Abstract
Purpose
Approximately 8% of autosomal recessive retinitis pigmentosa (RP) cases worldwide are due to defects in rod-specific phosphodiesterase PDE6, a tetramer consisting of catalytic (PDE6α and PDE6β) and two regulatory (PDE6γ) subunits. In mice homozygous for a nonsense Pde6brd1 allele, absence of PDE6 activity is associated with retinal disease similar to humans. Although studied for 80 years, the rapid degeneration Pde6brd1 phenotype has limited analyses and therapeutic modeling. Moreover, this model does not represent human RP involving PDE6B missense mutations. In the current study the mouse missense allele, Pde6bH620Q was characterized further.
Methods
Photoreceptor degeneration in Pde6bH620Q homozygotes was documented by histochemistry, whereas PDE6β expression and activity were monitored by immunoblotting and cGMP assays. To measure changes in rod physiology, electroretinograms and intracellular Ca2+ recording were performed. To test the effectiveness of gene therapy, Opsin::Pde6b lentivirus was subretinally injected into Pde6bH620Q homozygotes.
Results
Within 3 weeks of birth, the Pde6bH620Q homozygotes displayed relatively normal photoreceptors, but by 7 weeks degeneration was largely complete. Before degeneration, PDE6β expression and PDE6 activity were reduced. Although light-/dark-adapted total cGMP levels appeared normal, Pde6bH620Q homozygotes exhibited depressed rod function and elevated outer segment Ca2+. Transduction with Opsin::Pde6b lentivirus resulted in histologic and functional rescue of photoreceptors.
Conclusions
Pde6bH620Q homozygous mice exhibit a hypomorphic phenotype with partial PDE6 activity that may result in an increased Ca2+ to promote photoreceptor death. As degeneration in Pde6bH620Q mutants is slower than in Pde6brd1 mice and can be suppressed by Pde6b transduction, this Pde6bH620Q model may provide an alternate means to explore new treatments of RP.
Photoreceptor degenerations are a significantly large group of neurodegenerative disorders.1–3 In patients with retinitis pigmentosa (RP), early symptoms include night blindness and loss of peripheral vision due to rod photoreceptor degeneration. Cone death follows and results in loss of daytime central vision. Approximately 8% of cases of recessive RP worldwide are due to mutations in rod-specific PDE6 complex (Huang SH, et al. IOVS 1995;36:ARVO E-Abstract 3815),4–7 consisting of catalytic (PDE6α and PDE6β) and regulatory (PDE6γ) subunits.8 –26 Approximately 70% of rod-associated, PDE-based, RP cases are associated with missense mutations within the PDE6β catalytic domain, probably resulting in only partial loss of enzyme function.27,28
PDE6 is the effector enzyme of the G-protein-mediated visual transduction cascade. Light entering rod outer segments (OS) activates the photopigment that stimulates PDE6 enzyme activity via the photoreceptor-specific G-protein transducin. Activated PDE6 decreases the concentration of free cGMP approximately 10-fold on a millisecond time-scale.29 Prolonged exposure to light and PDE6 activity results in a two-fold difference between dark- and light-adapted total cGMP levels.9,30,31 Reductions in free cGMP concentration results in the closing of cGMP-gated cationic channels (CNG). A decrease in Na+ and Ca2+ influx through the CNG generates a membrane current change that hyperpolarizes the photoreceptor and decreases the release of neural transmitter at the synapse.14,18,32
The Pde6brd1 mouse has been used to study photoreceptor degeneration and as a therapeutic model for human RP.33–35 Pde6brd1 is a null allele and is not detectably expressed presumably due to nonsense-mediated mRNA decay.36,37 Homozygous mutant mice exhibit rapid rod and subsequent cone degeneration.35 As expected, PDE6 activity is absent, there is no difference in dark- and light-adapted total cGMP levels, which are dramatically increased over the control. It has been hypothesized that this high level of total cGMP promotes rod cell death.
Attempts to treat Pde6brd1 mice with gene therapy have had limited success.38 – 41 In particular, long-term functional restoration of retinal signaling has not been achieved, perhaps because of the rapid nature of the degeneration in the Pde6brd1 mouse, the therapeutic strategy, and/or the level of intervention in the neurodegeneration pathway.
A Pde6bH620Q mutant mouse was recently reported to have delayed photoreceptor degeneration relative to Pde6brd1 mice.42 Pde6bH620Q mice harbor a missense mutation in the catalytic domain. In the present study, Pde6bH620Q homozygous mice exhibited a hypomorphic degeneration phenotype. In contrast to Pde6brd1, we found Pde6bH620Q homozygotes to have normal photoreceptor differentiation and retain retinal function before degeneration onset. Degeneration etiology is likely to be similar in both mouse lines, as we found evidence of elevated free cGMP and Ca2+ in Pde6bH620Q retinas. Using Opsin::Pde6b lentiviral particles, we ameliorated the degeneration phenotype in this alternative mouse RP model.
Methods
Mouse Lines and Husbandry
Mice were used in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research, as well as the Policy for the Use of Animals in Neuroscience Research of the Society for Neuroscience. Pde6bH620Q mice were derived by oviduct transfer from morulae provided by the European Mouse Mutant Archive. All Pde6bH620Q mice analyzed in this study are homozygotes and will be referred to as Pde6bH620Q homozygotes, Pde6bH620Q mutants, or Pde6bH620Q mice. Pde6bH620Q and Pde6brd1 strains are in the C3H background, and age-matched DBA/2J or C57BL6 were used as the control.42– 45
Histochemical Analyses
Mice were euthanatized with an intraperitoneal injection of pentobarbital. Each eye was rapidly removed, punctured at the 12 o’clock position along the limbus, and placed in a separate solution of 3% glutaraldehyde in phosphate-buffered saline. After fixation for 1 to 2 days, the eyes were washed with saline, and the puncture was used to orient the eyes so that the posterior segment could be sectioned along the vertical meridian. Eyes were dehydrated and embedded in paraffin. Hematoxylin-eosin (H&E) staining of paraffin-embedded sections was conducted as described.46 Quantification of photoreceptor nuclei was conducted on several sections containing the optic nerve as follows: the distance between the optic nerve and the ciliary body was divided into quadrants, and three rows of nuclei were counted within each quadrant. Averages and standard deviations were calculated from 3 to 15 animals for each time-point.
For electron microscopy (EM), a rectangular piece of the retina, containing the superior to inferior ora serrata and optic nerve, was prepared for postfixation in osmic acid, dehydrated, and Epon embedded. A corner was cut out at the superior ora to allow identification of the upper retinal half of the segment. Sectioning proceeded along the long axis of the segment so that each section contained upper and lower retina as well as posterior pole. Semiserial sections were stained with either H&E or toluidine blue, mounted, and examined by light microscopy. Selected areas were trimmed for ultrathin sectioning, stained with uranyl acetate, and examined by EM.8,9,47,48
Total cGMP Assays
Direct retinal cGMP content and PDE assays were performed as described.8,9,12,46 For total cGMP, the retinas were isolated under infrared illumination or normal room light and homogenized in 0.1 N HCl. Measurement of cGMP was conducted in triplicate, and the results expressed in picomoles cGMP per nanogram total protein.
PDE6 Purification and cGMP Phosphodiesterase Assay
PDE6 was extracted from retinal membranes using a hypotonic buffer according to the method of Kühn.49 In brief, light-adapted retinas were homogenized in 100 mM Tris-HCl (pH 7.5), 1 mM MgCl2 supplemented with protease inhibitors (Calbiochem, San Diego, CA). After centrifugation for 2 minutes, the supernatant was discarded, and the pellet was washed. PDE6 was extracted twice from the pellet with 5 mM Tris-HCl (pH 7.5) supplemented with 1 mM MgCl2. These supernatants were pooled and concentrated (Centricon-3; Amicon, Beverly, MA) and the protein concentration determined according to Lowry. Samples were adjusted to 70 ng total protein/sample for the PDE assay. Samples (5 μL) were incubated with 20 μL cGMP (0.5 mM stock solution), 10 μL snake venom (3.7 mg/mL stock solution), and 15 μL assay buffer for 30 minutes at 37°C with gentle rotating.50 The reaction was stopped with 100 μL green reagent, and the color was allowed to develop for 30 minutes at room temperature (BioMol, Plymouth Meeting, PA). The OD was measured at 620 nm. The cleavage of cGMP to 5′-GMP by PDE was determined as phosphate release from 5′-GMP with the linkage of 5′-nucleotidase reaction.
