Abstract
Lipid bodies (LBs), multifunctional organelles present in most eukaryotic cells, are sites of eicosanoid formation in leukocytes; but little is known about the composition of leukocyte LBs or the biogenesis and internal structures of LBs from mammalian cells. Proteomic analyses of LBs purified from human monocytic U937 cells detected, common to LBs in other cells, proteins involved in cholesterol and triglyceride metabolism, Rab GTPases, and many membrane and endoplasmic reticulum (ER)-associated proteins. Newly lipid body (LB)-associated proteins included MRP-14, potentially involved in arachidonate transport, and ribosomal subunit proteins and translation regulatory proteins. Ultrastructurally, in U937 cells as well as human neutrophils and eosinophils, ribosomes are attached to and distributed within LBs, and LBs contain extensive ER-like membranes. The presence of ribosomes, ER-like membranes and many membrane-associated and ER luminal proteins within LBs, supports a new model by which enveloped ER-membranes and domains form LBs and indicates that LBs may be sites of protein synthesis.
Lipid bodies (LBs), also known as lipid droplets, are increasingly recognized as dynamic organelles (1, 2). In adipocytes and steroidogenic testicular, ovarian, and adrenal cells, LBs were recognized early for their roles in the storage and metabolism of fatty acids and cholesterol. The central core of LBs contains triglycerides. Lipid body (LB)-stored cholesterol in steroidogenic cells is a synthetic precursor of steroid hormones, and in other cells is important in atherogenic and other activities of cholesterol (3). LBs, rich in triglycerides and cho.lesterol (1), are surrounded not by a typical bilayer membrane but by a monolayer of phospholipids (4). Specific LB-associated proteins of the perilipin, adipophilin, TIP47 (PAT) family, perilipins, adipophilin, tail-interacting protein of 47 kDa (TIP47), and S3–12 (5–8) are involved in LB triglyceride metabolism (9).
Proteomic studies of LBs from Chinese hamster ovary K2 cells (10), squamous carcinoma A431 cells (11), hepatoma cells (12, 13), and 3T3-L1 adipocytes (14) provided broader insights into the functioning of LBs. For instance, recognition that several Rab GTPase proteins are associated with LBs (10–12, 14, 15) suggests roles for these LB-associated proteins in membrane trafficking or other regulated processes. Indeed, LBs have more roles than simply as depots of neutral lipids (16). While LBs are assumed to be derived from the endoplasmic reticulum (ER), critical aspects of LBs, including their internal structure and mode of derivation from ER membranes, have been unsettled. Although PAT proteins and caveolin were initially immunofluorescently localized to circumferential surfaces of LBs, recent freeze-fracture immunogold electron microscopy (EM) revealed that caveolin-1 in smooth muscle cells (17) and TIP47 and adipophilin (16, 18) in macrophages are present within LBs and localized to freeze-fractured lamellae within LB cores (16–18).
LBs have additional functions in leukocytes. Leukocyte LBs form in macrophages, neutrophils, and eosinophils associated with infectious and inflammatory responses (19). Specific pathogen- and ligand-initiated, receptor-mediated pathways activate intracellular signaling that leads to enhanced LB formation. For instance, Mycobacterium bovis bacillus Calmette-Guérin induces TLR2-mediated formation of LBs in macrophages (20), platelet-activating factor through its receptor induces LB formation in neutrophils and eosinophils (21, 22), and the chemokines eotaxin (CCL11) and RANTES (CCL5), acting via CCR3 receptors, stimulate LB formation in eosinophils (23). Leukocyte LBs contain esterified arachidonate (24, 25) and possess enzymes to liberate [e.g., cytosolic phospholipase (cPL) A2] (26) and metabolize [e.g., 5-lipoxygenase (5-LO), prostaglandin H synthases (PGHSs, cyclooxygenases), and leukotriene (LT) C4 synthase] (20, 22, 27, 28) arachidonate to synthesize eicosanoids. Increased leukocyte LB numbers have correlated with enhanced production of PGHS (PGE2) and 5-LO (LTB4 and LTC4) -derived eicosanoids (21, 22, 29). Moreover, as evidenced by immunolocalizations of newly synthesized eicosanoids, LBs are specific sites of de novo formation of LTC4 and PGE2 in leukocytes (20, 23). Roles for LBs in eicosanoid formation even in nonleukocyte cells have been indicated. For instance, cPLA2, PGHS-2, and microsomal PGE synthase were localized to LBs in human fetal epithelium and fibroblasts (30), and PGHS-1 and -2 were localized to LBs in the corpus luteum (31). In addition, neutrophil LBs rich in arachidonate are motile and transiently associate with phagosomes, potentially providing arachidonate to activate phagosome-associated NADPH oxidase (32).
