Abstract
In contrast to most stimulated lymphocytes, B cells exposed to Toll-like receptor 9 ligands are nonself-adherent, allowing individual cells and families to be followed in vitro for up to 5 days. These B cells undergo phases typical of an adaptive response, dividing up to 6 times before losing the impetus for further growth and division and eventually dying by apoptosis. Using long-term microscopic imaging, accurate histories of individual lymphocyte fates were collected. Quantitative analysis of family relationships revealed that times to divide of siblings were strongly related but these correlations were progressively lost through consecutive divisions. A weaker, but significant, correlation was also found for death times among siblings. Division cessation is characterized by a loss of cell growth and the division in which this occurs is strongly inherited from the original founder cell and is related to the size this cell reaches before its first division. Thus, simple division-based dilution of factors synthesized during the first division may control the maximum division reached by stimulated cells. The stochastic distributions of times to divide, times to die, and divisions reached are also measured. Together, these results highlight the internal cellular mechanisms that control immune responses and provide a foundation for the development of new mathematical models that are correct at both single-cell and population levels.
The coordinated regulation of cell proliferation and apoptosis is a striking feature of the adaptive response of lymphocytes after exposure to pathogens. The initially quiescent lymphocytes are stimulated to undergo a series of proliferation cycles that increase the number of reactive cells many hundredfold. After a period the response peaks, cells stop dividing, and ≈95% of the newly generated cells die by apoptosis (1, 2). Although these population kinetics are well understood, we have little knowledge of how the component single-cell fates add up to this outcome, nor how individual decisions of division, death, and quiescence are handled. Our current models of lymphocyte responses are strongly influenced by studies of tumor cells and fibroblasts undertaken by investigators in the 1960s and 1970s. At that time film and microscopy were used to measure the kinetics of cell division in vitro (3–5). These studies noted that intermitotic division times were different between cell types and that all eukaryotic cells, including both yeast and clonally derived tumor cell populations, exhibited significant variation within the population of dividing cells.
The variation observed in cell cycle times was incorporated into the widely used Smith and Martin mathematical model of cell growth that postulated a random time spent in an “A state” (assumed to be G1) that governed entry into a deterministic B phase (S, G2, and M) of the cell cycle (6, 7). Because the Smith–Martin model is relatively easy to implement, it has served as the backbone for numerous models of lymphocyte proliferation (6–10); however, there are many competing models based on alternative data and starting assumptions. Examples of alternative models include the rate model (11), continuum model (12), branching autoregression model (13), and size control model (14). Each of these models addresses systems in a steady-state growth phase that may not be suitable for an immune response where cell properties are modulated over time and by division-linked changes (15, 16). Furthermore, the regulation of cell death is a particularly important feature of lymphocyte responses, and to date there is no quantitative information on how lymphocyte survival is regulated and altered after the proliferative phases of the immune response. Recently, we found that variations in times to die for lymphocytes in vitro followed a skewed distribution akin to a lognormal, or similar, curve (17). Because times to divide also follow a similar distribution we proposed the cyton model of cellular proliferation and survival that assumed independent, and competing, age-dependent probabilities for division and death in each cell that were reset upon each mitotic event (17). Although this model provided excellent fits to population data obtained in vitro and in vivo, it suffered, in common with all previous models, from a lack of experimental information regarding the division and death times of cells once they have begun clonal expansion. Thus, despite our molecular understanding of division and death as independent processes, how they interleave over time at the single cell level to achieve the complex patterning of a population remains an outstanding question. To distinguish between current models and inform the development of new mathematical approaches further experimental information is critically needed. Here, we monitored division and death times in families of dividing lymphocytes in a representative system by using long-term video microscopy.
Results
Long-Term Parameter Measurements of CpG Containing Oligonucleotide (CpG) Stimulated B Lymphocytes.
In contrast to most tumor cells, lymphocytes cultured in vitro typically undergo homotypic adhesion and form 3D structures, preventing the tracking of single cells for long periods. However, we observed that primary naive B lymphocytes stimulated with the ligand for Toll-like receptor 9 (TLR-9), CpG, move only in the plane of the culture vessel, thus facilitating tracking and recording the fates of stimulated primary lymphocytes over many generations. Furthermore, CpG-stimulated naïve resting B cells have a strictly regulated division limit and will divide successfully a maximum of 6 times (18). Because CpG is typically associated with replicating foreign bacteria and is known to play a key role in driving B cell immune responses after infection, we consider that CpG stimulation of B cells is an ideal model system for studying single-cell fate within a population displaying proliferation, cessation of cell division, and contraction, typical of the immune response. A further advantage of this system is that CpG-stimulated B cells formed a highly homogenous effector population with little evidence of isotype switching and only weak unimodal expression of the plasmablast-associated transcription factor Blimp-1. For these reasons we undertook an extensive analysis of single-cell behavior during CpG stimulation.
