Abstract
Alveolar epithelial cells are among the first cells to encounter inhaled particles or organisms. These cells likely participate in the initiation and modulation of the inflammatory response by production of chemokines. However, there is little information on the extent or regulation of chemokine production by these cells. Rat type II cells were studied under differentiated and dedifferentiated conditions to determine their ability to express and secrete CXC chemokines. Both differentiated and dedifferentiated type II cells secreted MIP-2, MCP-1, and CINC-2 in response to a cytokine mixture of IL-1β, TNF-α, and IFN-γ or to IL-1β alone. The cytokine mixture also induced iNOS expression and nitrite secretion. Both differentiated and dedifferentiated type II cells expressed CINC-1 (GRO), CINC-2α, CINC-3 (MIP-2), and MCP-1 mRNA, and their expression was increased by the cytokine mixture or by IL-1β alone. However, CINC-2β, a splice variant of CINC-2, was only expressed under differentiated conditions stimulated by KGF and was not increased by the cytokine mixture or by IL-1β. In situ hybridization of normal lung and lung instilled with Ad-KGF demonstrated that CINC-2β was expressed by alveolar and bronchiolar epithelial cells in vivo. We conclude that CINC-2β is regulated differently from most other chemokines and that its expression is related to the state of alveolar type II cell differentiation.
Keywords: CCL2, chemokines, CXCL1, inflammation, type II cells
The pulmonary epithelium is one of the first lines of defense against inhaled particles, fumes, and organisms. One of the initial responses of the lung at sites of injury is to secrete a variety of chemokines to recruit neutrophils and monocytes. Previous research has focused on resident macrophages and dendritic cells as sources of these chemokines (1–3). However, it is probable that epithelial cells also secrete these chemokines. For example, bronchial epithelial cells have been reported to secrete IL-8 (CXCL8) (4), MIP-2 (CXCL2/3) (5), eotaxin (CCL11) (6), RANTES (CCL5) (7), and MCP-1 (CCL2) (8). The airway epithelium is thought to participate in initiating the inflammatory response to particles, ozone, and viral infections (9–12). The production of chemokines by bronchial epithelial cells is also important in persistent inflammation in chronic conditions such as asthma and cystic fibrosis (4, 13). However, the role of the alveolar epithelium in distal lung inflammation is less well defined. Alveolar type II cells have been reported to express MCP-1 (14, 15), MIP-2 (16), and RANTES (16). More recently, Vanderbilt and coworkers reported that alveolar type II cells express GRO (CINC-1) (CXCL1) and that the expression was markedly stimulated by instillation of Pseudomonas aeruginosa or acid (17). However, there are very little data on actual levels of secreted chemokines by type II cells so as to compare their capability to macrophages or other resident lung cells.
In vitro alveolar type II cells can be cultured as a polarized epithelium under conditions of differentiation as defined by expression of the surfactant proteins and phospholipid secretion or under conditions of dedifferentiation (18, 19). Under dedifferentiated conditions, the cells are flat, they spread, and they do not secrete surfactant components (20, 21). The exact phenotype of dedifferentiated type II cells in vitro and how closely these cells represent type I cells is controversial. However, they do increase their expression of type I cell markers, such as T1-α (22). In this study we sought to determine which chemokines were secreted by rat alveolar epithelial cells in primary culture and if the secretion of chemokines was related to their state of differentiation.
In the rat, the CXC chemokines that recruit neutrophils are termed cytokine-induced neutrophil chemoattractants (CINC) (23, 24). There are four members of the rat CINC family, namely CINC-1 (CXCL1, GRO), CINC-2α, CINC-2β, and CINC-3 (CXCL 2/3, MIP-2). CINC-1, CINC-2, and CINC-3 belong to the CXC chemokine family and are potent chemotactic factors for neutrophils (25, 26). There is one CINC-2 gene, and CINC-2α and CINC-2β arise by alternate RNA splicing (26). The only differences between CINC-2α and CINC-2β proteins are the three amino acids at the C-terminus (27). Both CINC-2α and CINC-2β are chemotactic for neutrophils with an effect at a 10 nM concentration (∼ 80 ng/ml) (27). CINC family members are thought to play a major role in the recruitment of neutrophils into rat lungs (28, 29). For example, CINC expression is thought to account for neutrophil accumulation in the lungs after instillation of LPS, Pseudomonas aeruginosa, or acid (17, 30). MIP-2 and MCP-1 are thought to be largely responsible for the neutrophil and macrophage infiltration in pneumococcal pneumonia (31). CINCs recruit neutrophils by signaling through the CXCR2 receptor (32).
Our hypothesis was that all CINC isoforms would be secreted by alveolar epithelial cells in response to cytokines but that differentiated type II cells would secrete much more chemokines than dedifferentiated alveolar epithelial cells. This hypothesis was based on the concept that type II cells play an important role in innate immunity (33). We measured protein levels as well as mRNA levels for those chemokines for which ELISA assays were available. The unexpected finding was that CINC-2β is regulated very differently from the other family members in that its expression appears to be restricted to differentiated type II cells and is not increased by cytokines.
MATERIALS AND METHODS
Isolation and Culture of Alveolar Type II Cells
Alveolar type II cells were isolated from specific pathogen–free adult male Sprague-Dawley rats (Harlan, Indianapolis, IN) by dissociation with porcine pancreatic elastase (Roche Diagnostics, Indianapolis, IN) and partial purification on discontinuous metrizamide gradients (34). Type II cells were plated on a filter insert (30-mm-diameter Millicell-CM; Millipore, Bedford, MA) that had been coated with 0.4 ml of a 4:1 (vol/vol) mixture of 0.8 mg/ml rat-tail collagen and 2 mg/ml EHS tumor matrix (Matrigel; BD Biosciences, Bedford, MA) as described previously (18, 19). The mixture was prepared at 4°C and allowed to gel at 37°C. Cell viability was determined with erythrosin B exclusion, and 2.5 × 106 viable cells were plated in 1 ml of DMEM containing 5% rat serum (Pel Freez Biologicals, Rogers, AK) plus 2 mM glutamine, 2.5 μg/ml amphotericin B, 100 μg/ml streptomycin, and 100 μg/ml penicillin G; and 2 ml of the same media was added to each well outside the insert. After attachment for 20 h, 0.4 ml of the specified medium was added to the apical surface, and 2 ml of the medium was placed outside the insert. In different experiments, the medium contained rat serum alone or rat serum plus 10 ng/ml KGF (R&D Systems, Minneapolis, MN). The wells were then placed on a rocking platform inside an incubator gassed with 10% CO2. The medium was changed every 48 h (18).
Experimental Design
The cells were stimulated by either a mixture of rat cytokines (IL-1β 10 ng/ml, TNF-α 10 ng/ml, and IFN-γ 10 ng/ml; R&D Systems) or by IL-1β (1, 10, or 100 ng/ml) alone on the seventh day of culture for chemokines expression. The cells were incubated for 4 h after addition of cytokines for mRNA determinations and for 24 h for protein secretion, iNOS protein expression, and nitrite measurement.
Measurement of SP-A and SP-D
SP-A and SP-D were measured by ELISA as described previously (35). Recombinant rat SP-A and SP-D produced in Chinese hamster ovary cells were used as the SP-A and SP-D standards. The IgG was purified on protein A sepharose. The standards and antibodies were generous gifts of Dennis R. Voelker and Amanda Evans (Denver, CO).
Measurement of Chemokines Secretion
MIP-2 and MCP-1 were measured with commercial ELISA kits according to the manufacturer's instructions (BioSource, Camarillo, CA). CINC-2 was measured with an ELISA based on standard techniques, with standards and antibodies provided by R&D Systems. The antibody for CINC-2β (R&D Systems) used in this ELISA shows significant crossreactivity to CINC-2α, and hence the product of this ELISA is referred to as simply CINC-2. The CINC-2 ELISA assay was established by Jay Westcott (ELISA Tech, Aurora, CO).