Western Blot Analysis
Retinas were homogenized in 10% sodium dodecyl sulfate (SDS) by brief sonication and denatured at 100°C for 5 minutes. After centrifugation, total protein content per sample was determined by a protein assay method (DC Protein Assay; Bio-Rad Laboratories, Hercules, CA). The proteins were separated by SDS–polyacrylamide gel electrophoresis. Samples were then transferred to nitrocellulose membranes, which were blocked in 3% BSA in 500 mM NaCl, 20 mM Tris (pH 7.6), and 0.1% Tween-20 (BSA-TTBS). Membranes were incubated with either rabbit anti-PDE6β (1 μg/mL, PAI-722; Affinity Bioreagents, Golden, CO) or mouse anti-OPSIN (1:1000, ID451) antibodies in BSA-TTBS. After washing in TTBS, filters were incubated with either anti-rabbit- or anti-mouse-conjugated horse radish peroxidase secondary antibodies (sc-2005 and sc-2004; Santa Cruz Biotechnology, Santa Cruz, CA). After washing, antibody complexes were visualized by chemiluminescence detection (Immobilon Western; Millipore Corp., Billerica, MA). Multiple exposures were obtained (BioMax film; Eastman Kodak, Rochester, NY) and signals were quantified (AlphaImager and AlphaImage software; Alpha Innotech Corp., San Leandro, CA). An integral density value (IDV) for each band was determined and normalized to total protein. Comparisons between mutant and control samples were performed under conditions where the normalized values were found to be in the linear range of the assay.
Measurement of Intracellular Calcium
Spot laser epifluorescence was used to determine the concentration of Ca2+ in living rod OS from retinas loaded with fluo5F (Invitrogen-Molecular Probes, Eugene OR).52,53 OS fluorescence was collected with 505-nm dichroic and 510-nm long-pass filters (Omega Optical, Brattleboro, VT).
Construction and Transduction of Lentiviral Vectors
The Opsin::Pde6b vector was constructed using 2 kb of the mouse Opsin promoter region and a full-length mouse Pde6b cDNA fragment.54 Self-inactivating (SIN) vectors consist of a 5′-long terminal repeat (LTR), a packaging signal ψ, a tRNA primer binding site, a lentiviral reverse response element, the mouse Opsin promoter linked to Pde6b, and 3′LTR. The vector also contains a central polypurine tract/DNA flap55 and a Woodchuck hepatitis virus posttranscriptional regulatory element. The Opsin::Pde6b, EF::Green Fluorescent Protein (GFP) vector contains a 5′LTR, a retroviral export element, a lentiviral Rev-response element, the mouse Opsin promoter linked to Pde6b, EF1::GFP, and the U5 region of the 5′LTR, and the 3′LTR was also used. GFP expression is driven by the EF1 promoter. Virus was generated by cotransfection of human kidney-derived 293T cells with vectors and packaging constructs using the Ca2+ phosphate method56 –58 (Bio-genova Corp., Ellicott City, MD and Lentigen Corp., Baltimore, MD). Anesthesia and surgery were preformed as described.59 Approximately 1 μL of virus particles (~2 × 107 transducing unit [TU]/mL) were injected subretinally in postnatal day 5 mice (P5). The contralateral control eye received saline, CMV::GFP, or Lenti-LacZ virus.
Electroretinograms
ERGs were performed as described.8,9,46,49,60 The mice were dark-adapted overnight, manipulations were conducted under dim red light illumination, and recordings were made (Espion ERG Diagnosys equipment; Diagnosys LLL, Littleton, MA). Adult C57BL/6 mice were tested at the beginning of each session as controls. Mice were anesthetized by intraperitoneal injection of (per 10 g body weight) 0.1 mL mix (1 mL ketamine 100 mg/mL and 0.5 mL xylazine 20 mg/mL in 8.5 mL PBS). The corneas were anesthetized with a drop of 0.5% proparacaine hydrochloride (Alcon Laboratories, Inc., Fort Worth, TX) and pupils were dilated with topical 2.5% phenylephrine hydrochloride and 1% tropicamide. Body temperature was maintained at 37°C with a heating pad. Burian-Allen bipolar mouse contact lens electrodes (Hansen Laboratories, Coralville, IA) were placed on each cornea, after application of gonioscopic prism solution (Alcon Laboratories). The reference electrode was placed SC in the anterior scalp between the eyes, and the ground electrode was inserted into the tail. Electrode impedance was balanced for each eye pair measured. The head of the mouse was placed in a standardized position in a Ganzfeld bowl illuminator that assured equal illumination of the eyes. Both eyes were recorded simultaneously.
For rod and mixed rod– cone responses, ERGs for dark-adapted mice were recorded using pulses of 0.001 30 and 3 cd-s/m2 (white 6500K), respectively. A total of 40 responses were averaged for each trial. For photopic responses, mice were light adapted in the Ganzfeld dome for at least 10 minutes. A rod-suppressing steady background of 30 cd/m2 (white 6500K) was continuously present throughout the trials. ERGs were recorded using pulses of 30 cd-s/m2 (xenon). A total of 40 responses (filtered from 0.03 to 300 Hz) were averaged for each trial.
Results
Hart et al.,42 described a new mouse Pde6b mutant line (Pde6bH620Q) that demonstrates delayed photoreceptor degeneration, compared to Pde6brd1 mice. We extended their studies by performing histologic, biochemical, and physiological measurements and by testing the ability of gene therapy to ameliorate degeneration. For our studies, age-matched DBA/2J or C57BL6 were used as control subjects,42– 45 which are not congenic with the C3H strain.
Delayed Photoreceptor Degeneration in Pde6bH620Q Homozygous Mutants
Previously, Pde6bH620Q photoreceptor degeneration was documented at P21 and P42.42 To further quantify the rate of degeneration, we performed a histologic analysis of Pde6bH620Q mutant photoreceptors between P9 and P64 (Fig. 1A, 1B). Although the number of photoreceptors between Pde6bH620Q mutant and control mice was similar at P9 and P14, nuclei numbers progressively diminished from P19 through P49. Afterward, one to two rows of nuclei remained from P56 through P77. The remaining layer contained cones, as demonstrated by peanut agglutinin staining (see Fig. 5E, bottom left). Modest differences in the rate of degeneration were observed between the optic nerve and ciliary body; degeneration of peripheral photoreceptors was slower than that of the central photoreceptors (Fig. 1B).
Figure 1.
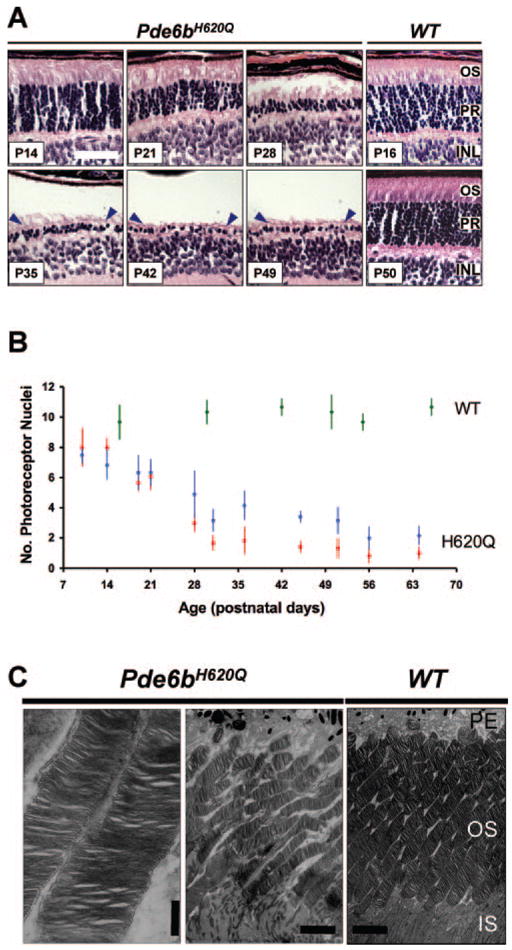
Pde6bH620Q homozygotes demonstrated delayed and progressive photoreceptor degeneration. (A) H&E-stained paraffin-embedded sections of Pde6bH620Q mutant (P14 –P49) and wild-type control (P16 and P50) mice. Shown are the outer segments (OS) photorecep-tors (PR) and inner nuclear layer (INL). Blue arrowheads: mutant photoreceptor layer after P35. (B) Rates of photoreceptor degeneration in Pde6bH620Q mutants. Average number of photoreceptor nuclei in mutants, counted near the optic nerve (red) and periphery (blue), and wild-type retinas (green). (C) Electron photomicrographs of Pde6bH620Q mutant (P21, left and middle) and wild-type control (right) OS discs. Portions of the retinal pigmented epithelium (PE) and inner segment (IS) are shown. Scale bar: (A) 50 mm; (C) left: 500 nm; middle, right: 55 nm.