Small resting B cells were seeded by serial dilution into Terasaki tissue culture plates in the presence of CpG and propidium iodide (PI) to monitor cell viability. After ≈24 h, before the first division had occurred [as determined by carboxyfluorescein succinimidyl ester (CFSE) dilution time-course analysis (Fig. S1 in SI Appendix)], Terasaki tissue culture plates were transferred to an environment-controlled microscope. Bright-field and CY3.5 (to detect PI) images were collected at 2-min intervals for 4 days. To ensure accuracy, cell division and death events were monitored frame by frame manually. Three independent sets of data were analyzed: one with 25 founder cells (Fam1) and two larger datasets of 107 founders (Fam2) and 89 founders (Fam3), respectively. A representative film is provided as Movie S1, and a representative family tree from a single founder cell is shown in Fig. S2A in SI Appendix.
Stochastic Features of Times to Divide and Die Are Division-Dependent.
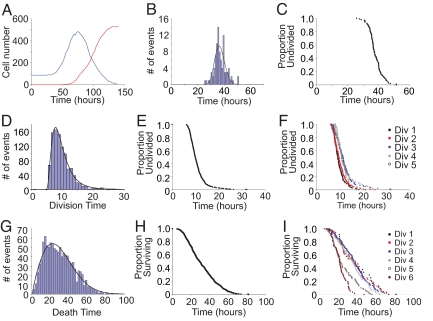
An overlay of total live and dead cell numbers recorded over time is shown for data from Fam2 (Fig. 1A). CpG-stimulated B cells remained viable (>95% ; Fig. S1 in SI Appendix) until the first cell division, then their numbers increased for ≈30 h, followed by a period of contraction when cell death results in the loss of a large fraction of the population. CFSE profiles indicate that during this contraction phase of the response cells failed to progress through further division rounds (Fig. S1 in SI Appendix). This cessation in division is internally regulated and not caused by the lack of stimulus, exhaustion of the cell culture medium, or the experiment time (18). The time to first division collated into 2-h time segments is shown in Fig. 1B. The same data are also shown in the time continuous alpha plot format introduced by Smith and Martin (19). These plots present the proportion of undivided cells against time from last mitosis and typically take the form of a time lag followed by an approximately exponential exit of cells to the next division (Fig. 1C). The time taken for all subsequent divisions after mitosis have been pooled and plotted in Fig. 1D and shown as an alpha plot in Fig. 1E. As observed previously, the time taken to enter the first division is much longer than for subsequent division rounds and conforms approximately to a lognormal distribution (17, 20). Although the level of variation among dividing primary lymphocytes was previously unknown, analysis of the data shown in Fig. 1 D and E revealed significant variation in division times in the range of 6 and 25 h. Furthermore, this observed variation is independent of division number (Fig. 1F).
Fig. 1.
Variation in division and death times of CpG-stimulated B cells. (A) Total numbers of live cells (blue) and dead cells (red) over time in CpG cultures. (B and C) The variation in entry to the first division of the cell population is represented as both a histogram collated into 1-h time intervals, with a fitted lognormal probability distribution (μ = 37.21, σ = 3.76, solid line) (B) and the time continuous alpha plot (C). (D–F) Data for variation in subsequent division times represented as a histogram collated in 1-h time intervals with a fitted lognormal probability distribution (μ = 9.30, σ = 2.54, solid line) (D), an alpha plot (E), and as separate alpha plots for each individual division (F). (G–I) Data for variation in death times in subsequent divisions represented as a histogram collated in 2-h time intervals with a fitted Weibull probability distribution (α = 1.86, β = 33.02, solid line) (G), an alpha plot (H), and alpha plots for each individual division (I).
To assist in the development of mathematical models of lymphocyte regulation and gain insight into the underlying stochastic process, we sought the optimal empirical fit to the division data by using a series of probability distributions. We found that skewed right distributions, such as the lognormal distribution, gamma distribution, and Weibull distribution, were excellent estimators of the underlying variation and were superior to Gaussian and delayed exponential distributions (Table S1 in SI Appendix). The introduction of a deterministic lag phase as an additional parameter increased the quality of fits (see Numerical Methods in the SI Appendix); however, lognormal and gamma distributions remained consistently superior (Table S1 in SI Appendix). The same conclusions were also reached when data were analyzed on a per-division basis (Table S2 in SI Appendix). Analysis of separate division rounds also demonstrated that in the later divisions (divisions 4 and 5), the mean time to divide was slightly delayed when compared with earlier divisions (Fig. 1F and Table S2 in SI Appendix).
Time to Die Is a Lymphocyte Age-Dependent Stochastic Event.