Measurement of DNA
To harvest type II cells for DNA assay, the collagen-EHS gel was teased off the insert and placed in a polypropylene tube. The matrix was digested by incubation with 1 ml of a 1:4 (vol/vol) mixture of 5 mg/ml type I collagenase (Worthington Biochemical, Lakewood, NJ) and dispase (BD Biosciences, Bedford, MA). The cells were collected in saline, sedimented, resuspended, and washed once before being resuspended in phosphate buffer for the DNA analysis. The suspension was frozen and stored at −20°C. After being thawed, the cells were sonicated and the DNA content was determined fluorometrically (Hoechst 33258) (36).
Western Blotting for iNOS
Immunoblotting was performed as previously described (18). The cells were washed with PBS three times, then lysed in 100 μl of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 8], 50 mM NaCl, 0.5% deoxycholate, 0.2% SDS, 1% NP-40) plus protease inhibitors (160 μg/ml benzoamidine, 100 μg/ml phenanthrolene, 100 μg/ml aprotinin, 100 μg leupeptin, 100 μg/ml pepstatin A; PharMingen, San Diego, CA), and a mixture of phosphatase inhibitors (Sigma, St. Louis, MO). Samples were triturated 15 times through 25-gauge needles and centrifuged at 4°C and 14,000 rpm to remove any insoluble membrane fraction. Protein that was equivalent to 100 ng of cellular DNA was separated by SDS-PAGE under reducing conditions using 8–16% gradient gels (Invitrogen, Carlsbad, CA). Gels were transferred to nitrocellulose or PDVF membranes. Blots were blocked in 5% nonfat dry milk in Tris-buffered salt solution and incubated with rabbit anti-iNOS antibody (Alexis Biochemicals, San Diego, CA) or goat anti-actin antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature. The immunoblots were washed and incubated with a secondary antibody, either horseradish peroxidase–conjugated anti-rabbit or anti-goat (Jackson ImmunoResearch Laboratories, West Grove, PA), for 30 min at room temperature. Antigens were detected by enhanced chemiluminescence according to manufacturer's instructions (Amersham Biosciences, Piscataway, NJ). Densitometry was used to quantify protein expression with the NIH image analysis program.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed as previously described (18). Type II cells on the gels were directly lysed into 4 M guanidinium isothiocyanate, 0.5% N-laurylsarcosine, and 0.1 M 2-mercaptoethanol in 25 mM sodium citrate buffer. Total cellular RNA was isolated by ultracentrifugation for 18 h at 150,000 × g through a 5.7 M CsCl cushion. Any genomic DNA was removed by treatment with DNase.
Quantitative real-time RT-PCR analyses for CINC-2α, CINC-2β, CINC-3, MCP-1, T1-α, and GAPDH mRNAs were performed using the ABI PRISM 7700 Sequence Detection System instrument and software (PE Applied Biosystems, Inc., Foster City, CA). The methods used have been reported previously (18, 37, 38). Primers and probes for the TaqMan system were designed using Primer Express software (Perkin Elmer, Foster City, CA). The sequences of the PCR primer pairs and fluorogenic probes used for each gene are shown in Table 1.
TABLE 1.
The sequences of the pcr primer pairs and fluorogenic probe
| Gene | Accession No. | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|---|
| CINC-1 | D11444 | GGGTGTCCCCAAGTAATGGA | TGTCAGAAGCCAGCGTTCAC | CAGACGCCATCGGTGCAATCTATCTTCTT |
| CINC-2α | D87927 | CCAGCTGAGCTGGGAAAGG | GGATCGCTGCTCTGCTTCA | AGGCAGGTCCTCCATCACCGTACAAGA |
| CINC-2β | D21095 | GAGACGGGAATGCAATTTGTTT | GGTCTGCTAGGAATGTTGTCGAT | CATCCGAATTCTACGTGCGTGAGGACTCT |
| CINC-3 | RNU45965 | CGGGCAGAATCAAAGAGAAAA | CTCAGACAGCGAGGCACATG | ACAAACTGCACCCAGGAAGCCTGG |
| MCP-1 | AF058786 | CTCACCTGCTGCTACTCATTCACT | CCTGCTGCTGGTGATTCTCTT | GTTCTCCAGCCGACTCATTGGGATCA |
| T1-α | RNU07797 | AACCGCTTCTTTCTGGACGAT | GGCTCTGGCATTTTGTGACA | CTCATCCCAGATGCTCAGAAAGTTTGTTGG |
Northern Analyses
Two probes for CINC-2β were generated by PCR for Northern analysis from type II cell RNA. The first was based on accession number BF553317 and used CCT CCCTGTGACA CTGAAGAGTT AC as the forward primer and GAA AAGCAGCTAG AGTTCCCCAG as the reverse primer. The second probe used accession number D21095. The forward primer was the same as that used for BF5553317 and the reverse primer was CCG TCCTGAGGCT CCATAAATG. The probe for CINC-2α was based on accession number BQ209915 and used GGAATTCTCC TGTGACGCTG TAAAAACCAC as the forward primer and CGGATCCATC CATCCAATGC TGCCTGG as the reverse primer. The PCR products were directionally cloned into plasmids and amplified using methods reported previously (18, 37). All probes were verified by sequencing.
RNA was isolated as stated for the real-time PCR measurements. Northern analyses used the probe isolated with primers from accession number D21095 and were performed as described previously (39). Type II cells were directly lysed into 4 M guanidinium isothiocyanate, 0.5% N-laurylsarcosine, and 0.1 m β-mercaptoethanol in 25 mM sodium citrate buffer. Total RNA was isolated by centrifugation through a 5.7 M CsCl cushion. Northern blots were probed with complementary DNAs that had been radiolabeled with [α-32P] deoxycytidine triphosphate. Hybridization, washing the blots, and imaging were performed as previously described (39). In another series of experiments, a nonradioactive method with DIG-labeled cDNAs (Roche Molecular Biochemicals, Indianapolis, IN) was used. This probe was constructed with primers from accession number BF553317 and produced the same result.
Microarray Analyses
Expression gene profiling was performed with Affymetrix rat microarrays and analyzed as reported in detail previously (18). Data from these experiments were added to document the state of gene expression of the differentiated and undifferentiated alveolar epithelial cells, and part of the data has been published previously (18).
In Situ Hybridization
Normal rats and rats that had been instilled with Ad-KGF were killed, and lungs were fixed with 4% paraformaldehyde as described in detail (34). In situ hybridization was performed as previously described (18, 40). Tissue sections (4–6 μM) were mounted on Super Frost II glass slides (Fisher Scientific, St. Louis, MO) and hybridized with 33P-labeled sense or antisense RNA Probes transcribed from rat cDNA for CINC-2α and CINC-2β. After a series of high-stringency washes, the slides were dipped in Kodak NTB-2 nuclear track emulsion (Eastman Kodak, Rochester, NY). Autoradiograms were exposed in light tight boxes for 17 d at 4°C, developed, and then counterstained with hematoxylin.
Statistics
t Tests were used to compare cytokine treatments to control values, when a single treatment was used. The t test was also used to compare the effect of KGF plus rat serum to rat serum alone in the absence of cytokines. The Kruskal-Wallis test and Dunn's multiple comparison tests were used to determine if any IL-1β treatments were different from control. Statistical significance was defined as P < 0.05, and values are presented as means ± SE.