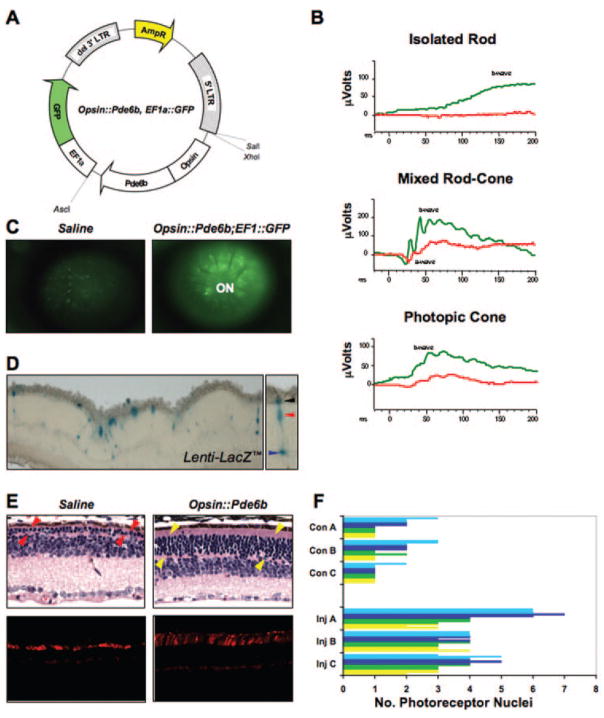
Figure 5.
Transduction of Pde6bH620Q homozygous photoreceptors with Opsin::Pde6b lentiviral particles. (A) The Opsin::Pde6b, EF1a::Green Fluorescent Protein (GFP) vector used for generation of lentiviral particles. (B) ERG responses of mutants subretinally injected with Opsin:Pde6b; EF1::GFP virus (right eye, green trace) and saline (left eye, red trace). Injections were performed at P5, whereas ERGs were measured at P38. b-Wave: inner retinal responses; a-wave: photoreceptor responses. (C) Live retinal fluorescence imaging of Pde6bH620Q eyes injected with saline (left) or Opsin:Pde6b; EF1::GFP (right). Images were obtained at 8 weeks, 54 days after injection. (D) Left: β-galactosidase activity detected in photoreceptors from Lenti-LacZ -transduced Pde6bH620Q retinas at 2 weeks, 9 days after injection. Right: black, red, and blue arrowheads indicate photoreceptor OS/IS, cell body, and synapse, respectively. (E) Histologic analysis of Opsin::Pde6b-transduced retinas. H&E stained (top, P33) and peanut agglutinin-stained (bottom, P77) retinal sections from (left) saline- and (right) Opsin::Pde6b-injected Pde6bH620Q eyes. Images were obtained from similar retinal locations: superior to the optic nerve head, toward the posterior side of the retina. Arrowheads: photo-receptor nuclear layer. (F) Quantitative analysis of photoreceptors in Pde6bH620Q homozygous eyes injected with saline (Con A, B, C) or Opsin:Pde6b; EF1::GFP (Inj A, B, C). The number of photoreceptor nuclei were counted from retinas divided equally into four quadrants from optic nerve to the periphery (yellow, green, dark blue, light blue bars). Within each quadrant, three different locations were randomly selected for nucleus counting.
Our H&E analysis suggests, before extensive degeneration, mutant OS are present and intact. To confirm this finding, we examined mutant retinas by transmission EM (Fig. 1C). We find at P21, the inner segments (IS) and OS of Pde6bH620Q photo-receptors segments are mildly disorganized but have normally spaced discs.
Progressive Retinal Dysfunction in Pde6bH620Q Homozygous Mutants
Our histologic analyses suggest the mutant retinas are functional before extensive degeneration. We therefore used ERG analyses to assess global retinal function in Pde6bH620Q mutant mice from P20 through P55 (Fig. 2).
Figure 2.
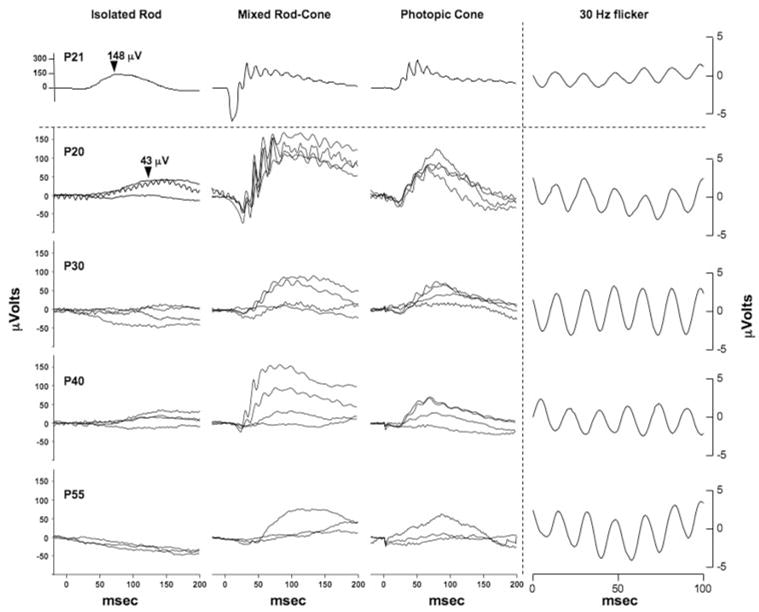
Progressive reduction of ERG responses in Pde6bH620Q homozygotes. Rod-specific (left), mixed rod-cone (middle), and transient photopic cone (right) responses in wild-type (above dotted line) and Pde6bH620 mutant (below dotted line) mice. Ages of mice are indicated in the left column. Traces from three to four mice are shown for each time-point. Arrowheads: the rod b-wave peaks for 3-week-old wild-type and mutant mice. Far right: average bright light stimulation (30 Hz) responses of wild-type and mutant mice are shown.
Rods
Compared with wild-type, P20 mutant maximum rod-specific b-wave peaks were depressed and delayed (e.g., 148 μV vs. 43 μV; 70 ms vs. 119 ms; Fig. 2). In mutants older than P20, rod responses are weaker or extinguished.
Mixed Rod–Cone
In P20 mutants, the maximum rod-cone responses, a- and b-wave peaks, are smaller in amplitude and delayed compared to WT mice, although the differences are not as striking as the isolated rod responses (Fig. 2). Reductions in rod– cone responses and increased variation between animals were observed in older mutants.
Transient Photopic Cone
Cone responses in P20 mutant retinas also showed amplitude reductions and delays in implicit time, compared with WT controls (Fig. 2). Changes in photopic cone responses over time and between animals were similar to the mixed rod– cone responses.
Photopic 30-Hz Flicker Responses
We observed no significant delay in the implicit time up to P55 (Fig. 2, right).
These results indicate that the Pde6bH620Q mutant mice had detectable photoreceptor function that progressively decreased. Initially, both rod- and cone-specific ERGs were depressed and delayed, possibly due to low PDE6 activity, photoreceptor loss, and/or inner retinal changes. Rod and cone responses were reduced in progressively older mice, with losses in rod function preceding cone changes.
Loss of PDE6 Function in Pde6bH620Q Homozygous Mutant Mice
The Pde6bH620Q allele is defined as hypomorphic, as the phenotype of the Pde6bH620Q/Pde6brd1 compound heterozygotes is intermediate between Pde6bH620Q homozygotes and Pde6brd1 homozygotes.42 As the H620Q missense mutation is located within the H-loop of the PDE6β catalytic domain, loss of function may be due to a decrease in PDE6 activity.61 Alternatively, loss of function may be attributable to the reduced level of the enzyme. To test these hypotheses, we analyzed PDE6β expression and PDE6 biochemical function in Pde6bH620Q mutant and control mice.