We next sought to test our hypothesis that timing of death events would follow a probability distribution clocked from last mitosis (17). We tracked apoptosis of single lymphocytes via morphology and the uptake of PI (see Materials and Methods and Movie S1) and measured the distribution of death times in dividing lymphocyte cultures and how these parameters were affected by division number (Fig. 1 G–I). Consistent with our previous analysis of variation in death times of undivided cells (17), distributions such as lognormal, gamma, and Weibull gave excellent fits to death data (Table S3 in SI Appendix). Exponential times to die, with and without a lag, were clearly unsuitable, strongly arguing for a stochastic age-sensitive internal mechanism. Importantly, the average time to die was ≈18 h longer when compared with the average time to divide (Fam2 − division = 10.66 h, death = 31.86; Fam3 − division = 10.82 h, death = 27.54; Fig. 1 E and H). It was also clear that the average time to die diminished progressively with successive divisions (Fig. 1I and Table S4 in SI Appendix). Fig. S2B in SI Appendix summarizes these data as progressive cyton plots (17) that illustrate the best estimate for probability of dividing and dying for each division as determined above. The fact that division times are so early compared with death times suggests that cells that go on to die must have lost their capacity to undergo division.
Lymphocytes Do Not Grow in Their Last Division Before Dying.
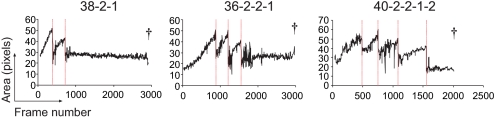
As cells usually increase in size as a prelude to mitosis we tested the hypothesis that cells that die had previously also lost the impetus to grow. Cell size was measured in a series of founder families by automatically recording the pixel area of individual cells by using Metamorph software in combination with manual cell tracking (see Materials and Methods). Typically, at the last division number reached, cells displayed very weak, if any, evidence of cell growth (Fig. 2). An in-depth history of 9 representative founders and their 70 subsequent progeny is shown in Fig. S3 in SI Appendix. The lack of growth before death is striking and is consistent with cell-cycle analysis of CpG-stimulated B cell cultures, which demonstrates that a large proportion of cells are stalled in the G1 phase of the cell cycle during the contraction phase of the response (after 90 h) (Fig. S4 and Table S5 in SI Appendix). Thus, these data strongly support the hypothesis that cells are dropping out of the cell cycle as a prelude to death. These data cannot distinguish between the possibilities that arrest from cell cycle triggers the death machinery, or an underlying default time to die, usually hidden by earlier entry to division, is revealed by division cessation as proposed by Hawkins et al. (17).
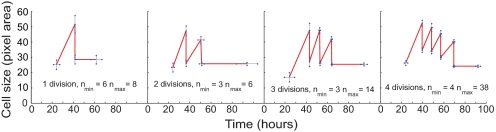
Fig. 2.
Analysis of cell size shows a reduction through divisions and lack of growth after the final cell mitosis. Cell size measurements (solid black lines) for three progeny from Fam3 are shown at the top. Cell divisions are denoted by dashed reds lines. Note the reduction in cell size at final mitosis, reduction in the rate of growth, and the lack of growth by cells in their last division number before death. More detail is shown in Fig. S3 in SI Appendix, which contains plots for 9 representative founders and all 70 progeny. † indicates time of cell death.
Founder Cells Control the Division Destiny of Their Descendants.
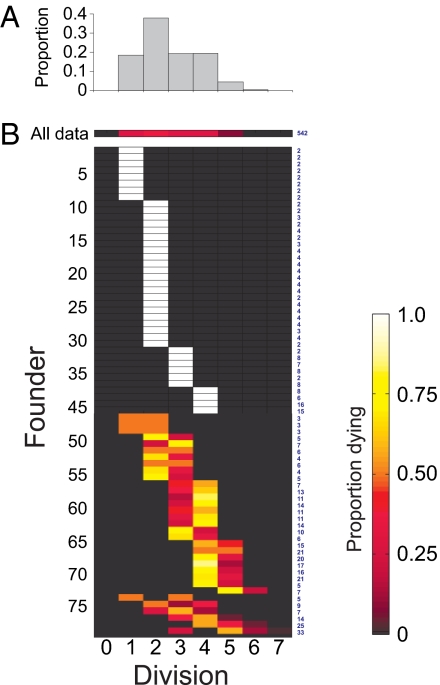
The lack of growth before cell death indicates that the division number in which cells die can also be taken as a surrogate measure of the maximum division limit for that cell. The number of times a cell divides before undergoing this arrest we have termed the cell's division destiny (17). Although it is clear that this process is a fundamental property of lymphocytes responses that can be regulated to affect the final outcome of an immune response we have little understanding of the mechanisms and phenotypic markers of this process. To investigate how division destiny operates within related cells we first plotted the proportion of all cells that died in each possible division number. Fig. 3A and Fig. S5 in SI Appendix show this plot for all founder cells, normalized by dividing by 2i (where i = the division number) to remove the distorting effect of cell division. Fig. 3A reveals the significant range of division destiny exhibited by the progeny of the founders (Fig. 3A and Fig. S5 in SI Appendix). We next conducted this analysis for each individual founder. Remarkably, the division destinies of progeny from single founders were extremely closely aligned and far less variable than the general population (Fig. 3B and Fig. S5 in SI Appendix). In 45.1% of cases all of the progeny of single founders died in the same division number, whereas in 90.2% of cases all progeny died within adjacent division rounds. Given the population data we calculate the probability that, by chance, 8 or more of the 75 founder cells would have all of their progeny die in division 1 and at the same time 13 or more founders would have all of their progeny die in division 2 is 1.6 × 10−14 (Numerical Methods in SI Appendix). Thus, the division destiny of each cell is determined internally and is strongly inherited from the original founder.