Cytokines and signaling kinases are present in leukocyte LBs (26, 33–35). Although immunofluorescent localizations of PAT proteins to surfaces of LBs under-detect PAT proteins within LBs (18), immunolocalizations of eicosanoid-forming enzymes to LBs have consistently documented labelings within LBs (22, 23). By immunogold, EM 5-LO localized throughout eosinophil LBs (22). Moreover, EM has at times revealed honeycomb-like internal ultrastructures within eosinophil LBs (22), and transmembrane spanning proteins, such as LTC4 synthase, were localized throughout eosinophil LBs (22). Thus, at least in leukocytes, LBs likely contain as yet undefined membranous structures and membrane-associated proteins within LB cores.
Given the broader functional roles recognized for LBs in leukocytes, we used human monocyte U937 line cells to evaluate proteins associated with isolated LBs doubly purified by subcellular fractionation. We complemented proteomic analyses of isolated LBs with transmission EM studies of LBs in U937 cells as well as native blood-derived human eosinophils and neutrophils. We find that leukocyte LBs share a number of proteins associated with LBs in other cells and also contain proteins involved in protein translation, a finding corroborated by EM studies showing ribosomes associated with and within leukocyte LBs, contain ER luminal proteins and possess membranous internal structures within LB cores. These findings support a novel model for the formation of LBs from the ER.
Materials and Methods
Purification of lipid body proteins
Human U937 cells (CRL-1593.2, American Type Culture Collection; Manassas, VA, USA) cultured at ≤2 × 105 cells/ml overnight in RPMI 1640 medium, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin (37°C, 5% CO2) contained numerous LBs. Disrupted U937 cells were subjected to two cycles of subcellular fractionation for isolation of buoyant LBs by modifications of prior methods (24). Cells were resuspended at 108 cells/ml in cold disruption buffer (25 mM Tris-HCl pH 7.4, 5 mM ethylenediaminetetraacetic acid, 1 mM ethylene glycol-bis-tetraacetic acid, 0.2 mM phenylmethylsulfonyl fluoride, 50 μg/ml Nα-p-tosyl-l-lysine-chloromethyl ketone, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 1 μg/ml aprotinin). After nitrogen cavitation (800 psi, 10 min, 4°C), disrupted cells were mixed with sucrose to a concentration of 0.54 M sucrose. Nuclei and intact cells were removed by centrifugation (1000 g, 30 min, 4°C). The cavitate was overlaid with 0.27, 0.135, and 0 M sucrose solutions and subjected to ultracentrifugation (150,500 g, 3–4 h, 4°C). Cytosolic and buoyant LB-containing fractions were harvested from the 0.54 M sucrose and uppermost layers, respectively.
To minimize contamination of LBs, initially isolated LB fractions were adjusted to contain 0.35 M sucrose and 0.15 M NaCl and placed under layers of 0/0.135 M/0.27 M sucrose solutions for a second ultracentrifugation. As controls, initially isolated cytosolic fractions containing equal amounts of protein were likewise placed under the same layered sucrose solutions. After ultracentrifugations as above, sucrose-free upper layers containing buoyant LBs or control proteins were collected. Fractions from multiple isolations derived from >2 × 109 cells were pooled and mixed with chloroform and methanol at volume ratios of 1:1.25:2.5 (sample:chloroform: methanol). Proteins were pelleted by centrifugation, resolved on 4–12% NuPAGE SDS reducing gels (Invitrogen, Carlsbad, CA, USA), and stained with GelCode Blue (Pierce, Rockford, IL, USA). Purity of doubly isolated LBs was ascertained. Specific organelle markers—nuclear lamin B, endosomal annexin VI, and lysosomal cathepsin D—were not detectable in isolated LB fractions by Western blot, nor were enzyme activities of cytosolic lactate dehydrogenase and microsomal sulfatase C.