We first measured PDE6β expression in Pde6bH620Q mutants. Western blot analysis of total retinal homogenates using anti-PDE6β antibodies demonstrated that PDE6β was expressed at lower levels in Pde6bH620Q mutant retinas than in wild-type retinas (Figs. 3A, 3B). For these experiments, retinas from P16 or P18 were analyzed. Equivalent amounts of total protein were loaded for mutant and control mice, as confirmed by similar levels of OPSIN expression in the samples. Densito-metric analysis of the Western signals indicated that PDE6β was expressed at ~22-fold lower levels in mutants than in controls (Figs. 3A, 3B).
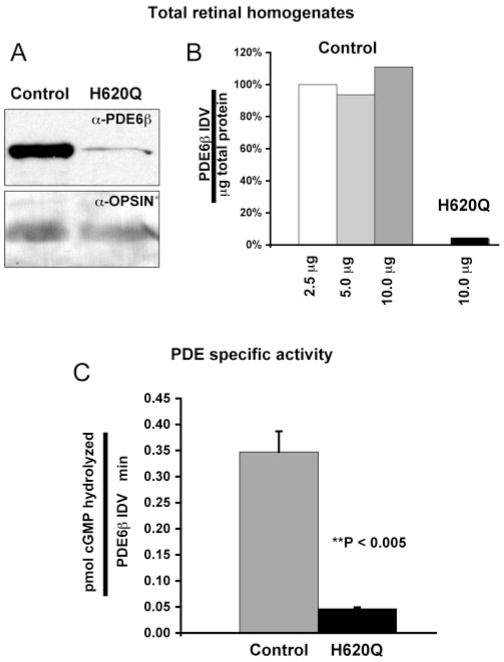
Figure 3.
Reduced PDE6β expression and PDE6 activity in Pde6bH620Q homozygous retinas. (A) Immunoblot detection of PDE6β and opsin in P16 to P18 mutant and wild-type in total retinal homogenates. (B) Quantitative analysis of PDE6β signals from total retinal homogenates. Signal intensities were determined by densitometry to calculate an IDV for each band. The values were normalized for the amount of protein loaded and expressed as a percentage relative to PDE6β signal in 2.5 mg of control lysate. Similar normalized values are obtained at different loading amounts. (C) Light-adapted PDE6 activity in P20 control and mutant retinal extracts. PDE activity from control and mutant hypotonic extracts were normalized to PDE6β content (IDV).
We then investigated whether Pde6bH620Q homozygous mutant retinas exhibit reduced PDE6 activity. PDE6 was extracted according to the method of Kühn.49 After homogenization, PDE was isolated from washed membranes by extraction in hypotonic buffer and concentrated, and the total protein content was determined. PDE activity was then measured in P20 control and mutant extracts. In wild-type samples, average light-activated PDE was 0.392 picomoles cGMP hydrolyzed/ng protein/min, whereas Pde6bH620Q homozygote extracts were 0.006 pmol cGMP hydrolyzed/ng protein/min (~65 times more wild-type activity per nanogram extract protein than mutant activity). Since PDE6β was expressed at lower levels in retinal homogenates, we measured the amount of PDE6β present in these crude hypotonic extracts by densito-metric analysis of immunoblots. We loaded amounts of wild-type (0.3 μg) and mutant (20 μg) extracts estimated to induce nearly identical activity (~3.6 nanomoles cGMP hydrolysis in 30 minutes) and detected ~7.7 times more PDE6β in the mutant samples than in the wild-type. Normalizing wild-type and mutant PDE activity to PDE6β content gave 0.347 pico-moles cGMP hydrolyzed/PDE6β IDV/minute and 0.046 pico-moles cGMP hydrolyzed/PDE6β IDV/minute, respectively (~7.5 times more wild-type than mutant; Fig. 3C). Thus, in these experiments, we can estimate the specific activity of mutant PDE6 at ~13% of wild-type PDE. Together, these biochemical and expression results are consistent with Pde6bH620Q’s being a hypomorphic allele.
Measurement of Total cGMP Levels in Pde6bH620Q Homozygous Mutant Mice
Reduced PDE6 activity may result in the abnormal accumulation of total cGMP levels in Pde6bH620Q mutant retinas. Indeed, Pde6brd1 null mice, which contain no detectable PDE6β expression or PDE6 activity, exhibit an abnormal increase in light-adapted total cGMP levels before onset of degeneration.43– 45
To explore this possibility, we measured and compared total cGMP levels between Pde6brd1 mutant, Pde6bH620Q mutant, and control retinas (Fig. 4). In Pde6brd1 mutants (C3H strain), we found changes in total cGMP (rise-and-fall profile and light-adapted cGMP levels) that were consistent with those in previous reports, supporting the validity of our methodology (Fig. 4A).43– 45 In Pde6bH620Q mutants, we observed differences in cGMP levels between light- and dark-adapted retinas (compare gray and black lines) and total cGMP levels that decreased in parallel with photoreceptor degeneration (compare Fig. 4A with Fig. 1B). In addition, peak total cGMP levels in light-adapted Pde6bH620Q retinas (at any time point) does not reach the same level as light-adapted Pde6brd1 retinas at P14 (compare solid gray line to dashed line). Finally, to test whether partial loss of PDE6 function results in abnormal cGMP accumulation in Pde6bH620Q mutant retinas, we compared cGMP levels between mutant and control retinas (Fig. 4B). However, at three time points, P14, P18, and P21, we found no significant differences in total cGMP in light- and dark-adapted retinas. These results complement our biochemical findings that Pde6bH620Q mutant retinas contain light-induced PDE6 activity. Our data also suggest that this lower than normal level of PDE6 enzymatic activity is not sufficient to result in abnormal light- or dark-adapted total cGMP levels. However, these measurements of steady state total cGMP do not address the possibility that there may be differences in free cGMP between mutant and controls.
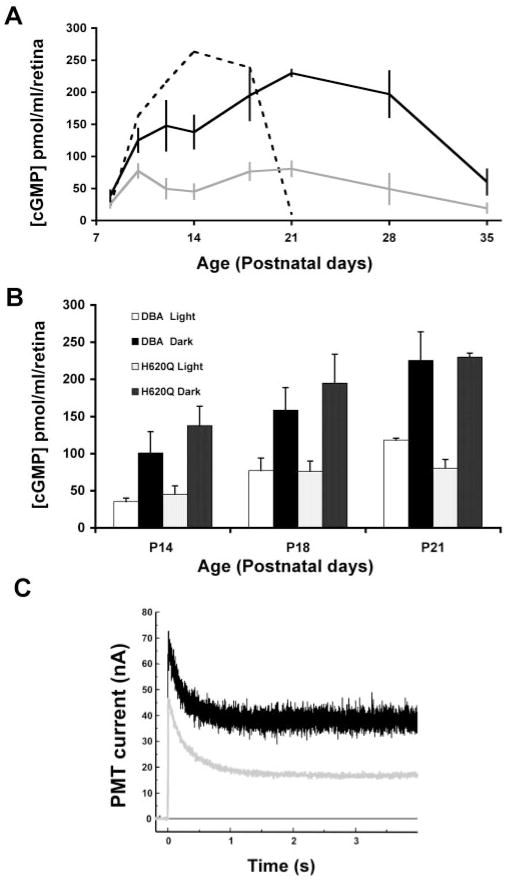
Figure 4.
Light stimulated changes in cGMP and Ca2+ levels in Pde6bH620Q homozygotes. (A) Total cGMP levels measured in (gray line) light- and (black line) dark-adapted Pde6bH620Q mice (1–7 weeks of age). Dashed line: changes in cGMP levels in light-adapted Pde6brd1 null mutants (C3H strain). (B) Comparison of light- and dark-adapted cGMP levels between Pde6bH620Q mutants and DBA/2J control mice at P14, P18, and P21 (see also Refs. 43,44). White columns: light-adapted control mice; black columns: dark-adapted control; light stippled columns: light-adapted mutant mice; dark stippled columns: dark-adapted mutants. Averages and standard deviations are based on measurements from at least four mice per time point. (C) Intracellular Ca2+ measurements in Pde6bH620Q mutant (black trace) and control (gray trace) rod OS by spot confocal recording. Fluorescence from rods loaded with Ca2+ indicator fluo5F are detected as photomultiplier tube current (PMT). Decreases in fluorescence reflect changes in intracellular Ca2+ after membrane channel closure (laser illumination at time 0). Mean dark and light intracellular [Ca2+] levels are then calculated.