Fig. 3.
Maximum division number is highly correlated for progeny of single founders. The division number in which cells died was taken to represent their maximum division potential. Data were normalized to remove the effects of cell division by dividing by 2i, where i = the division number. The proportion of cells in each division number was normalized to exclude cells lost through tracking. (A) Histogram and corresponding heat map illustrate the result of all founders in a population, and therefore, the population variation. (B) Traces the progeny of 75 individual founders from Fam3. The data are ranked to illustrate founders where progeny die in 1 division, followed by founders where progeny die over 2 divisions, and founders where progeny die in ≥3 divisions. Founders that produced only 1 trackable progeny were omitted from analysis. Numbers in blue to the right of the heatmap indicate the number of progeny counted for each founder. A representative experiment is shown at the top (Fam3). Results for both datasets are shown in Fig. S5 in SI Appendix.
Size of Founder Cells at Division Is Predictive of Progeny Division Destiny.
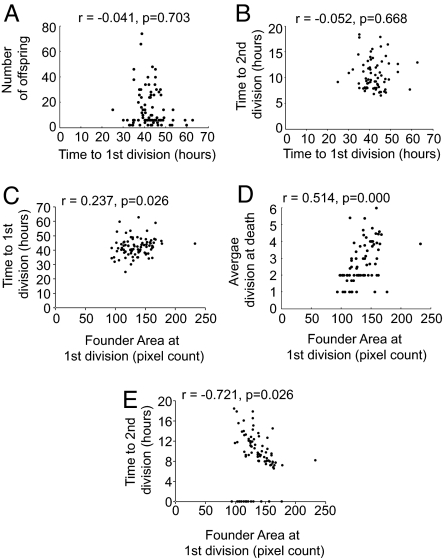
Given that division destiny was strongly transmitted to all progeny, we searched for measurable properties of the founder cell that might give clues to the possible mechanism. Lymphocytes display broad variation in the time taken to enter the first division. Therefore, we tested the hypothesis that cells entering the first division earlier progress through more divisions and generate more offspring. Spearman's rank correlation coefficient analysis of the data illustrated that the time taken to enter the first division did not correlate significantly with the number of offspring produced or the division time of cells in their second division (Fig. 4 A and B and Fig. S6 in SI Appendix). We next investigated cell size as this property has been previously found to correlate with the division time of daughter cells (21). We found a striking and highly significant correlation between cell size and the division destiny of progeny (Fig. 4D). In general, the larger the cell at first division, the more divisions the offspring progressed through before division ceased. In contrast, there was no correlation between the time taken to enter first division and the size of founders at first division (Fig. 4C and Fig. S6 in SI Appendix). We also observed that the division time taken by progeny in their second division were significantly faster in the daughters of larger cells compared with smaller ones, revealing that inherited cell components influence both the time to divide and the division destiny (Fig. 4E and Fig. S6 in SI Appendix).
Fig. 4.
Correlations between cell size, time to first division, and progeny behavior: Analysis was performed by using Spearman's rank correlation coefficient on scatter plots to measure the correlation between the time to first division and number of offspring (A); time to first division and time to second division (B); time to first division and pixel area per founder cell at first division (C); area at first division and division destiny (as represented by average division at death of progeny) (D); and time to second division and pixel area per founder cell at first division (E). A representative experiment is shown (Fam3). Data for both experiments are shown in Fig. S6 in SI Appendix.
Correlations in Division and Death Times of Related Cells.