RESULTS
Cytokines Increase CINC-2, MIP-2, and MCP-1 Secretion and iNOS Expression
To determine if alveolar epithelial cells could secrete CXC chemokines, alveolar cells were stimulated by a cytokine mixture of IL-1β, TNF-α, and IFN-γ (all 10 ng/ml). Isolated type II cells were cultured in rat serum plus KGF to produce differentiated type II cells or in rat serum alone to produce dedifferentiated type II cells (19) (Table 2). Cells were cultured for 6 d with KGF, since KGF produces reproducible differentiated alveolar type II cells, but it takes time to achieve this effect (19). Type II cells cultured with KGF proliferate and are differentiated as demonstrated by the expression and secretion of surfactant proteins as reported previously (18, 19) (Table 2). In the current experiments, KGF stimulated proliferation as measured by DNA per well (21.9 ± 1.8 μg DNA per well for KGF plus rat serum versus 5.0 ± 0.5 μg DNA per well for rat serum alone [n = 7]). KGF also stimulated differentiation as measured by SP-A secreted into the apical medium. The SP-A level in the medium collected from Day 7 to Day 8 of culture was 1,504 ± 350 μg/ml for KGF plus rat serum and 23 ± 8 μg/ml for rat serum alone (n = 7). The dedifferentiated type II cells cultured in rat serum alone have a very low level of surfactant protein gene expression (18), but they have an increased level of expression of T1-α, a marker of the type I cell phenotype. By real-time PCR the dedifferentiated type II cells (5% rat serum) expressed 3.1 ± 0.5 times more T1-α mRNA than the differentiated type II cells (KGF + rat serum), when the mRNA values were normalized to GAPDH (n = 5). By immunoblotting there was a similar increase in T1-α protein in cells cultured in rat serum alone (data not shown). Table 1 shows some additional gene profiling data on these culture conditions to document the two phenotypes. By oligonucleotide microarray analyses the differentiated cells cultured with KGF had an increased expression of type II cell markers (surfactant proteins) and a decreased expression of type I cell markers (T1α and α-crystallin B). The cytokine mixture was added from Day 7 to Day 8 of culture, and the cells and media were harvested at 24 h after the addition of the cytokines. Although the cytokine mixture caused no apparent morphologic change at 4 h, at 24 h some of the cells cultured with KGF plus the cytokine mixture rounded up and detached. There was no significant effect of the cytokine mixture on cell attachment or DNA per well in the absence of KGF (rat serum alone). The cytokine mixture increased MIP-2, CINC-2, and MCP-1 secretion in both phenotypes, and this was apparent if the data were expressed per ml, per μg DNA, or per Millicell (Figure 1). For example, the secretion of MIP-2 was 0.50 ± 0.09 ng/μg DNA without stimulation and 4.64 ± 1.55 ng/μg DNA with cytokine stimulation in the dedifferentiated alveolar epithelial cells and 1.13 ± 0.10 ng/μg DNA without stimulation and 3.50 ± 0.50 ng/μg DNA with cytokine stimulation in the differentiated type II cells (n = 7). The secretion of MCP-1 was greater in the dedifferentiated alveolar epithelial cells (5% rat serum alone) (173 ± 34 pg/μg DNA without cytokines and 522 ± 11 pg/μg DNA with cytokines compared with those differentiated with KGF) (18 ± 7 pg/μg DNA without cytokines and 198 ± 65 pg/μg DNA with cytokines[n = 7]) (Figure 1). This observation is also supported by the mRNA data in Table 1. To document the response to the cytokine mixture further, nitrite in the medium and iNOS protein in the cells were also measured. The cytokine mixture stimulated nitrite level in the medium and iNOS protein level in the cells, and the response was independent of the state of differentiation (Figure 2).
TABLE 2.
Comparison of gene expression in alveolar type II cell cultured in kgf plus rat serum versus rat serum alone
| Name | Accession No. | Fold Change KGF+RS/RS |
|---|---|---|
| CINC-2β | D 21095 | 32.0 ± 1.76 |
| CINC-1 (CXCL1) | D 111445 | 1.7 ± 0.28 |
| MCP-1 (CCL2) | X 17053 | 0.28 ± 0.02 |
| Surfactant Protein A | M 33201 | 18.0 ± 1.7 |
| Surfactant Protein B | Af 170350 | 11.8 ± 0.7 |
| Surfactant Protein C | X 14221 | 6.3 ± 0.5 |
| Surfactant Protein D | M 81231 | 2.5 ± 0.2 |
| Glycoprotein 38 (T1α) | U 92081 | 0.50 ± 0.04 |
| Alpha-Crystallin B | M 55534 | 0.29 ± 0.07 |
Some of these data have been published previously (18). The data are expressed as fold change (mean ± SEM, N = 3).
Figure 1.
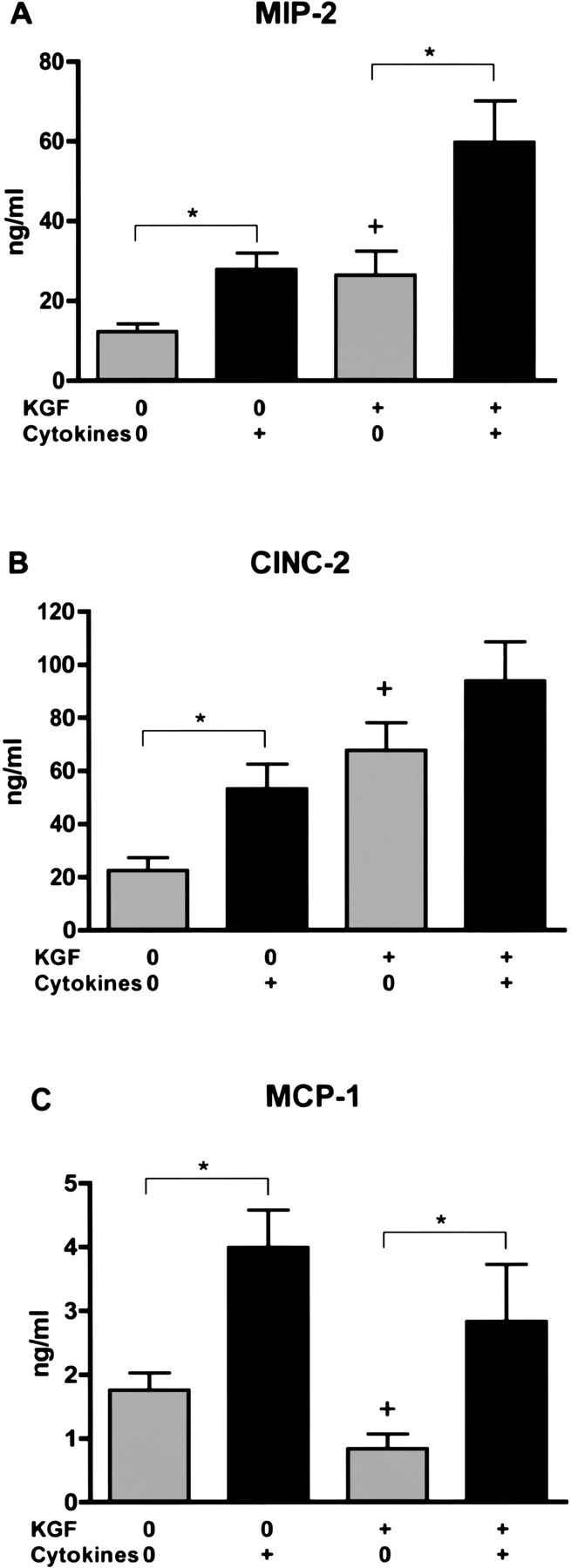
The combination of IL-1β, TNF-α, and IFN-γ increase chemokine secretion. Alveolar type II cells were cultured in rat serum with or without KGF. The mixture of cytokines (10 ng/ml IL-1β, 10 ng/ml TNF-α, and 10 ng/ml IFN-γ) was added on Day 7 of culture, and the cells and the apical media were collected 24 h later. The cytokine mixture increased the secretion of MIP-2 and MCP-1 in both differentiated and dedifferentiated alveolar type II cells. The results are shown as ng/ml and data per μg DNA are stated in the text. The means ± SE of seven independent experiments is shown. The asterisk signifies a statistically significant change (P < 0.05) compared with the controls without cytokines, and the plus sign signifies a significant change (P < 0.05) effect of KGF in the absence of cytokines.