Abnormal Intracellular Ca2+ Levels in Pde6bH620Q Homozygous Mutant Outer Segments
In dark-adapted retinas, exposure to light results in activation of PDE6 and a rapid reduction of a small fraction of total cGMP (free cGMP) in the OS. These changes in free cGMP produce closure of cGMP-gated CNG channels and a reduction in the influx of Ca2+. Because Ca2+ continues to be removed by the Na+,K+,Ca2+ exchanger in the rod OS, Ca2+ levels fall. As direct measurement of free cGMP is not tractable, we measured intracellular Ca2+ directly in live Pde6bH620Q rod OS, using laser spot confocal microscopy.53,62
Eight mutant OS were evaluated: Dark-adapted Ca2+ levels were 375 ± 94 nM, whereas light-exposed levels were 92 ± 18 nM. In contrast, in wild-type OS, dark-adapted and light-exposed Ca2+ levels were 241 ± 15 nM and 36 ± 3 nM, respectively (Fig. 4C). Student’s t-test demonstrates that both dark-adapted and light-exposed Ca2+ levels are significantly different between mutant and wild-type rod OS (P = 0.01035 and P < 0.00005, respectively).53 The rates of Ca2+ decreased in wild-type and mutant rods are similar. These data suggest that Ca2+ levels (and, by extension, free cGMP) are significantly elevated in Pde6bH620Q rod OS, at least during a period after exposure to light of dark-adapted retinas.
Somatic Rescue of Photoreceptor Degeneration with an Opsin::Pde6b Lentivirus
Because Pde6bH620Q homozygous mice exhibit a hypomorphic degeneration phenotype, they may be amenable to gene therapy. We therefore introduced lentiviral vectors encoding wild-type Pde6b into Pde6bH620Q retinas and performed histologic and functional analyses.
Two vectors, Opsin::Pde6b and Opsin::Pde6b, EF1::GFP, were constructed (Fig. 5A), and viral particles were prepared and injected subretinally into P5 Pde6bH620Q mice. Contralateral eyes received CMV::GFP lentivirus or saline injection as controls. To test transduction efficiency, Opsin::Pde6b, EF1::GFP-injected retinas were examined by live imaging.63,64 We found GFP fluorescence throughout the eye indicating widespread transduction (Fig. 5C). Injection of Lenti-LacZ particles resulted in detectable β-galactosidase activity within Pde6bH620Q photoreceptors (Fig. 5D).
To determine the effect of Opsin::Pde6b transduction on the Pde6bH620Q degeneration phenotype, we performed a histologic study of 6-week-old transduced and control retinas (Figs. 5E, 5F). Examination of H&E-stained serial sections showed a larger number of photoreceptor nuclei present in the Opsin::Pde6b-transduced retinas, compared to controls (Fig. 5E, top). Peanut agglutinin staining showed that some of these rescued photoreceptors are cones (Fig. 5E, bottom). Although control sections contain cones with shortened OS, in the rescued retinas cone OS are greater in length.
Quantitative analysis indicated an increased number of nuclei present along the optic nerve-to-periphery axis (Fig. 5F). Although control mutants contained one to three photoreceptor nuclei, contralateral transduced retinas contained two to seven. These differences are not attributable to variation between animals, as the comparisons were performed on the same animals. In addition, the nuclei distribution in these control samples are representative of the range observed in 10 additional mutant eyes (Fig. 1B). These results indicate that transduction of wild-type Pde6b into Pde6bH620Q mutant retinas can rescue the photoreceptor degeneration phenotype.
To test if Pde6b gene transfer improves retinal function, we performed serial ERGs on injected and control eyes between P14 and P90 (Figs. 5B, 6). In general, we observed significant differences in maximum b-wave amplitudes in rod and mixed rod– cone responses between injected and control eyes (Figs. 6A, 6B; Table 1). These differences became apparent after P55 and were sustained for several weeks. By P90, rod-specific injected ERGs were indistinguishable from control ERGs. In contrast, photopic cone responses were not significantly improved in injected eyes compared with the control (Fig. 6C). Together, our histology and ERG data show that gene therapy can rescue the Pde6bH620Q degeneration phenotype.
Figure 6.
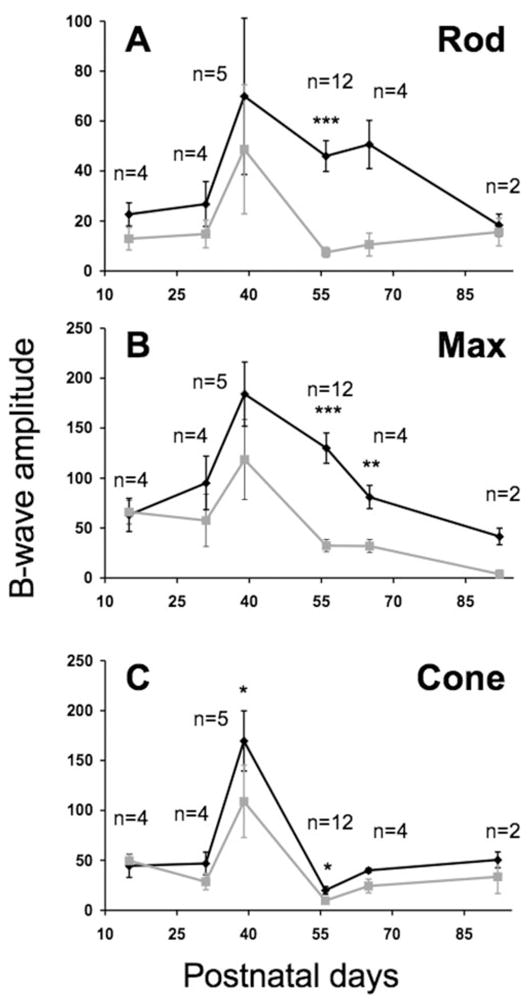
Functional rescue of phototransduction in Opsin::Pde6b transduced Pde6bH620Q homozygous retinas. Subretinal injections of Opsin::Pde6b into the right eye (black line) or control injections of CMV::GFP lentivirus or saline into the left eye (gray line) of P5 mutant mice. ERGs were performed on both eyes simultaneously, between P15 and P92. Shown are the mean and SEM of b-wave amplitudes (Vmax) for (A) dark-adapted rod isolated, (B) maximum mixed rod-cone, and (C) light-adapted cone responses. Student’s t-test was performed to determine probabilities for paired differences in b-wave peaks between injected and controls (see also Table 1). Significance: *P < 0.05; **P < 0.01; ***P < 0.001. The number of mice analyzed per time point (n) is indicated.
Table 1.
Student’s t-Test of Paired Differences of b-Wave Peaks
| Age (d) | Rod | Max | Cone | n |
|---|---|---|---|---|
| 15 | 0.0707 | 0.9048 | 0.7768 | 4 |
| 31 | 0.1890 | 0.0656 | 0.1505 | 4 |
| 39 | 0.4629 | 0.0912 | 0.0168* | 5 |
| 56 | 0.0002*** | 0.0001*** | 0.0187* | 12 |
| 65 | 0.0546 | 0.0064** | 0.1670 | 4 |
| 92 | 0.2427 | 0.1998 | 0.3086 | 2 |
P < 0.05,
P < 0.01;
P < 0.001.
Discussion
We extended the initial phenotypic analysis of Pde6bH620Q homozygous mutant mice conducted by Hart et al.42 and found that Pde6bH620Q mutants exhibited reduced PDE6 β expression and PDE6 activity and delayed photoreceptor degeneration, relative to Pde6brd1 mice. Pde6bH620Q mice demonstrated relatively normal photoreceptor histology and physiology, within 3 weeks of birth that was not observed in Pde6brd1 mice. Finally, using Opsin::Pde6b lentiviral particles, we demonstrated, after subretinal injection, that the Pde6bH620Q phenotype can be histologically and functionally rescued.