MS/LS analyses
Six protein-containing zones from doubly isolated LB fractions, excised from gels, were subjected to trypsin digestion (36). Digested samples reconstituted in 5% acetonitrile, 0.005% heptafluorobutyric acid, 0.4% acetic acid were loaded on nanoscale C18 reverse-phase HPLC capillary columns (37). Peptides, eluted with increasing concentrations of 95% acetonitrile, 0.005% heptafluorobutyric acid, 0.4% acetic acid were subjected to electrospray ionization and entered into a LCQ DECA ion trap mass spectrometer (ThermoFinnigan, San Jose, CA, USA). Peptide sequences were determined by matching protein or translated nucleotide databases with acquired fragmentation patterns by the Sequest program (ThermoFinnigan). Proteins were identified based on two or, more commonly, multiple peptide sequences (except where noted in Table 1). Potential membrane insertion domains were assessed through TMAP prediction (http://bioinfo.limbo.ifm.liu.se./tmap) based on single sequence alignment (38).
Table 1. U937 monocyte lipid body-associated proteins identified by MS/LS.
| Location or function TM: predicted membrane insertion |
Band no. | Protein name | M.W. (kDa) | Accession ID in Swiss-Prot database | Found with LBs in mammalian cells | Ref. |
|---|---|---|---|---|---|---|
| Related to lipid bodies | ||||||
| 2 | Adipophilin (ADRP) | 48.0 | Q99541 | yes | (6, 10–14) | |
| 2 | TIP47 (mannose-6-phosphate receptor binding protein) | 47.0 | O60664 | yes | (11–14, 63, 64) | |
| TM1:281–309 | 2 | CGI-49 (saccharopine dehydrogenase) | 46.9 | Q9Y363 | yes | (12–14) |
| TM2: 309–385 | ||||||
| TM:77–95 | 4 | CGI-58 | 38.8 | Q9Y369 | yes | (10, 11, 65, 66) |
| Lipid metabolism | ||||||
| TM: 1810–1833 | 1 | Acetyl CoA carboxylase | 265 | Q13085 | yes | (10, 11) |
| TM: 95–123 | 1 | Lanosterol synthase | 83.3 | P48449 | yes | (6, 10–14) |
| TM1: 16–44 | 1 | Long chain fatty acid CoA synthase 3 | 80.3 | O95573 | yes | (6, 10–12, 14) |
| TM2: 172–190 | ||||||
| TM1: 19–44 | 2 | Squalene epoxidasea | 63.9 | Q14534 | yes | (10, 11) |
| TM2: 54–82 | ||||||
| TM3:125–147 | ||||||
| TM4: 513–533 | ||||||
| TM5: 546–566 | ||||||
| 2 | RIKEN cDNA 0610039C21, Triglyceride lipase | 47.4 | AK002826 (nucleotide sequence) | yes | (67, 68) | |
| TM: 296–324 | 3 | NAD(P)H-dependent steroid dehydrogenase | 41.9 | Q15738 | yes | (11–14, 69, 70) |
| TM1: 86–112 | 4 | 17-β-hydroxysteroid dehydrogenase 7 | 38.2 | P56937 | yes | (10–12, 14) |
| TM2: 230–258 | ||||||
| TM: 1355–1379 | 4 | Retinal short chain dehydrogenase, ret SDR1 | 33.5 | AF061741 | yes | SDR families (10–12, 14) |
| TM1: 13–33 | 4 | Retinal short chain dehydrogenase, ret SDR2 | 32.9 | Q8TDV9 | yes | SDR families (10–14) |