Figure 2.
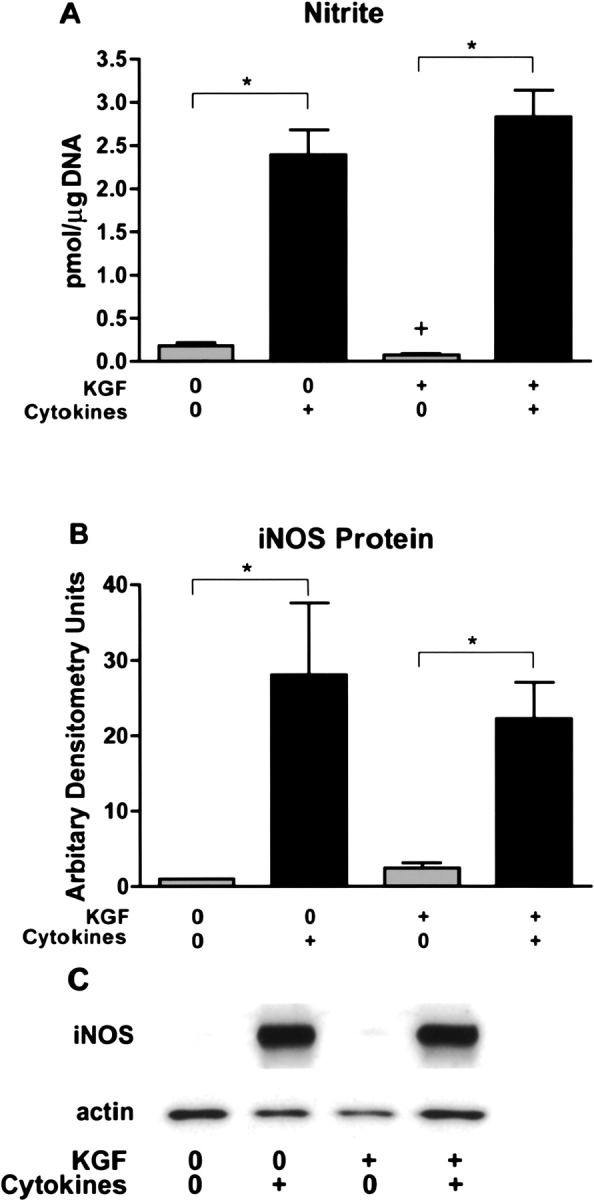
The combination of IL-1β, TNF-α, and IFN-γ increase nitrite production and iNOS expression. The conditions are the same as in Figure 1 and nitrite measurement in the media and iNOS immunoblotting were performed as stated in MATERIALS AND METHODS. The immunoblotting was normalized to actin. A shows nitrite secretion into the media, B shows the effect of the mixture of cytokines on iNOS protein levels as measured by immunoblotting, and C shows a representative immunoblot for iNOS and actin. The results are the mean ± SE of six experiments. The asterisk signifies P < 0.05 compared with the matched control without cytokines. The cytokine mixture of IL-1β, TNF-α, and IFN-γ increased iNOS protein and nitrite secretion independent of the state of differentiation.
IL-1β Stimulates MIP-2, CINC-2, and MCP-1 but Not SP-A or SP-D
Because of some apparent toxicity induced by the mixture of cytokines in the differentiated cells (KGF plus rat serum), the experiments were repeated with IL-1β alone. Under these conditions, there was no detachment. IL-1β alone (100 ng/ml) stimulated secretion of MIP-2, CINC-2, and MCP-1 (Figure 3). In data not shown, IL-1β alone also stimulated nitrite secretion in both phenotypes but less than with the cytokine mixture. IL-1β did not stimulate cell proliferation or secretion of SP-A or SP-D (data not shown). Hence, both differentiated and dedifferentiated alveolar type II cells secreted chemokines in response to a mixture of cytokines or IL-1β alone.
Figure 3.
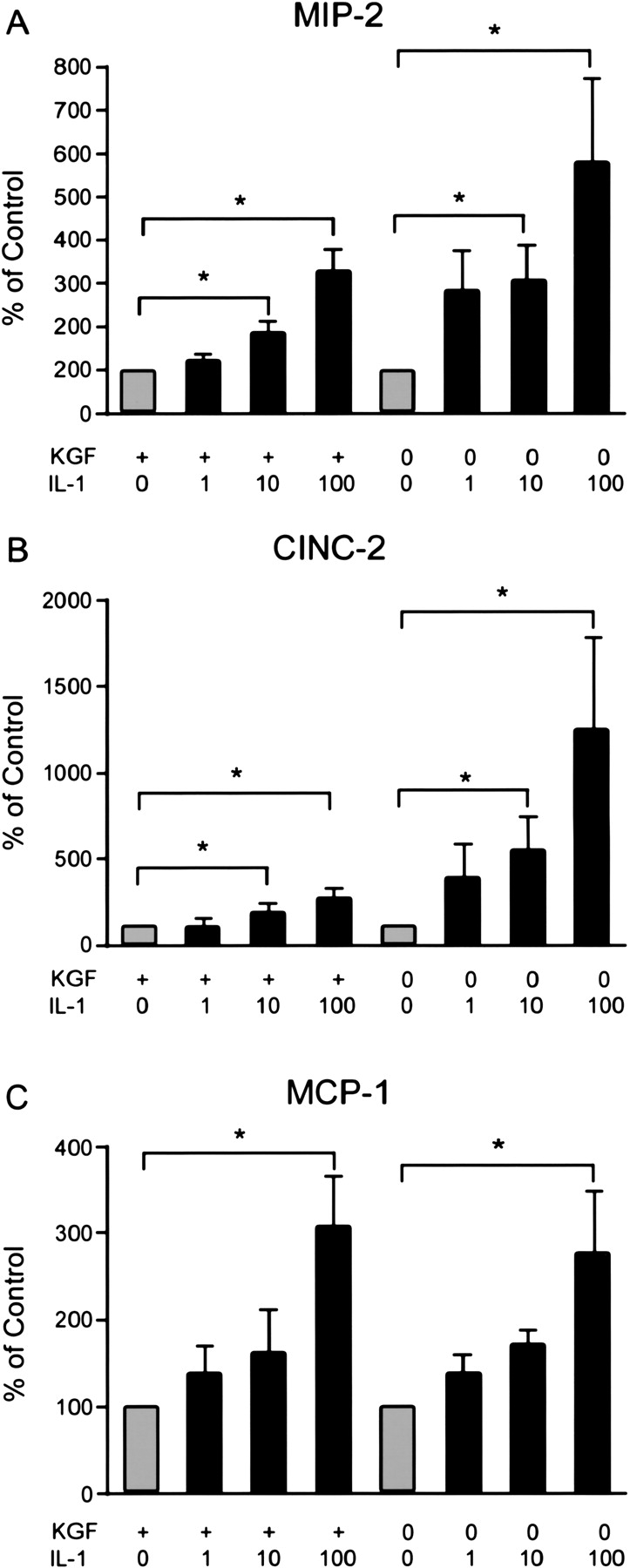
IL-1β alone increased chemokine secretion. The cells were cultured and harvested as stated in Figure 1. Varying concentrations of IL-1β were added on Day 7 of culture, and the media and cells were harvested 24 h later. IL-1β (100 ng/ml) increased the secretion of MIP-2, CINC-2, and MCP-1. The original values are ng/ml and have been normalized in each experiment to the value of the control without IL-1β for each phenotype and expressed as % of control. The results are the means ± SE of six independent experiments. The asterisk signifies an increase compared with the control without addition of IL-1β (P < 0.05).