Comparison of Pde6bH620Q and Pde6brd1 Homozygous Mutant Mice
Pde6brd1 is a null allele, due to a nonsense mutation, and mutant mice exhibit abnormal photoreceptor differentiation and subsequent rapid rod degeneration.35,65 In particular, Pde6brd1 OS are sparsely populated, disorganized, and truncated as early as P11, and rod degeneration is complete by 3 weeks.9,66 Before degeneration PDE6β expression, PDE6 activity, and retinal function are undetectable in these mice.35,43,44 Associated with absence of PDE6 activity, Pde6brd1 mice show no light-dependent changes in retinal total cGMP levels.43– 45 Moreover, since cGMP levels are abnormally high, relative to the control, it has been proposed that Ca2+ influx through CNG channels promotes rod cell death in Pde6brd1 mice.
Although Pde6bH620Q mice also demonstrate progressive rod degeneration, our studies indicated phenotypic and biochemical features that are distinguishable from Pde6brd1 mice. In particular, Pde6bH620Q mice initially exhibited relatively normal photoreceptor histology and physiology. Indeed, within 3 weeks after birth, Pde6bH620Q mutants expressed PDE6βand showed light-dependent PDE6 activity and detectable rod and cone function. However, residual Pde6b function detected in these mutants was not sufficient to maintain rod photoreceptor health indefinitely.
Partial Loss of PDE6 Function in Pde6b Homozygous Mutants
Consistent with the Pde6bH620Q being a hypomorphic allele, we observed PDE6 β expression and PDE6 activity reduced in Pde6bH620Q mutants. Surprisingly, PDE6β expression was ~22-fold lower, and PDE6 specific activity was ~7.5-fold lower in Pde6bH620Q mutants than in the control mice. Others have documented changes in PDE6β expression in mice homozygous for another Pde6b missense allele, Pde6bR560C.67 Although both H620Q and R560C occur within the catalytic domain and may alter PDE6 activity, these mutations, and possibly others, may also affect PDE6β expression/stability. For our studies, age-matched DBA/2J or C57BL6 were used as the control, which were used previously for the analysis of the Pde6brd1 allele.42– 45 As these are not congenic with the C3H strain, which is the background for the Pde6brd1 and Pde6bH620Q alleles, there may be some differences in the phenotypic parameters measured between these backgrounds.
Despite these expression and activity changes, we found light-adapted total cGMP level between Pde6bH620Q mutants and controls are not significantly different. We estimated the level of total PDE activity in the mutant retinas as ~0.6% of wild-type. This was due to the combined effect of reduced expression (1/22) and lower specific activity (1/7.7) in the mutants. How do we reconcile the fact that total light-adapted cGMP levels are the same between mutants and wild-type, while there is only 0.6% of PDE activity present in the retinas? One possibility is that this level of PDE activity, given enough time, is sufficient to reduce total cGMP to nominal levels under these conditions. Wild-type PDE activity is increased 300-fold by transducin activation and rapidly drives down total cGMP levels on the millisecond time-scale.29 Since, in these experiments, we light adapted the mice at least 3 hours before isolation of total cGMP, mutant PDE may have had enough time to reduce total cGMP to nominal levels.
On a shorter-time scale, the effect of Pde6b loss of function may be more readily detectable. Indeed, our rod-specific ERGs suggest depressed and delayed photoreceptor hyperpolarization milliseconds after light stimulation. Similarly, we found abnormal reductions in intracellular Ca2+ in mutant rod OS after light exposure. A common denominator between these assays is a changing concentration of free cGMP on a millisecond time-scale.
It has been proposed that high cGMP increases Ca2+ influx through CNG channels to induce cell death. Regulation of intracellular Ca2+ by cGMP is an important factor in maintaining photoreceptor health.52,68,69 Our results showed for the first time in a Pde6b mutant model of RP, that Ca2+ levels in living rod OS are significantly elevated. This finding was suspected to be the case in Pde6brd1 mice, but was not testable because of malformation of rod OS in these mice.70,71 It is also a formal possibility that Ca2+ independent factors are involved in initiating degeneration.68,72
Retroviral Gene Therapy of Retinal Degeneration
Using lentiviral-mediated gene transfer, we can increase the number and significantly improve the function of Pde6bH620Q photoreceptors, which is a result not observed in previous attempts using Pde6brd1 mice.38 – 41 Nevertheless, our studies showed localized morphologic and only partial functional restoration. The incomplete efficacy was probably due to focal viral transduction, immune clearance, vector inactivation, and/or nonautonomous degeneration effects of nontransduced cells.73–76 Recently, AAV and adenoviral vectors have been shown to delay photoreceptor degeneration in Pde6bR560C mice (Kumar-Singh R, et al. IOVS 2007;48:ARVO E-Abstract 1980).77 These findings also support our hypothesis that the previous attempts at viral gene therapy were, in part, limited by the severe null degeneration phenotype in Pde6brd1 mice.
As most human PDE6B mutations are missense alleles, features of Pde6b partial loss of function may more closely resemble human RP. The testing of pharmacologic and gene therapy strategies in Pde6bH620Q mice may yield tools for intervention in human photoreceptor specific diseases. Lessons learned in the Pde6bH620Q mouse may be translated into strategies for future treatments.
Acknowledgments
Supported by a Charles E. Culpeper Scholarship in Medical Science, The Foundation Fighting Blindness, the Hirschl Trust, the Crow-ley Research Fund, the Schneeweiss Stem Cell Fund, a Joel Hoffmann Scholarship, the Donald and Barbara Jonas Family Fund, the Hartford/American Geriatrics Society, the Eye Surgery Fund, the Bernard Becker-Association of University Professors in Ophthalmology-Research to Prevent Blindness (RPB), and National Eye Institute Grants K08-EY004081 (SHT) and EY01844 (GLF). SHT is a Burroughs-Wellcome Program in Biomedical Sciences Fellow. CSL is a Homer Rees scholar.
The authors thank the members of the Allikmets, Fain, Gouras, and Nagasaki laboratories for sharing of advice, ideas, and equipment; Siu-Hang Ngan for help in the initial studies; Chai Lin Chou for help with the lentiviral figures; Debora B. Farber and Peter Gouras for critical reading of the manuscript; and the anonymous reviewers’ useful and constructive comments.