| TM2: 44–64 | ||||||
| TM3: 192–208 | ||||||
| TM4: 267–295 | ||||||
| Cytoskeleton protein | ||||||
| TM:127–155 | 2 | Tubulin-α | 50.0 | Q13748 | yes | (10, 14) |
| 3,4 | Actin | 41.7 | P02570 | yes | (11, 71) | |
| Endoplasmic reticulum proteins/glycosylation | ||||||
| TM1:4–25, TM2:408–433 | 2 | Oligosaccharyl transferase subunita | 48.0 | P39656 | — | |
| TM: 60–85 | 4 | Dehydrodolichyl diphosphate synthase | 38.7 | Q86SQ9 | — | |
| TM1: 8–36, | 4 | Dolichyl-phosphate-glucosyltransferase | 36.9 | Q9Y673 | yes | (10) |
| TM2: 207–232, | ||||||
| TM3: 259–278 | ||||||
| TM: 4–26 | 4 | NADH-cytochrome b5 reductase | 34.1 | P00387 | yes | (11–13) |
| Chaperones/folding enzymes | ||||||
| 1 | Heat shock protein 70 | 75 | P38646 | yes | (11, 14, 72) | |
| 1 | BiP (glucose regulated-protein 78) | 72.3 | P11021 | yes | (10, 11, 13, 14, 73) | |
| TM1: 3–20 | 1 | Calnexin | 67.6 | P27824 | yes | (13, 14, 73) |
| TM2: 479–503 | ||||||
| TM:4–21 | 2 | Protein disulfide isomerase P5 | 48.1 | Q15084 | yes | PDI family (10, 11, 13) |
| TM: 4–21 | 5 | Cyclophilin B | 22.7 | P23284 | — | |
| Ribosome/translation proteins | ||||||
| TM: 98–124 | 2 | Eukaryotic elongation factor 1-α | 50 | P04720 | — | |
| 2 | Eukaryotic elongation factor 1-γ | 50 | P26641 | — | ||
| 2 | Eukaryotic initiation factor 4A-1 | 46.2 | P04765 | — | ||
| TM:117–145 | 5 | 60S ribosomal protein L11a | 20.1 | P39026 | — | |
| 6 | 60S ribosomal protein L23a | 14.9 | P23131 | — | ||
| 6 | 40S ribosomal protein S20a | 13.4 | P17075 | — | ||
| TM: 41–61 | 6 | 60S acidic ribosomal proteina | 11.5 | P05386 | — | |
| Vesicular trafficking | ||||||
| TM1: 162–184 | 3 | VAT-1 (synaptic vesicle membrane protein) homolog | 32.6 | Q99536 | — | |
| TM2: 190–206 | ||||||
| 4 | SNAP29, vesicle-membrane fusion protein | 29 | Q95721 | yes | SNAP family (10) | |
| TM1: 9–37 | 5 | Transmembrane traffic protein | 25 | P49755 | — | |
| TM2: 184–210 | ||||||
| 5 | Rab 1A (Ras-related protein) | 22.7 | P11476 | yes | (10, 13) | |
| 5 | Rab 1B | 22.2 | Q9H0U4 | yes | (10, 11, 13) | |
| 5 | Rab 7 | 23.5 | P51149 | yes | (10, 11, 13, 14) | |
| 5 | Rab 10 | 22.5 | Q88386 | yes | (10, 11, 13) | |
| TM: 71–99 | 5 | Rab 11 | 24.4 | P24410 | yes | (10, 13) |
| TM: 12–37 | 5 | Rab 14 | 23.9 | P35287 | yes | (10, 14) |
| 5 | Rab 18 | 23 | Q9NP72 | yes | (10, 11, 13–15, 44) | |
| 5 | GTP-binding protein SAR 1a | 22.4 | Q9NR31 | — | ||
| 5 | Rap-1a | 21 | P10113 | yes | Rap-1b(12) | |
| Nuclear proteins | ||||||
| 2 | BAT-1 (HLA-B associated transcript 1) | 49.0 | Q13838 | — | ||
| TM: 56–76 | 4 | HnRNP E1a | 37.5 | Q15365 | — | |
| 4 | HnRNP C1/C2a | 33.7 | P07910 | — | ||