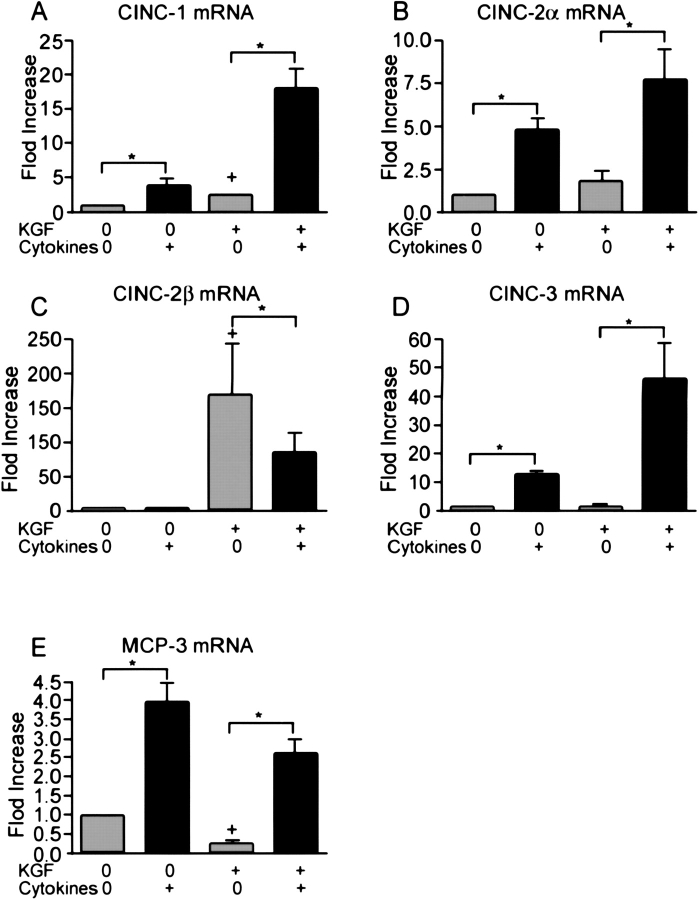
Cytokines Increase mRNA Levels of CINC-1, CINC-2α, CINC-3, and MCP-1 but Not CINC-2β
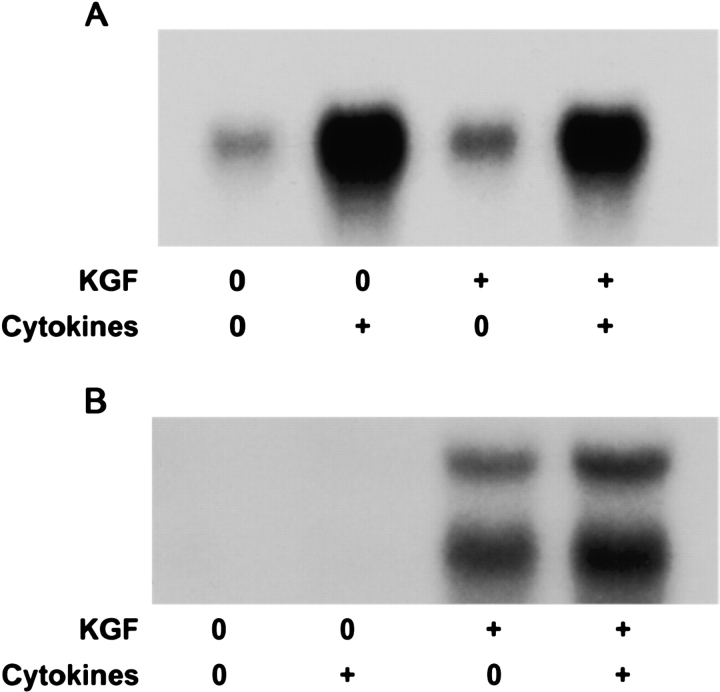
To define CXC chemokine expression more precisely, real-time quantitative PCR was used to measure chemokine mRNA levels after 4 h of incubation with cytokines. Primers and probes were constructed to measure the individual CINC chemokines. Because CINC-2α and CINC-2β are very similar and arise by alternative RNA splicing, primers and probes were designed to match the 3′ untranslated portions of the mRNA transcripts. The expression of CINC-1, CINC-2α, CINC-3, and MCP-1, were increased by the cytokine mixture independent of the state of differentiation (Figure 4). Interestingly, CINC-2β mRNA was only detected in the differentiated type II cell phenotype, and the expression was not stimulated by the cytokine mixture. In additional studies, IL-1β alone also increased expression of CINC-1, CINC-2α, CINC-3, and MCP-1, but not CINC-2β (data not shown). To confirm the real-time PCR data, we also performed Northern analyses. The cytokine mixture increased CINC-2α mRNA levels in both phenotypes (Figure 5A). However, expression of CINC-2β was detected only in the differentiated phenotype, and there was no increase with the cytokine mixture (Figure 5B). Another unexpected finding was that there were consistently two mRNA transcripts for CINC-2β (2.7 kb and 4.1 kb). Because of these unexpected results, a second probe was generated and another method of Northern analysis was used. Both sets of experiments confirmed the two transcripts, the increase in mRNA level of CINC-2α with cytokines, the decrease in CINC-2β mRNA level with cytokines, and detectable expression of CINC-2β only in differentiated type II cells.
Figure 4.
The cytokine mixture increased mRNA levels of CINC-1, CINC-2α, CINC-3, and MCP-1 but not CINC-2β. Type II cells were cultured as described in Figure 1, and the cytokine mixture (10 ng/IL-1β, 10 ng/ml TNF-α, and 10 ng/ml IFN-γ) was added on Day 7 of culture. The cells were harvested and the RNA was extracted 4 h later. The mRNA values were measured by quantitative real-time PCR as stated in MATERIALS AND METHODS. The results are the means ± SE for four independent experiments. The asterisk signifies an increase compared with the control without addition of cytokines, P < 0.05.
Figure 5.
The mRNA levels of CINC-2α and CINC-2β show different responses to cytokine stimulation. Differentiated and dedifferentiated cultures of alveolar type II cells were cultured as defined in MATERIALS AND METHODS. The cytokine mixture (10 ng/ml IL-1β, 10 ng/ml TNF-α, and 10 ng/ml IFN-γ) was added on Day 7 of culture, and 4 h later the mRNA was extracted and processed for Northern analyses. A shows results for CINC-2α and B shows results for CINC-2β. Cytokines increased mRNA levels for CINC-2α in both phenotypes. The estimated size for the single CINC-2α transcript is 0.8 kb. Cytokines did not increase the mRNA level for CINC-2β. The estimated sizes of the two transcripts for CINC-2β are 2.7 kb and 4.1 kb. The results are from one experiment but are representative of three independent experiments.
To confirm these observations, the Day 7 cultures were also tested for expression of CINC-2β and CINC-2α by in situ hybridization (18). CINC-2β was detected in the differentiated type II cells (rat serum plus KGF) but not the dedifferentiated type II cells (rat serum alone) (data not shown). Expression of CINC-2α was below the level of detection in these experiments.
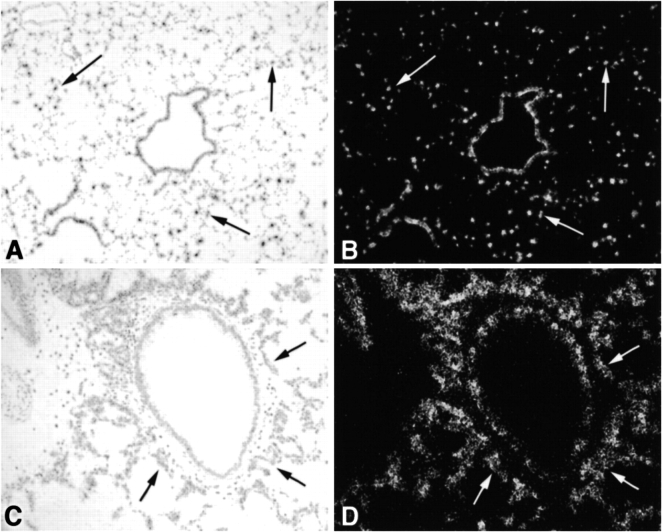
CINC-2β Is Expressed in Type II Cells In Vivo
To confirm the expression in vitro, in situ hybridization was used to demonstrate expression in vivo. Identifying individual type II cells in vivo in normal lung is difficult with 33P probes (18). However, hyperplastic type II cells can be readily identified in lungs previously instilled with an adenovirus expressing KGF (Ad-KGF) (35). CINC-2β was observed in airway epithelial cells (Clara cells) and alveolar cells (presumably alveolar type II cells) in the normal lung and in the hyperplastic type II cells in rats previously instilled with Ad-KGF (Figures 6C and 6D). Expression of CINC-2α was below the level of detection in these experiments.