Footnotes
Disclosure: R.J. Davis, None; J. Tosi, None; K.M. Janisch, None; J.M. Kasanuki, None; N.-K. Wang, None; J. Kong, None; I. Tsui, None; M. Cilluffo, None; M.L. Woodruff, None; G.L. Fain, None; C.-S. Lin, None; S.H. Tsang, None
References
- 1.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119 –1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- 2.Klein R, Klein BE, Jensen SC, Mares-Perlman JA, Cruickshanks KJ, Palta M. Age-related maculopathy in a multiracial United States population: the National Health and Nutrition Examination Survey III. Ophthalmology. 1999;106:1056 –1065. doi: 10.1016/S0161-6420(99)90255-5. [DOI] [PubMed] [Google Scholar]
- 3.Klein R, Rowland ML, Harris MI. Racial/ethnic differences in age-related maculopathy. Third National Health and Nutrition Examination Survey. Ophthalmology. 1995;102:371–381. doi: 10.1016/s0161-6420(95)31012-3. [DOI] [PubMed] [Google Scholar]
- 4.Hahn LB, Berson EL, Dryja TP. Evaluation of the gene encoding the gamma subunit of rod phosphodiesterase in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1994;35:1077–1082. [PubMed] [Google Scholar]
- 5.Huang SH, Pittler SJ, Huang X, Oliveira L, Berson EL, Dryja TP. Autosomal recessive retinitis pigmentosa caused by mutations in the alpha subunit of rod cGMP phosphodiesterase. Nat Genet. 1995;11:468 – 471. doi: 10.1038/ng1295-468. [DOI] [PubMed] [Google Scholar]
- 6.Dryja TP, Rucinski DE, Chen SH, Berson EL. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1999;40:1859 –1865. [PubMed] [Google Scholar]
- 7.Danciger M, Blaney J, Gao YQ, et al. Mutations in the PDE6B in autosomal recessive retinitis pigmentosa. Genomics. 1995;3:1–7. doi: 10.1006/geno.1995.0001. [DOI] [PubMed] [Google Scholar]
- 8.Tsang SH, Burns ME, Calvert PD, et al. Role of the target enzyme in deactivation of photoreceptor G protein in vivo. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 9.Tsang SH, Gouras P, Yamashita CK, et al. Retinal degeneration in mice lacking the γ subunit of rod cGMP phosphodiesterase. Science. 1996;272:1026 –1029. doi: 10.1126/science.272.5264.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang SH, Woodruff ML, Chen CK, et al. GAP-independent termination of photoreceptor light response by excess gamma subunit of the cGMP-phosphodiesterase. J Neurosci. 2006;26:4472– 4480. doi: 10.1523/JNEUROSCI.4775-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsang SH, Woodruff ML, Janisch KM, Cilluffo MC, Farber DB, Fain GL. Removal of phosphorylation sites of γ subunit of phosphodi-esterase 6 alters rod light response. J Physiol. 2007;579:303–312. doi: 10.1113/jphysiol.2006.121772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang SH, Yamashita CK, Doi K, et al. In vivo studies of the gamma subunit of retinal cGMP-phosphodiesterase with a substitution of tyrosine-84. Biochem J. 2001;353:467– 474. doi: 10.1042/0264-6021:3530467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsang SH, Yamashita CK, Lee WH, et al. The positive role of the carboxyl terminus of the gamma subunit of retinal cGMP-phospho-diesterase in maintaining phosphodiesterase activity in vivo. Vision Res. 2002;42:439 – 445. doi: 10.1016/s0042-6989(01)00213-9. [DOI] [PubMed] [Google Scholar]
- 14.Fain GL, Matthews HR, Cornwall MC, Koutalos Y. Adaptation in vertebrate photoreceptors. Physiol Rev. 2001;81:117–151. doi: 10.1152/physrev.2001.81.1.117. [DOI] [PubMed] [Google Scholar]
- 15.Stryer L. Visual excitation and recovery. J Biol Chem. 1991;266:10711–10714. [PubMed] [Google Scholar]
- 16.Yarfitz S, Hurley JB. Transduction mechanisms of vertebrate and invertebrate photoreceptors. J Biol Chem. 1994;269:14329 –14332. [PubMed] [Google Scholar]
- 17.Burns ME, Baylor DA. Activation, deactivation, and adaptation in vertebrate photoreceptor cells. Annu Rev Neurosci. 2001;24:779 –805. doi: 10.1146/annurev.neuro.24.1.779. [DOI] [PubMed] [Google Scholar]
- 18.Burns ME, Arshavsky VY. Beyond counting photons: trials and trends in vertebrate visual transduction. Neuron. 2005;48:387–401. doi: 10.1016/j.neuron.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Fung BK-K, Hurley JB, Stryer L. Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc Natl Acad Sci USA. 1981;78:152–156. doi: 10.1073/pnas.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arshavsky VY, Lamb TD, Pugh EN., Jr G proteins and phototrans-duction. Annu Rev Physiol. 2002;64:153–187. doi: 10.1146/annurev.physiol.64.082701.102229. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Cote RH. cGMP signaling in vertebrate retinal photoreceptor cells. Front Biosci. 2005;10:1191–1204. doi: 10.2741/1612. [DOI] [PubMed] [Google Scholar]
- 22.Baehr W, Devlin MJ, Applebury ML. Isolation of bovine ROS phosphodiesterase. J Biol Chem. 1979;254:11669 –11677. [PubMed] [Google Scholar]
- 23.Fung BKK, Young JH, Yamane HK, Griswold-Prenner I. Subunit stoichiometry of retinal rod cGMP phosphodiesterase. Biochemistry. 1990;29:2657–2664. doi: 10.1021/bi00463a006. [DOI] [PubMed] [Google Scholar]
- 24.Tsang SH, Gouras P. Photoreceptors and photoreceptor dysfunctions. In: Adelman G, Smith B, editors. Encyclopedia of Neuroscience. Amsterdam: Elsevier Science; 2004. pp. 1633–1644. [Google Scholar]
- 25.Guo LW, Grant JE, Hajipour AR, et al. Asymmetric interaction between rod cyclic GMP phosphodiesterase gamma subunits and alpha/beta subunits. J Biol Chem. 2005;280:12585–12592. doi: 10.1074/jbc.M410380200. [DOI] [PubMed] [Google Scholar]
- 26.Guo LW, Muradov H, Hajipour AR, Sievert MK, Artemyev NO, Ruoho AE. The inhibitory gamma subunit of the rod cGMP phosphodiesterase binds the catalytic subunits in an extended linear structure. J Biol Chem. 2006;28:15412–22. doi: 10.1074/jbc.M600595200. [DOI] [PubMed] [Google Scholar]
- 27.McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the b subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Nat Acad Sci. 1995;92:3249 –3253. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danciger M, Blaney J, Gao Y, et al. Mutations in the PDE6B gene in autosomal recessive retinitis pigmentosa. Genomics. 1995;30:1–7. doi: 10.1006/geno.1995.0001. [DOI] [PubMed] [Google Scholar]
- 29.Bownds MD, Arshavsky VY. What are the mechanisms of photo-receptor adaptation? Behav Brain Sci. 1995;18:415– 424. [Google Scholar]
- 30.Ionita MA, Pittler SJ. Focus on molecules: rod cGMP phosphodiesterase type 6. Exp Eye Res. 2007;84:1–2. doi: 10.1016/j.exer.2005.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stryer L. Transducin and the cyclic GMP phosphodiesterase: amplifier proteins in vision. Cold Spring Harbor Symp Quant Biol. 1983;48(Pt 2):841–852. doi: 10.1101/sqb.1983.048.01.087. [DOI] [PubMed] [Google Scholar]
- 32.Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. J Physiol (Lond) 1979;288:613– 634. [PMC free article] [PubMed] [Google Scholar]
- 33.Keeler CE. The inheritance of a retinal abnormality in white mice. Proc Nat Acad Sci USA. 1924;10:329 –333. doi: 10.1073/pnas.10.7.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keeler C. Retinal degeneration in the mouse is rodless retina. J Hered. 1966;57:47–50. doi: 10.1093/oxfordjournals.jhered.a107462. [DOI] [PubMed] [Google Scholar]
- 35.Farber DB, Flannery JG, Bowes-Rickman C. The rd mouse story: seventy years of research on an animal model of inherited retinal degeneration. Prog Retin Eye Res. 1994;13:31– 64. [Google Scholar]
- 36.Bowes C, Li T, Danciger M, Baxter LC, Applebury ML, Farber DB. Retinal degeneration in the rd mouse is caused by a defect in the b subunit of rod cGMP phosphodiesterase. Nature (Lond) 1990;347:677– 680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- 37.Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase b-subunit gene of the rd mouse. Proc Nat Acad Sci USA. 1991;88:8322– 8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar-Singh R, Farber DB. Encapsidated adenovirus mini-chromosome-mediated delivery of genes to the retina: application to the rescue of photoreceptor degeneration. Hum Mol Genet. 1998;7:1893–1900. doi: 10.1093/hmg/7.12.1893. [DOI] [PubMed] [Google Scholar]
- 39.Bennett J, Tanabe T, Sun D, et al. Photoreceptor cell rescue in retinal degeneration (rd) mice by in vivo gene therapy. Nat Med. 1996;2:649. doi: 10.1038/nm0696-649. [DOI] [PubMed] [Google Scholar]