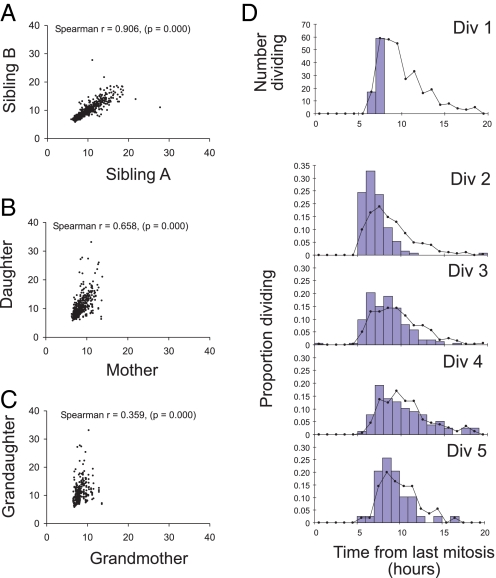
After discovering the strong founder effects above, we explored more closely the inheritance of division times and how they transmit through generations. We constructed time-based family trees through multiple generations. These data revealed a very strong correlation in division times between sibling cells (Fig. 5A and Figs. S7 and S8 in SI Appendix). This high level of correlation indicates that division times are strongly heritable from the mother and argues against the operation of an independent transition probability in each daughter cell (19). Despite the strong correlation in division times by siblings, there is a progressively weaker correlation in division times between mothers and daughters (Fig. 5B and Fig. S7 in SI Appendix). Furthermore, the correlation continues to dissipate through successive division rounds (Fig. 5 A–C and Fig. S7 in SI Appendix). Thus, at some point in the cell cycle, the division time passed on to the next generation is randomized. This pattern of progressive randomization of division times through subsequent rounds of mitosis is illustrated in Fig. 5D. Here, a cohort of cells that enters division within 1 h of each other is followed through divisions 2–5. Clearly the tendency is for the population of progeny to return to the variation distribution of the whole population within a few divisions. We also examined correlations between siblings for times to die (Fig. S8 in SI Appendix). A weak positive correlation was found, suggesting some shared regulation of death times is passed to daughter cells from the mother.
Fig. 5.
Inheritance and randomization of division times through generations. (A–C) Scatter plots illustrating the relationship between division times of siblings (A), mother/daughters (B), and grandmother/granddaughters (C). Significance of correlations was determined using Spearman's rank correlation coefficient. A representative experiment is shown (Fam3). Data for both experiments are shown in Fig. S7 in SI Appendix. (D) The division times of progeny from a 2-h cohort of cells (blue histogram bars) in division 1 were followed through subsequent division rounds. As the progeny progress through subsequent divisions, the variation of the cohort resembles that of the entire population of cells (solid black line) even after as few as 2 divisions.
Size at Division Gets Smaller with Successive Division Cycles.
These studies reveal that heritable cell components contribute to the decision of whether a cell will divide or not. Furthermore, they indicate that the strength of the effect of these components are in some way correlated with cell size at division. Because all cells eventually reach division cessation these data were best explained by the hypothesis that critical cell-cycle components made by the founder during the slow first-division cycle were diluted in families through consecutive divisions. If correct we might also expect that cell size would progressively diminish as cells passage through division rounds. To test this possibility we made accurate measurements of the individual cell size pedigrees presented earlier (see Materials and Methods and Fig. 6). By fitting lines to the growth curves we avoided the noise in size measurements and gained estimates of cell size during division rounds, rates of growth, and division times. The data presented in Fig. 6 show a clear trend for cells to diminish in size as they progress through divisions (Table S6 in SI Appendix). We also found that cells with a larger size at time of division corresponded with a faster progression through the subsequent cell cycle although this correlation was weak (Fig. S9). More strikingly, the time of passage through the penultimate division was always slow when compared with the preceding division rounds (≈12 compared with 9 h) (Fig. 6 and Table S6 in SI Appendix).
Fig. 6.
Analysis of cell size pedigrees illustrates a changes in cell size, rate of growth, and lengthening in cell cycle times through divisions. A simple growth and division fitting tool was developed to analyze the cell size pedigree plots shown in Fig. S3 in SI Appendix. Data points represent the means of measurements. Error bars (blue) represent the SEM of division times (x axis) and cell size (y axis).
Mothers of Nondividing Cells Take Longer to Divide.
The above data reveal that the decision to enter a round of division and the time taken to divide both are influenced by founder-derived heritable factors. If the same heritable factors control both results we would expect that mothers that give rise to daughters that do not grow must themselves have been close to the threshold required to motivate division and would, therefore, always divide slowly. This prediction is consistent with the data in Fig. 6. We explored this possible correlation further by comparing the division times for mothers that gave rise to daughters that both divided (type 1), one divided and one died (type 2), and both died (type 3). This analysis confirmed our expectation that average division times of type 3 mothers were longer than for type 2 (12.17 h compared with 10.07 h), which in turn were longer than for type 1 mothers (8.13 h) (Fig. S10A in SI Appendix). Following a similar line of reasoning, we predicted that siblings that both divided would have a shorter division time than a cell that had a sibling that did not divide and went on to die. Confirmation of this prediction is shown in Fig. S10 in SI Appendix (9.73 h compared with 16.13 h). We also noted that the frequency that one daughter cell divided and one died was very rare (Fig. S10 in SI Appendix), suggesting that unequal distribution of cellular components necessary for division is an unlikely event. Furthermore, cells that take >15 h to divide show much poorer time correlation with their siblings and have a higher probability that their partner will not divide (Fig. S11 in SI Appendix).