Figure 6.
Expression of CINC-2 β mRNA in normal and Ad-KGF instilled rat lung. The expression of CINC-2β in vivo was determined by in situ hybridization in normal and Ad-KGF instilled rat lung. The in situ hybridization was performed as stated in MATERIALS AND METHODS. In A and B, normal lung is shown in brightfield and darkfield exposures (original magnification: ×100). In C and D, similar exposures of lung that had been instilled 48 h previously with Ad-KGF (35) are shown (original magnification: ×200). The arrows point to alveolar epithelial cells that express CINC-2β.
DISCUSSION
Our study demonstrates that cultured type II cells secrete MIP-2, CINC-2, and MCP-1 in response to IL-1β alone or the combination of IL-1β, TNF-α, and IFN-γ. Secretion of MIP-2, CINC-2, and MCP-1 occurred in both differentiated and dedifferentiated type II cells in response to cytokines and was unrelated to the usual differentiation markers of type II cells. In addition, the dedifferentiated phenotype secreted more MCP-1 than the differentiated type II cell phenotype, especially when the data are expressed per μg DNA. In contrast, CINC-2β was expressed much more abundantly in differentiated rat type II cells. In addition, CINC-2β mRNA level was not stimulated by the cytokines mixture of IL-1β, TNF-α, and IFN-γ, or by IL-1β alone. Hence, CINC-2β appears to be restricted to the alveolar type II cell phenotype, whereas expression of the other CINC isoforms and MCP-1 was expressed by both differentiated and dedifferentiated alveolar type II cells.
The most striking unexpected finding was the different level of expression and regulation of CINC-2α and CINC-2β. The first studies on mRNA expression of CINC-2 isoforms were done by quantitative real-time PCR. The primers and probes used for real-time PCR were designed to differentiate CINC-2α from CINC-2β transcripts by targeting the 3′ untranslated portions of these transcripts. These studies showed that CINC-2α was expressed in both phenotypes and was increased with cytokines, whereas expression of CINC-2β was restricted to the type II cell phenotype and was not stimulated by cytokines. These data were also confirmed in independent experiments by microarray analyses on mRNA from rat type II cells cultured under similar conditions. We have done several series of microarray analyses with rat type II cells, and CINC-2β is consistently increased in differentiated type II cells compared with dedifferentiated type II cells (data not shown). Similar results were recently reported by Gonzales and colleagues (41).
To confirm the real-time PCR measurements, we also performed Northern analyses. One set used a probe designed from the EST (BF553317) and the other used a probe generated from CINC-2β (D21095). Initially we were concerned that the probe generated from accession number BF553317 might identify another related CXC chemokine, because it is not totally identical to CINC-2β. However, both probes identified two CINC-2β mRNA transcripts, one of 2.7 kb and the other 4.1 kb. These were larger than that of CINC-2α (0.8 kb). KGF increased the expression of CINC-2β, whereas the cytokine mixture or IL-1β alone did not. In murine lung, another related CXC chemokine, CXCL15 (lungkine or WECHE), has been identified (42, 43). CXCL15 is a 20-kD protein, much larger than the CINC family members. However, we could not find this gene in rat databases, and our probes do not match the murine sequence.
To evaluate the CINC-2β expression in the lung, in situ hybridization was performed using normal and Ad-KGF instilled rat lung. The CINC-2β was highly expressed in the lung stimulated with Ad-KGF and weakly in the normal lung. The cells that express CINC-2β in vivo appear to be type II cells and the Clara cells. Unfortunately, we were not able to distinguish CINC-2α and CINC-2β at the protein level. We used a commercially available antibody against CINC-2β for our ELISA measurements. However, it was not specific for CINC-2β and also cross-reacted 100% with CINC-2α. Thus, the CINC-2 that was measured by the ELISA is likely both CINC-2α and CINC-2β, especially with the type II cell phenotype. The CINC-2 protein measurement tracks well with the CINC-2α mRNA in the dedifferentiated, type I-like cells but less well in the differentiated type II cells. This is presumably due to the fact that in the differentiated type II cells there is a mixture of CINC-2α and CINC-2β. CINC-2α is reported to play a major role for neutrophil influx into the lung in response to Pseudomonas aeruginosa, and in this model CINC-2β was not detected in the BALF or the lung tissue (44). Similar results were reported for CINCs in lavage fluid after instillation of lipopolysaccharide (30). More recently, Vanderbilt and coworkers reported that rat type II cells express CINC-1 (GRO), CINC-2α, and CINC-3 (MIP-2) and that their expression increased after instillation of HCl or P. aeruginosa (17). In their report, the expression of these chemokines decreased rapidly when cultured on plastic but was maintained albeit at a lower level on Matrigel. CINC-2β was also detected by RT-PCR but not reported in detail. Macrophage secretion of CINC-1 and CINC-3 is thought to play an important role in the recruitment of neutrophils to the lung in LPS-induced acute lung injury (45). We conclude expression of CINC-2β mRNA is apparently restricted to differentiated alveolar type II cells and that it is not regulated by the usual inflammatory cytokines. In contrast, CINC-2α increases with cytokine stimulation, is found in lavage during acute inflammation, and likely plays an important role in neutrophil chemotaxis in vivo.
CINC-2β may have functions in addition to its putative role as a neutrophil chemotactic protein. Specifically, CINC-2β may be involved in wound healing. Since CINC-2β is expressed at least at the mRNA level in the normal lung and the normal lung does not have abundant neutrophil infiltration, either the mRNA is not processed into a secreted protein or the processed secreted CINC-2β has other functions and presumably is not a strong neutrophil chemoattractant in vivo. In the stomach, CINC-2β is expressed in the gastric mucosa and has been suggested to be involved in epithelial healing after ulcer formation (46). Since epithelial cells express the CXCR2 receptor, CINC-2β could serve as an autocrine regulator and have functions independent of neutrophil chemotaxis. In the rat stomach, CINC-2β has been reported to be expressed as a single mRNA transcript, although it is not clear that the probe used would distinguish CINC-2β from CINC-2α (47). In the lung, CINC-2β may be derived from different RNA splicing than observed in the stomach. Additional studies on the regulation of differential splicing and RNA processing of CINC-2β transcripts are warranted as well as means of measuring protein levels. These issues are important and remain for future studies. Currently, we do not know the function of CINC-2β in the normal lung.
Our results on chemokine production by alveolar epithelial cells are similar to those reported previously by others. Vanderbilt and colleagues reported expression of GRO (CINC-1) and MIP-2 (CINC-3) in type II cells response to P. aeruginosa in vivo and that expression decreased in type II cells cultured in vitro (17). Paine and coworkers reported that type II cells secrete MIP-1 in response to IL-1β, and their studies were done under conditions in which the type II cells were dedifferentiated (14). They showed preferential apical secretion of MCP-1. Crippen and colleagues had reported that LPS, IL-1β, and TNF-α increase total CINC secretion by rat type II cells (48). Several groups have reported rat type II cells cultured on plastic secrete MIP-2 in response to LPS and TNF-α (49, 50).