- 40.Jomary C, Vincent KA, Grist J, Neal MJ, Jones SE. Rescue of photoreceptor function by AAV-mediated gene transfer in a mouse model of inherited retinal degeneration. Gene Ther. 1997;4:683–690. doi: 10.1038/sj.gt.3300440. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi M, Miyoshi H, Verma IM, Gage FH. Rescue from photo-receptor degeneration in the rd mouse by human immunodeficiency virus vector-mediated gene transfer. J Virol. 1999;73:7812–7816. doi: 10.1128/jvi.73.9.7812-7816.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart AW, McKie L, Morgan JE, et al. Genotype-phenotype correlation of mouse pde6b mutations. Invest Ophthalmol Vis Sci. 2005;46:3443–3450. doi: 10.1167/iovs.05-0254. [DOI] [PubMed] [Google Scholar]
- 43.Farber DB, Lolley RN. Cyclic guanosine monophosphate: elevations in degenerating photoreceptor cells of the C3H mouse retina. Science. 1974;186:449 – 451. doi: 10.1126/science.186.4162.449. [DOI] [PubMed] [Google Scholar]
- 44.Farber DB, Lolley RN. Enzyme basis for cyclic GMP accumulation in degenerative photoreceptor cells of mouse retina. J Cyclic Nucleotide Res. 1976;2:139 –148. [PubMed] [Google Scholar]
- 45.Lolley R, Farber D, Rayborn M, Hollyfield J. Cyclic GMP accumulation causes degeneration of photoreceptor cells: simulation of an inherited disease. Science. 1977;196:664 – 666. doi: 10.1126/science.193183. [DOI] [PubMed] [Google Scholar]
- 46.Tsang SH, Woodruff ML, Jun L, et al. Transgenic mice carrying the H258N mutation in the gene encoding the beta-subunit of phosphodiesterase-6 (PDE6B) provide a model for human congenital stationary night blindness. Hum Mutat. 2007:243–254. doi: 10.1002/humu.20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ortega S, Ittmann M, Tsang SH, Ehrlich M, Basilico C. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc Natl Acad Sci U S A. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsang SH, Chen J, Kjeldbye H, et al. Retarding photoreceptor degeneration in Pdegtm1/Pdegtm1 mice by an apoptosis suppressor gene. Invest Ophthalmol Vis Sci. 1997;38:943–950. [PubMed] [Google Scholar]
- 49.Kuhn H. Light- and GTP-regulated interaction of GTPase and other proteins with bovine photoreceptor membranes. Nature. 1980;283:587–589. doi: 10.1038/283587a0. [DOI] [PubMed] [Google Scholar]
- 50.Artemyev NO. Interactions between catalytic and inhibitory subunits of PDE6. Methods Mol Biol. 2005;307:277–288. doi: 10.1385/1-59259-839-0:277. [DOI] [PubMed] [Google Scholar]
- 51.Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653– 660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 52.Olshevskaya EV, Calvert PD, Woodruff ML, et al. The Y99C mutation in guanylyl cyclase-activating protein 1 increases intracellular Ca2+ and causes photoreceptor degeneration in transgenic mice. J Neurosci. 2004;24:6078 – 6085. doi: 10.1523/JNEUROSCI.0963-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodruff ML, Sampath AP, Matthews HR, Krasnoperova NV, Lem J, Fain GL. Measurement of cytoplasmic calcium concentration in the rods of wild-type and transducin knock-out mice. J Physiol. 2002;542:843– 854. doi: 10.1113/jphysiol.2001.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baehr W, Champagne M, Kee AK, Pittler SJ. Complete cDNA sequences of mouse rod photoreceptor cGMP phosphodiesterase alpha- and beta-subunits, and identification of beta′-, a putative beta-subunit isozyme produced by alternative splicing of b-subunit gene. FEBS Lett. 1991;278:107–114. doi: 10.1016/0014-5793(91)80095-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 56.Doi K, Hargitai J, Kong J, et al. Lentiviral transduction of green fluorescent protein in retinal epithelium: evidence of rejection. Vision Res. 2002;42:551–558. doi: 10.1016/s0042-6989(01)00237-1. [DOI] [PubMed] [Google Scholar]
- 57.Lai CC, Gouras P, Doi K, et al. Tracking RPE transplants labeled by retroviral gene transfer with green fluorescent protein. Invest Ophthalmol Vis Sci. 1999;40:2141–2146. [PubMed] [Google Scholar]
- 58.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gouras P, Kong J, Tsang SH. Retinal degeneration and RPE transplantation in Rpe65(−/−) mice. Invest Ophthalmol Vis Sci. 2002;43:3307–3311. [PubMed] [Google Scholar]
- 60.Salchow DJ, Gouras P, Doi K, Goff SP, Schwinger E, Tsang SH. A point mutation (W70A) in the rod PDE-gamma gene desensitizing and delaying murine rod photoreceptors. Invest Ophthalmol Vis Sci. 1999;40:3262–3267. [PMC free article] [PubMed] [Google Scholar]
- 61.Muradov H, Boyd KK, Artemyev NO. Structural determinants of the PDE6 GAF A domain for binding the inhibitory gamma-subunit and noncatalytic cGMP. Vision Res. 2004;44:2437–2444. doi: 10.1016/j.visres.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 62.Woodruff M, Wang Z, Chung HY, Redmond T, Fain GL, Lem J. Spontaneous activity of opsin apoprotein is a cause of Leber congenital amaurosis. Nat Genet. 2003;35(2):158 – 64. doi: 10.1038/ng1246. [DOI] [PubMed] [Google Scholar]
- 63.Nagasaki T, Zhao J. Centripetal movement of corneal epithelial cells in the normal adult mouse. Invest Ophthalmol Vis Sci. 2003;44:558 –566. doi: 10.1167/iovs.02-0705. [DOI] [PubMed] [Google Scholar]
- 64.Nagasaki T, Zhao J. Uniform distribution of epithelial stem cells in the bulbar conjunctiva. Invest Ophthalmol Vis Sci. 2005;46:126 –132. doi: 10.1167/iovs.04-0356. [DOI] [PubMed] [Google Scholar]
- 65.Lolley RN, Rong H-M, Craft CM. Linkage of photoreceptor degeneration by apoptosis with inherited defect in phototransduction. Invest Ophthalmol Vis Sci. 1994;35:358 –362. [PubMed] [Google Scholar]
- 66.Carter-Dawson LD, LaVail MM, Sidman RL. Differential effect of the rd mutation on rods and cones in the mouse retina. Invest Oph-thalmol Vis Sci. 1978;17:489 – 498. [PubMed] [Google Scholar]
- 67.Chang B, Hawes NL, Pardue MT, et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res. 2007;47:624 – 633. doi: 10.1016/j.visres.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fain GL. Why photoreceptors die (and why they don’t) Bioessays. 2006;28:344 –354. doi: 10.1002/bies.20382. [DOI] [PubMed] [Google Scholar]
- 69.Woodruff M, Olshevskaya E, Savchenko A, et al. Constitutive excitation by Gly90Asp rhodopsin rescues rods from degeneration caused by elevated production of cGMP in the dark. J Neurosci. 2007;27:8805– 8815. doi: 10.1523/JNEUROSCI.2751-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fain GL, Lisman JE. Light, Ca2+, and photoreceptor death: new evidence for the equivalent- light hypothesis from arrestin knockout mice. Invest Ophthalmol Vis Sci. 1999;40:2770 –2772. [PubMed] [Google Scholar]
- 71.Lisman J, Fain G. Support for the equivalent light hypothesis for RP. Nat Med. 1995;1:1254 –1255. doi: 10.1038/nm1295-1254. [DOI] [PubMed] [Google Scholar]
- 72.Rohrer B, Pinto FR, Hulse KE, Lohr HR, Zhang L, Almeida JS. Multidestructive pathways triggered in photoreceptor cell death of the rd mouse as determined through gene expression profiling. J Biol Chem. 2004;279:41903– 41910. doi: 10.1074/jbc.M405085200. [DOI] [PubMed] [Google Scholar]
- 73.Sahel JA. Saving cone cells in hereditary rod diseases: a possible role for rod-derived cone viability factor (RdCVF) therapy. Retina. 2005;25:S38 –S39. doi: 10.1097/00006982-200512001-00015. [DOI] [PubMed] [Google Scholar]
- 74.Huang PC, Gaitan AE, Hao Y, Petters RM, Wong F. Cellular interactions implicated in the mechanism of photoreceptor degeneration in transgenic mice expressing a mutant rhodopsin gene. Proc Natl Acad Sci U S A. 1993;90:8484 – 8488. doi: 10.1073/pnas.90.18.8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.West JD. Distortion of patches of retinal degeneration in chimaeric mice. J Embryol Exp Morphol. 1976;36:145–149. [PubMed] [Google Scholar]
- 76.LaVail MM, Mullen RJ. Experimental chimeras: a new approach to the study of inherited retinal degeneration in laboratory animals. Adv Exp Med Biol. 1977;77:153–173. doi: 10.1007/978-1-4899-5010-9_13. [DOI] [PubMed] [Google Scholar]
- 77.Pang J, Boye S, Deng W, et al. AAV-mediated gene therapy delays retinal degeneration in the rd10 mouse containing a recessive PDEβ mutation. Invest Ophthalmol Vis Sci. 2008 June 27; doi: 10.1167/iovs.07-1622. Published online on. [DOI] [PMC free article] [PubMed] [Google Scholar]