Discussion
One of the primary motivations for this study was to support the development of mathematical models of lymphocyte growth and death that can serve as a bridge to translate molecular knowledge to cellular operation in a way that can predict population outcomes. The development of scaleable models with this capability is a key goal of systems biology. To provide the framework for models with this capacity we have analyzed primary murine resting B lymphocytes stimulated through the TLR-9 receptor that undergo a typical, although abbreviated, immune response. CpG is a T-independent response that is usually characterized by short-term proliferation without leading to long-lived plasma cells or memory responses (18). Our results give some insight into the regulation of this outcome and the in-built programming that results in the self-limiting behavior.
The stimulated cells take an average of 35 h to divide the first time and then go through cycles of cell growth and division up to 5 times at an average division time of 9 h. After a variable number of divisions daughter cells no longer grow and will eventually die. A minimal model to describe these features of lymphocyte growth and death must include quantitative tuning of three interacting mechanical controls governing the fate of each cell. One regulates the decision to enter a growth phase that leads to cell division. A second regulates the time to divide if initiated, and, the third regulates the time to die if growth is not initiated. All three controls showed evidence that their operation can be altered by passage through cell division. In addition each mechanical control displayed an extraordinary degree of variation that ensured the experience of every cell was unique. This level of variation was consistent between experiments and appeared intrinsic to the operation of the lymphocytes. Incorporated within these stochastic features were surprisingly strong family correlations. These observations provide an insight into the internal molecular regulation of cell growth cessation.
The ability of B cells to undergo a limited number of divisions in vitro has been observed for cells stimulated by CpG, LPS, and anti-CD40 (17, 18). Here, we found that most progeny of a single founder divided and exhibited a similar division destiny. Furthermore, this property, the average division destiny of the progeny cells, was correlated with the size of the founder at first division. As a result we proposed the hypothesis that an internal factor synthesized during the slow first division and divided among daughter cells regulated entry to cell growth leading to division. We also argued that this factor contributed to setting of division times by the dividing lymphocytes. This hypothesis explains the strong correlation in division destiny among families and the tendency for mothers that give rise to daughters that do not divide themselves take longer than average to divide. The variation in growth rate and cell size reached by founder cells suggests that they vary in their sensitivity to the stimulus provided, presumably because of stochastic variation in the cellular machinery for detecting CpG, such as the TLR9 receptor number or levels of downstream signaling components. Although we are unsure of the identity of heritable factors promoting entry to cell cycle, cyclin, cyclin-dependent kinases (CDK), and CDK inhibitors serve as excellent initial candidates for investigation. The CDK inhibitors P18 and P27 and cyclins D2 and E are known to contribute to humoral immune responses by modifying cell-cycle entry and plasma cell differentiation (22). Biologically based models that capture effects on division destiny and attempt to reproduce all of the correlative features of our data will be presented elsewhere.
Earlier studies of correlations in division times by siblings and mother–daughters for dividing mammalian cells in a steady-state growth phase have given a range of answers. All studies found positive sibling correlations (23–25), while both positive and negative mother–daughter correlations have been reported (23, 24). Further investigation as to whether our high sibling correlations will be repeated for T lymphocytes or B lymphocytes stimulated in other ways are warranted. However, the data provide a very convincing argument that division times can be internally regulated by constituents of a cell handed to it by the mother. An alternative explanation for sibling correlations is that division times are influenced by external factors such as the local microenvironment. However, we reject this hypothesis for two reasons. First, many of the sibling pairs are highly motile and are capable of migrating a significant distance from each other while sharing a related division time. And second, despite the considerable variation in the number of cells seeded in each terasaki well, we observe no statistically significant differences in division times, death times, pedigree sizes or division destiny between wells (Numerical Methods in the SI Appendix).
Recently Chang et al. (26) have observed that the first division of T cells in response to infection can induce an asymmetric division that leads to daughter cells with different proliferative and differentiation fates. In contrast, we found a remarkable similarity in subsequent behavior of each daughter produced upon the first division of founder cells. Although unable to track differentiation, the times to divide and division destiny of the first siblings were strongly correlated, suggesting there is no asymmetric first cell division in this system. This result is analogous to lymphocytes undergoing homeostatic T cell proliferation where asymmetric cell division is also not observed (26).
Although division times in families have been studied in cell culture before, there have been little data reported for death times in siblings during a regulated phase of division followed by death. We found a strong correlation among siblings for propensity to die (that is, if one sibling died, the other was likely to die and not divide). In instances where both siblings were followed, only 11% and 7% of cell pairs were observed to have different fates (i.e., one sibling died while the other divided) in Fam2 and Fam3, respectively (Fig. S10 in SI Appendix). We also observed a correlation in times to die (Fig. S8 in SI Appendix), although it was substantially weaker than that seen for sibling division times. Nevertheless, shared death times provide evidence that the highly variable death times seen in the population are the result of changes and fluctuations in internal molecular events and that the control of this variation can play an important role in the regulation of population growth and decline (17).