Most of our studies were done in vitro, and our dedifferentiated type II cells in vitro may be different from type I cells in vivo. The phenotype markers for type I and type II cells can be readily changed by culture conditions, and there is plasticity in their expression in vitro. Expression of these markers depends on the matrix on which the cells are grown and on the soluble factors in the media. Hence, extrapolation of the observations of the dedifferentiated alveolar epithelial cells in this report to type I cells in vivo should be done with caution.
In summary, cytokines stimulate chemokine secretion from rat alveolar epithelial cells in primary culture, and secretion of CINC-1, CINC-2α, and CINC-3 appear to be independent of the state of differentiation. CINC-2β is expressed only in differentiated alveolar type II cells and bronchial epithelial cells and appears to be inhibited by cytokines. MCP-1 appears to be preferentially expressed in the dedifferentiated alveolar epithelial cells.
Acknowledgments
The authors are grateful for help in preparation of this manuscript by Shirley Pearce and Teneke Warren. Shuanglin Wang prepared the figures and did the statistical analyses. Mandy Evans and Dennis Voelker kindly provided the antibody and surfactant standards used in the SP-A ELISA.
This work was supported by grants from NIH (HL-29891 and HL-67671) and from the EPA (R825702 and X83084601).
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Snella MC. Production of a neutrophil chemotactic factor by endotoxin stimulated alveolar macrophages in vitro. Br J Exp Pathol 1986;67:801–807. [PMC free article] [PubMed] [Google Scholar]
- 2.Hunninghake GW, Gadek JE, Lawley TJ, Crystal RG. Mechanisms of neutrophil accumulation in the lungs of patients with idiopathic pulmonary fibrosis. J Clin Invest 1981;68:259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer FR, Luo Y, Luo M, Santambrogio L, Dorf ME. RANTES-induced chemokine cascade in dendritic cells. J Immunol 2001;167:1637–1643. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura H, Yoshimura K, McElvaney NG, Crystal RG. Neutrophil elastase in respiratory epithelial lining fluid of individuals with cystic fibrosis induces interleukin-8 gene expression in a human bronchial epithelial cell line. J Clin Invest 1992;89:1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driscoll KE, Howard BW, Carter JM, Asquith T, Johnston C, Detilleux P, Kunkel SL, Isfort RJ. α-quartz-induced cheomokine expression by rat lung epithelial cells. Am J Pathol 1996;149:1627–1637. [PMC free article] [PubMed] [Google Scholar]
- 6.Lilly C, Nakamura H, Kesselman H, Nagler-Anderson C, Asano K, Garcia-Zepeda E, Rothenberg M, Drazen J, Luster A. Expression of eotaxin by human lung epithelial cells: induction by cytokines and inhibition by glucocorticoids. J Clin Invest 1997;99:1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JH, Devalia JL, Xia C, Sapsrod RJ, Davies RJ. Expression of RANTES by human bronchial epithelial cells in vitro and in vivo and the effect of corticosteroids. Am J Respir Cell Mol Biol 1996;14:27–35. [DOI] [PubMed] [Google Scholar]
- 8.Becker S, Quay J, Koren HS, Haskill JS. Constitutive and stimulated MCP-1, GRO alpha, beta, and gamma expression in human airway epithelium and bronchoalveolar macrophages. Am J Physiol 1994;266:L278–L286. [DOI] [PubMed] [Google Scholar]
- 9.Driscoll KE, Carter JM, Hassenbein DG, Howard B. Cytokines and particle-induced inflammatory cell recruitment. Environ Health Perspect 1997;105:1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N, Wang M, Oberley TD, Sempf JM, Nel AE. Comparison of the pro-oxidative and proinflammatory effects of organic diesel exhaust particle chemicals in bronchial epithelial cells and macrophages. J Immunol 2002;169:4531–4541. [DOI] [PubMed] [Google Scholar]
- 11.Matsukura S, Kokubu F, Noda H, Tokunaga H, Adachi M. Expression of IL-6, IL-8, and RANTES on human bronchial epithelial cells, NCI-H292, induced by influenza virus A. J Allergy Clin Immunol 1996;98:1080–1087. [DOI] [PubMed] [Google Scholar]
- 12.Leikauf GD, Simpson LG, Santrock J, Zhao Q, Abbinante-Nissen J, Zhou S, Driscoll KE. Airway epithelial cell responses to ozone injury. Environ Health Perspect 1995;103:91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marini M, Vittori E, Hollemborg J, Mattoli S. Expression of the potent inflammatory cytokines, granulocyte-macrophage-colony-stimulating factor and interleukin-6 and interleukin-8, in bronchial epithelial cells of patients with asthma. J Allergy Clin Immunol 1992;89:1001–1009. [DOI] [PubMed] [Google Scholar]
- 14.Paine R III, Rolfe MW, Standiford TJ, Burdick MD, Rollins BJ, Strieter RM. MCP-1 expression by rat type II alveolar epithelial cells in primary culture. J Immunol 1993;150:4561–4570. [PubMed] [Google Scholar]
- 15.Jaspers I, Chen LC, Flescher E. Induction of interleukin-8 by ozone is mediated by tyrosine kinase and protein kinase A, but not by protein kinase C. J Cell Physiol 1998;177:313–323. [DOI] [PubMed] [Google Scholar]
- 16.Barrett EG, Johnston C, Oberdorster G, Finkelstein JN. Silica-induced chemokine expression in alveolar type II cells is mediated by TNF-alpha. Am J Physiol 1998;275:L1110–L1119. [DOI] [PubMed] [Google Scholar]
- 17.Vanderbilt JN, Mager EM, Allen L, Sawa T, Wiener-Kronish J, Gonzalez R, Dobbs LG. CXC chemokines and their receptors are expressed in type II cells and upregulated following lung injury. Am J Respir Cell Mol Biol 2003;29:661–668. [DOI] [PubMed] [Google Scholar]
- 18.Mason RJ, Pan T, Edeen KE, Nielsen LD, Zhang F, Longphre M, Eckart MR, Neben S. Keratinocyte growth factor and the transcription factors C/EBP{alpha}, C/EBP{delta}, and SREBP-1c regulate fatty acid synthesis in alveolar type II cells. J Clin Invest 2003;112:244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason RJ, Lewis MC, Edeen KE, McCormick-Shannon K, Nielsen LD, Shannon JM. Maintenance of surfactant protein A and D secretion by rat alveolar type II cells in vitro. Am J Physiol Lung Cell Mol Physiol 2002;282:L249–L258. [DOI] [PubMed] [Google Scholar]
- 20.Shannon JM, Mason RJ, Jennings SD. Functional differentiation of alveolar type II epithelial cells in vitro: effects of cell shape, cell-matrix interactions, and cell-cell interactions. Biochim Biophys Acta 1987;931:143–156. [DOI] [PubMed] [Google Scholar]
- 21.Shannon JM, Jennings SD, Nielsen LD. Modulation of alveolar type II cell differentiated function in vitro. Am J Physiol 1992;262:L427–L436. [DOI] [PubMed] [Google Scholar]
- 22.Borok Z, Danto SI, Lubman RL, Cao Y, Williams MC, Crandall ED. Modulation of t1alpha expression with alveolar epithelial cell phenotype in vitro. Am J Physiol 1998;175:L155–L164. [DOI] [PubMed] [Google Scholar]
