–82. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Soares MP, Guan Y, Matheravidathu S, Wnek R, Johnson KE, Meisher A, Iliff SA, Mudgett JS, Springer MS, et al. Functional expression and characterization of macaque C–C chemokine receptor 3 (CCR3) and generation of potent antagonistic anti-macaque CCR3 monoclonal antibodies. J Biol Chem 2002;277:33799–33810. [DOI] [PubMed] [Google Scholar]
- 21.Lauweryns JM, Van Ranst L. Protein gene product 9.5 expression in the lungs of humans and other mammals. Immunocytochemical detection of neuroepithelial bodies, neuroendocrine cells and nerves. Neurosci Lett 1988;85:311–316. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura M, Suzuki N, Imai T, Takagi S, Suzuki R, Nakajima T, Hirai K, Nomiyama H, Yoshie O. Molecular cloning of a novel human CC chemokine (Eotaxin-3) that is a functional ligand of CC chemokine receptor 3. J Biol Chem 1999;274:27975–27980. [DOI] [PubMed] [Google Scholar]
- 23.Uguccioni M, Mackay CR, Ochensberger B, Loetscher P, Rhis S, LaRosa GJ, Rao P, Ponath PD, Baggiolini M, Dahinden CA. High expression of the chemokine receptor CCR3 in human blood basophils. Role in activation by eotaxin, MCP-4, and other chemokines. J Clin Invest 1997;100:1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heath H, Qin S, Rao P, Wu L, LaRosa G, Kassam N, Ponath PD, Mackay CR. Chemokine receptor usage by human eosinophils. The importance of CCR3 demonstrated using an antagonistic monoclonal antibody. J Clin Invest 1997;99:178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sallusto F, Mackay CR, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science 1997;277:2005–2007. [DOI] [PubMed] [Google Scholar]
- 26.Beaulieu S, Robbiani DF, Du X, Rodrigues E, Ignatius R, Wei Y, Ponath P, Young JW, Pope M, Steinman RM, et al. Expression of a functional eotaxin (CC chemokine ligand 11) receptor CCR3 by human dendritic cells. J Immunol 2002;169:2925–2936. [DOI] [PubMed] [Google Scholar]
- 27.Hirata T, Morii E, Morimoto M, Kasugai T, Tsujimura T, Hirota S, Kanakura Y, Nomura S, Kitamura Y. Stem cell factor induces outgrowth of c-kit-positive neurites and supports the survival of c-kit-positive neurons in dorsal root ganglia of mouse embryos. Development 1993;119:49–56. [DOI] [PubMed] [Google Scholar]
- 28.Jin K, Mao XO, Sun Y, Xie L, Greenberg DA. Stem cell factor stimulates neurogenesis in vitro and in vivo. J Clin Invest 2002;110:311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukacs NW, Miller AL, Hogaboam CM. Chemokine receptors in asthma: searching for the correct immune targets. J Immunol 2003;171:11–15. [DOI] [PubMed] [Google Scholar]
- 30.Ying S, Robinson DS, Meng Q, Rottman J, Kennedy R, Ringler DJ, Mackay CR, Daugherty BL, Springer MS, Durham SR, et al. Enhanced expression of eotaxin and CCR3 mRNA and protein in atopic asthma: association with airway hyperresponsiveness and predominant co-localization of eotaxin mRNA to bronchial epithelial and endothelial cells. Eur J Immunol 1997;27:3507–3516. [DOI] [PubMed] [Google Scholar]
- 31.Zeibecoglou K, Ying S, Meng Q, Robinson D, Kay A, Macfarlane A, Barnes N, Pavord I. Increases in eotaxin-positive cells in induced sputum from atopic asthmatic subjects after inhalational allergen challenge. Allergy 1999;54:730–735. [DOI] [PubMed] [Google Scholar]
- 32.Gauvreau GM, Watson RM, O'Byrne PM. Kinetics of allergen-induced airway eosinophilic cytokine production and airway inflammation. Am J Respir Crit Care Med 1999;160:640–647. [DOI] [PubMed] [Google Scholar]
- 33.Humbles AA, Conroy DM, Marleau S, Rankin SM, Palframan RT, Proudfoot AEI, Wells TNC, Li D, Jeffery PK, Griffiths-Johnson DA, et al. Kinetics of eotaxin generation and its relationship to eosinophil accumulation in allergic airways disease: analysis in a guinea pig model in vivo. J Exp Med 1997;186:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown JR, Kleimberg J, Marini M, Sun G, Bellini A, Mattoli S. Kinetics of eotaxin expression and its relationship to eosinophil accumulation and activation in bronchial biopsies and bronchoalveolar lavage (BAL) of asthmatic patients after allergen inhalation. Clin Exp Immunol 1998;114:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalo J-A, Lloyd CM, Kremer L, Finger E, Martinez-A C, Siegelman MH, Cybulsky M, Guitierrez-Ramos J-C. Eosinophil recruitment to the lung in a murine model of allergic inflammation: the role of T cells, chemokines, and adhesion receptors. J Clin Invest 1996;98:2332–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothenberg M, MacLean J, Pearlman E, Luster A, Leder P. Targeted disruption of the chemokine eotaxin partially reduces antigen-induced tissue eosinophilia. J Exp Med 1997;17:785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berkman N, Ohnona S, Chung FK, Breuer R. Eotaxin-3 but not eotaxin gene expression is upregulated in asthmatics 24 hours after allergen challenge. Am J Respir Cell Mol Biol 2001;24:682–687. [DOI] [PubMed] [Google Scholar]
- 38.Chae S-C, Lee Y-C, Park Y-R, Shin J-S, Song J-H, Oh G-J, Hong S-T, Pae H-O, Choi B-M, Chung H-T. Analysis of the polymorphisms in eotaxin gene family and their association with asthma, IgE, and eosinophil. Biochem Biophys Res Commun 2004;320:131–137. [DOI] [PubMed] [Google Scholar]
- 39.Curran DR, Walsh MT, Costello RW. Interactions between inflammatory cells and nerves. Curr Opin Pharmacol 2002;2:243–248. [DOI] [PubMed] [Google Scholar]
- 40.Jacoby DB, Costello RM, Fryer AD. Eosinophil recruitment to airway nerves. J Allergy Clin Immunol 2001;107:211–218. [DOI] [PubMed] [Google Scholar]
- 41.Kradin R, MacLean J, Duckett S, Schneeberger EE, Waeber C, Pinto C. Neuroimmune interactions promote the recruitment of dendritic cells to the lung and the cellular immune response to inhaled antigen. Am J Pathol 1997;150:1735–1743. [PMC free article] [PubMed] [Google Scholar]
- 42.Dunzendorfer S, Kaser A, Meierhofer C, Tilg H, Wiedermann CJ. Cutting edge: peripheral neuropeptides attract immature and arrest mature blood-derived dendritic cells. J Immunol 2001;166:2167–2172. [DOI] [PubMed] [Google Scholar]
- 43.Petkovic V, Moghini C, Paoletti S, Uguccioni M, Gerber B. Eotaxin-3/CCL26 is a natural antagonist for CC chemokine receptors 1 and 5: a human chemokine with a regulatory role. J Biol Chem 2004;279:23357–23363. [DOI] [PubMed] [Google Scholar]