Collectively these mechanical features of cell control can be captured into mathematical frameworks that attach chosen probability distributions around each mechanical operation. Without a molecular model of the cellular operations that result in the observed randomness we can make no fundamental prediction of the “correct” probability distribution surrounding division and death times. However, by fitting known distributions we found a number of excellent candidates that provide an extremely accurate quantitative framework for future models of lymphocyte responses. In particular, we noted that the simple lag-exponential of Smith and Martin (19) can be greatly improved with other skewed-right distributions for both division and death times (Tables S1–S4 in SI Appendix). Furthermore, the shape of these skewed right distributions is consistent with the predicted and measured variation in single gene expression in a cell population (27, 28) that may contribute to the cell–cell differences (17). Adopting these distributions into a general framework such as the cyton model provides a valuable tool for dissecting and calculating the effect of multiple parameter changes on the population response (17, 29). A potential weakness of this model is that it does not assume the high level of correlation in division times of siblings or the strong founder effect on division destiny revealed here. However, further calculations have shown that these correlations do not alter the predictions of the mean number of cells at any time (29). However, these correlations will affect the higher moments of the calculation, serving to increase the expected variation for a given starting cell number (29).
The cell volume growth, proliferation, cessation of cell division, and gradual loss of CpG-stimulated cells is a simple, highly reproducible model system for studying the regulation of growth and survival at the molecular, cellular, and population levels. The response is a condensed form of that seen for all adaptive immune responses, whether CD8-, CD4-, or T-dependent B cell activation. As a model it will be useful for studying the molecular controls that put a brake on cell division after a nominated number of cycles and the molecular source of the remarkable variation inherent in every kinetic process. Furthermore, this model can shed light on other biological processes that shape a population response by simultaneous manipulation of cell division and cell death.
Materials and Methods
Mice.
Inbred 6- to 8-week-old male C57BL/6 mice were used for all experiments. These mice were bred at the Walter and Eliza Hall Institute (WEHI) animal facility (Kew, Australia) and maintained in specific pathogen-free conditions at WEHI (Parkville, Australia) in accordance with institutional animal ethics committee regulations.
B Cell Isolation and Microscopy Cell Culture.
B cells preparation and CFSE labeling, where performed, were conducted as described (17). Cell numbers were determined by ratio to a known number of added identifiable beads as described (24). Small resting B cells were cultured in B cell media (20) and seeded into Terasaki tissue culture plates in the presence of 1 μM CpG (sequence, 5′-TCCATGACGTTCCTGATGCT-3′; Geneworks) and 1 μg/mL PI (Sigma). B cells were initially cultured for 24 h at 37 °C and 10% CO2 before transfer to an environment-controlled microscope (Carl Ziess) maintained at these conditions. Bright-field and CY3.5-filtered images were taken every 2 min for 4–5 days.
Analysis of Division and Death Times (Cell Tracking).
All cell tracking was conducted manually to ensure accuracy. Cells were followed until they either divided, died (as determined by uptake of PI and fluorescence in the CY3.5 channel and morphology in the bright field), or were lost because of interactions with neighboring cells, or the experiment ended. For cell size tracking analysis of pedigrees, images were compiled in stack format and processed to detect a threshold by using Metamorph software (Molecular Devices) and measurements taken frame by frame according to original manual tracking trajectories.
Supplementary Material
Acknowledgments.
We thank G. Belz, L. Corcoran, K. Duffy, B. Heath, S. Nutt, A. Strasser, and D. Tarlinton for critical reading of the manuscript and helpful suggestions; C. Wellard for help with implementation of numerical methods, discussions, and proofreading; and A. Perelson and R. de Boer for challenging us to measure the distributions of cell cycle times in primary cells. This work was supported by the National Health and Medical Research Council and the Human Frontiers Science Program. E.D.H was supported by an Australian Post Graduate Award.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905629106/DCSupplemental.