- 23.Bacon K, Baggiolini M, Broxmeyer H, Horuk R, Lindley I, Mantovani A, Maysushima K, Murphy P, Nomiyama H, et al., the IUIS/WHO Subcommittee on Chemokine Nomenclature. Chemokine/chemokine receptor nomenclature. J Interferon Cytokine Res 2002;22:1067–1068. [DOI] [PubMed] [Google Scholar]
- 24.Modi WS, Yoshimura T. Isolation of novel GRO genes and a phylogenetic analysis of the CXC chemokine subfamily in mammals. Mol Biol Evol 1999;16:180–193. [DOI] [PubMed] [Google Scholar]
- 25.Shibata F, Konishi K, Kato H, Komorita N, al-Mokdad M, Fujioka M, Nakagawa H. Recombinant production and biological properties of rat cytokine-induced neutrophil chemoattractants, GRO/CINC-2 alpha, CINC-2 beta and CINC-3. Eur J Biochem 1995;231:306–311. [DOI] [PubMed] [Google Scholar]
- 26.Shibata F, Konishi K, Nakagawa H. Gene structure, cDNA cloning, and expression of the rat cytokine-induced neutrophil chemoattractant-2 (CINC-2) gene. Cytokine 1998;10:169–174. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa H, Komorita N, Shibata F, Ikesue A, Konishi K, Fujioka M, Kato H. Identification of cytokine-induced neutrophil chemoattractants (CINC), rat GRO/CINC-2 alpha and CINC-2 beta, produced by granulation tissue in culture: purification, complete amino acid sequences and characterization. Biochem J 1994;301:545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frevert CW, Huang S, Danaee H, Paulauskis JD, Kobzik L. Functional characterization of the rat chemokine KC and its importance in neutrophil recruitment in a rat model of pulmonary inflammation. J Immunol 1995;154:335–344. [PubMed] [Google Scholar]
- 29.Driscoll KE, Hassenbein DG, Carter JM, Kunkel SL, Quinlan TR, Mossman BT. TNF alpha and increased chemokine expression in rat lung after particle exposure. Toxicol Lett 1995;82–83:483–489. [DOI] [PubMed] [Google Scholar]
- 30.Mitsuhashi H, Hata J, Asano S, Kishimoto T. Appearance of cytokine-induced neutrophil chemoattractant isoforms and immunolocalization of them in lipopolysaccharide-induced acute lung inflammation in rats. Inflamm Res 1999;48:588–593. [DOI] [PubMed] [Google Scholar]
- 31.Fillion I, Ouellet N, Simard M, Bergeron Y, Sato S, Bergeron MG. Role of chemokines and formyl peptides in pneumococcal pneumonia-induced monocyte/macrophage recruitment. J Immunol 2001;166:7353–7361. [DOI] [PubMed] [Google Scholar]
- 32.Shibata F, Konishi K, Nakagawa H. Identification of a common receptor for three types of rat cytokine-induced neutrophil chemoattractants (CINCs). Cytokine 2000;12:1368–1373. [DOI] [PubMed] [Google Scholar]
- 33.Mason RJ, Williams MC. Type II alveolar cell: defender of the alveolus. Am Rev Respir Dis 1977;115:81S–91S. [DOI] [PubMed] [Google Scholar]
- 34.Dobbs LG, Mason RJ. Pulmonary alveolar type II cells isolated from rats. Release of phosphatidylcholine in response to beta-adrenergic stimulation. J Clin Invest 1979;63:378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morikawa O, Walker TA, Nielsen LD, Pan T, Cook JL, Mason RJ. Effect of adenovector-mediated gene transfer of keratinocyte growth factor on the proliferation of alveolar type II cells in vitro and in vivo. Am J Respir Cell Mol Biol 2000;23:626–635. [DOI] [PubMed] [Google Scholar]
- 36.Labarca C, Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem 1980;102:344–352. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F, Pan T, Nielsen LD, Mason RJ. Lipogenesis in fetal rat lung: importance of C/EBPalpha, SREBP-1c, and stearoyl-CoA desaturase. Am J Respir Cell Mol Biol 2004;30:174–183. [DOI] [PubMed] [Google Scholar]
- 38.Portnoy J, Curran-Everett D, Mason RJ. Keratinocyte growth factor stimulates alveolar type II cell proliferation through the extracellular signal-regulated kinase and phosphatidylinositol 3-OH kinase pathways. Am J Respir Cell Mol Biol 2004;30:901–907. [DOI] [PubMed] [Google Scholar]
- 39.Shannon JM, Pan T, Nielsen LD, Edeen KE, Mason RJ. Lung fibroblasts improve differentiation of rat type II cells in primary culture. Am J Respir Cell Mol Biol 2001;24:235–244. [DOI] [PubMed] [Google Scholar]
- 40.Deterding RR, Havill AM, Yano T, Middleton SC, Jacoby CR, Shannon JM, Simonet WS, Mason RJ. Prevention of bleomycin-induced lung injury in rats by keratinocyte growth factor. Proc Assoc Am Physicians 1997;109:254–268. [PubMed] [Google Scholar]
- 41.Gonzalez R, Yang YH, Griffin C, Allen L, Tigue Z, Dobbs L. Freshly isolated rat alveolar type I cells, type II cells, and cultured type II cells have distinct molecular phenotypes. Am J Physiol Lung Cell Mol Physiol 2005;288:L179–L189. [DOI] [PubMed] [Google Scholar]
- 42.Ohneda O, Ohneda K, Nomiyama H, Zheng Z, Gold SA, Arai F, Miyamoto T, Taillon BE, McIndoe RA, Shimkets RA, et al. WECHE: a novel hematopoietic regulatory factor. Immunity 2000;12:141–150. [DOI] [PubMed] [Google Scholar]
- 43.Rossi DL, Hurst SD, Xu Y, Wang W, Menon S, Coffman RL, Zlotnik A. Lungkine, a novel CXC chemokine, specifically expressed by lung bronchoepithelial cells. J Immunol 1999;162:5490–5497. [PubMed] [Google Scholar]
- 44.Amano H, Oishi K, Sonoda F, Senba M, Wada A, Nakagawa H, Nagatake T. Role of cytokine-induced neutrophil chemoattractant-2 (CINC-2) alpha in a rat model of chronic bronchopulmonary infections with Pseudomonas aeruginosa. Cytokine 2000;12:1662–1668. [DOI] [PubMed] [Google Scholar]
- 45.Yamasawa H, Ishii Y, Kitamura S. Cytokine-induced neutrophil chemoattractant in a rat model of lipopolysaccharide-induced acute lung injury. Inflammation 1999;23:263–274. [DOI] [PubMed] [Google Scholar]
- 46.Kazumori H, Ishihara S, Hoshino E, Kawashima K, Moriyama N, Suetsugu H, Sato H, Adachi K, Fukuda R, Watanabe M, et al. Neutrophil chemoattractant 2 beta regulates expression of the Reg gene in injured gastric mucosa in rats. Gastroenterology 2000;119:1610–1622. [DOI] [PubMed] [Google Scholar]
- 47.Okada A, Kinoshita Y, Waki S, Fukui H, Maekawa T, Matsushima Y, Kawanami C, Kishi K, Nakata H, Wang HY, et al. Rat gastric mucosal cells express ICAM-1 and proinflammatory cytokines during indomethacin-induced mucosal injury. J Lab Clin Med 1998;131:538–547. [DOI] [PubMed] [Google Scholar]
- 48.Crippen TL, Klasing KC, Hyde DM. Cytokine-induced neutrophil chemoattractant production by primary rat alveolar type II cells. Inflammation 1995;19:575–586. [DOI] [PubMed] [Google Scholar]
- 49.Xavier AM, Isowa N, Cai L, Dziak E, Opas M, McRitchie DI, Slutsky AS, Keshavjee SH, Liu M. Tumor necrosis factor-α mediates lipopolysaccharide-induced macrophage inflammatory protein-2 release from alveolar epithelial cells: autoregulation in host defense. Am J Respir Cell Mol Biol 1999;21:510–520. [DOI] [PubMed] [Google Scholar]
- 50.Isowa N, Keshavjee SH, Liu M. Role of microtubules in LPS-induced macrophage inflammatory protein-2 production from rat pneumocytes. Am J Physiol Lung Cell Mol Physiol 2000;279:L1075–L1082. [DOI] [PubMed] [Google Scholar]