References
- 1.Antia R, Ganusov VV, Ahmed R. The role of models in understanding CD8+ T cell memory. Nat Rev Immunol. 2005;5:101–111. doi: 10.1038/nri1550. [DOI] [PubMed] [Google Scholar]
- 2.Borghans JA, de Boer RJ. Quantification of T cell dynamics: From telomeres to DNA labeling. Immunol Rev. 2007;216:35–47. doi: 10.1111/j.1600-065X.2007.00497.x. [DOI] [PubMed] [Google Scholar]
- 3.Dawson KB, Madoc-Jones H, Field EO. Variations in the generation times of a strain of rat sarcoma cells in culture. Exp Cell Res. 1965;38:75–84. doi: 10.1016/0014-4827(65)90429-5. [DOI] [PubMed] [Google Scholar]
- 4.Marin G, Bender MA. Radiation-induced mammalian cell death: Lapse-time cinemicrographic observations. Exp Cell Res. 1966;43:413–423. doi: 10.1016/0014-4827(66)90068-1. [DOI] [PubMed] [Google Scholar]
- 5.Froese G. The distribution and interdependence of generation times of Hela cells. Exp Cell Res. 1964;35:415–419. doi: 10.1016/0014-4827(64)90108-9. [DOI] [PubMed] [Google Scholar]
- 6.De Boer RJ, Ganusov VV, Milutinovic D, Hodgkin PD, Perelson AS. Estimating lymphocyte division and death rates from CFSE data. Bull Math Biol. 2006;68:1011–1031. doi: 10.1007/s11538-006-9094-8. [DOI] [PubMed] [Google Scholar]
- 7.De Boer RJ, Homann D, Perelson AS. Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J Immunol. 2003;171:3928–3935. doi: 10.4049/jimmunol.171.8.3928. [DOI] [PubMed] [Google Scholar]
- 8.Ganusov VV, et al. Quantifying cell turnover using CFSE data. J Immunol Methods. 2005;298:183–200. doi: 10.1016/j.jim.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Pilyugin SS, Ganusov VV, Murali-Krishna K, Ahmed R, Antia R. The rescaling method for quantifying the turnover of cell populations. J Theor Biol. 2003;225:275–283. doi: 10.1016/s0022-5193(03)00245-5. [DOI] [PubMed] [Google Scholar]
- 10.Yates A, et al. Reconstruction of cell population dynamics using CFSE. BMC Bioinformatics. 2007;8:196. doi: 10.1186/1471-2105-8-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castor LN. A G1 rate model accounts for cell-cycle kinetics attributed to transition probability. Nature. 1980;287:857–859. doi: 10.1038/287857a0. [DOI] [PubMed] [Google Scholar]
- 12.Cooper S. The continuum model: Statistical implications. J Theor Biol. 1982;94:783–800. doi: 10.1016/0022-5193(82)90078-9. [DOI] [PubMed] [Google Scholar]
- 13.Cowan R, Staudte R. The bifurcating autoregression model in cell lineage studies. Biometrics. 1986;42:769–783. [PubMed] [Google Scholar]
- 14.Koch AL. Does the variability of the cell cycle result from one or many chance events? Nature. 1980;286:80–82. doi: 10.1038/286080a0. [DOI] [PubMed] [Google Scholar]
- 15.Hasbold J, Corcoran LM, Tarlinton DM, Tangye SG, Hodgkin PD. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat Immunol. 2004;5:55–63. doi: 10.1038/ni1016. [DOI] [PubMed] [Google Scholar]
- 16.Hodgkin PD, Lee JH, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J Exp Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawkins ED, Turner ML, Dowling MR, van Gend C, Hodgkin PD. A model of immune regulation as a consequence of randomized lymphocyte division and death times. Proc Natl Acad Sci USA. 2007;104:5032–5037. doi: 10.1073/pnas.0700026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner ML, Hawkins ED, Hodgkin PD. Quantitative regulation of B cell division destiny by signal strength. J Immunol. 2008;181:374–382. doi: 10.4049/jimmunol.181.1.374. [DOI] [PubMed] [Google Scholar]
- 19.Smith JA, Martin L. Do cells cycle? Proc Natl Acad Sci USA. 1973;70:1263–1267. doi: 10.1073/pnas.70.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deenick EK, Gett AV, Hodgkin PD. Stochastic model of T cell proliferation: A calculus revealing IL-2 regulation of precursor frequencies, cell cycle time, and survival. J Immunol. 2003;170:4963–4972. doi: 10.4049/jimmunol.170.10.4963. [DOI] [PubMed] [Google Scholar]
- 21.Tyson JJ, Diekmann O. Sloppy size control of the cell division cycle. J Theor Biol. 1986;118:405–426. doi: 10.1016/s0022-5193(86)80162-x. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, et al. Homeostatic cell-cycle control by BLyS: Induction of cell-cycle entry but not G1/S transition in opposition to p18INK4c and p27Kip1. Proc Natl Acad Sci USA. 2004;101:17789–17794. doi: 10.1073/pnas.0406111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Absher M, Cristofalo VJ. Analysis of cell division by time-lapse cinematographic studies of hydrocortisone-treated embryonic lung fibroblasts. J Cell Physiol. 1984;119:315–319. doi: 10.1002/jcp.1041190309. [DOI] [PubMed] [Google Scholar]
- 24.Colly-d'Hooghe M, Valleron AJ, Malaise EP. Time-lapse cinematography studies of cell cycle and mitosis duration. Exp Cell Res. 1977;106:405–407. doi: 10.1016/0014-4827(77)90190-2. [DOI] [PubMed] [Google Scholar]
- 25.Minor PD, Smith JA. Explanation of degree of correlation of sibling generation times in animal cells. Nature. 1974;248:241–243. doi: 10.1038/248241a0. [DOI] [PubMed] [Google Scholar]
- 26.Chang JT, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 27.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 28.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian VG, Duffy KR, Turner ML, Hodgkin PD. Determining the expected variability of immune responses using the cyton model. J Math Biol. 2008;56:861–892. doi: 10.1007/s00285-007-0142-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.