| TM:222–246 | 5 | Emerin | 29.0 | P50402 | — | |
| TM: 46–66 | 6 | Histone 2Az | 13.4 | P17317 | — | |
| 6 | Histone 2Bb | 13.8 | P58876 | — | ||
| 6 | Histone 4 | 11.2 | P02304 | — | ||
| Serum proteins | ||||||
| TM1: 1–24 | 1 | Albumin | 69.3 | P02768 | yes | (72) |
| TM2: 32–56 | ||||||
| TM: 105–133 | 6 | Hemoglobin γ | 16.0 | P02096 | — | |
| 6 | Hemoglobin β | 15.9 | P02023 | — | ||
| TM: 98–116 | 6 | Hemoglobin ζ | 15.5 | P02008 | — | |
| TM: 98–15 | 6 | Hemoglobin α | 15.1 | P69905 | — | |
| Miscellaneous | ||||||
| EF-hand Motif | ||||||
| 5 | Calcium binding protein p22 | 22.3 | Q99653 | yes | (11) | |
| 6 | S100 calcium binding protein A9, Migration inhibitory related protein (MRP)-14 | 13.2 | P06702 | — | ||
| Apoptosis | ||||||
| TM:8–36 | 4 | AMID, P53-responsive gene | 40.5 | Q9BRQ8 | yes | (11, 13) |
| Other | ||||||
| 5 | VDAC-1 (Voltage-dependent anion-selective channel protein-1) | 30.6 | P21796 | yes | (10) | |
| TM1: 16–44 | 2 | AUP-1 (Ancient ubiquitous protein-1) | 53.0 | Q9Y679 | yes | (13, 14) |
| TM2: 149–177 | ||||||
| TM3: 269–288 | ||||||
| TM1: 60–85 | 2 | HSPC028, basic leu zipper and W2 domain 2 | 48.2 | Q9Y6E2 | — | |
| TM2: 151–170 | ||||||
| TM3: 355–383 | ||||||
| 2 | Carboxypeptidase A2 | 46.4 | P48052 | — | ||
| 2 | Polyubiquitin | 25.7 | Q9BWD6 | — | ||
| TM: 178–195 | 4 | Hypothetical protein FLJ21820 | 22.3 | Q9H6V9 | yes | (13) |
| 4 | GAPDH | 37.3 | P04406 | yes | (12) | |
| TM1: 78–96 | 4 | Hypothetical protein MGC10084 gene | 36 | BC015168 | — | |
| TM2: 1288–1308 | ||||||
| TM3: 1323–1343 | ||||||
| TM: 8–36 | 5 | TD 54 | 35 | O43399 | yes | (11, 14) |
Only a single matching peptide sequence.
Electron microscopy
U937 cells
Cultured U937 cells were fixed in 1% paraformaldehyde/1.25% glutaraldehyde, processed for EM and examined as detailed before (39). Seventy-five electron micrographs ranging from 8000× to 75,000× were evaluated.
Neutrophil and eosinophil isolation and stimulation
As approved by the Committee on Clinical Investigation, blood granulocytes were isolated and eosinophils were purified by negative selection as described (39). Eosinophils (106 cells/ml) were stimulated with recombinant human eotaxin or RANTES (100 ng/ml, R&D Systems, Minneapolis, MN, USA) or medium alone (RPMI 1640, 0.1% ovalbumin) at 37°C for 1 h. Cells were processed for EM, as above.
Results
U937 lipid body protein composition
To minimize contamination, initially isolated LB fractions were resuspended in higher concentration (0.15M NaCl) salt and reisolated from buoyant fractions on second ultracentrifugations. The stained protein profiles of doubly isolated LBs (Fig. 1, lane 1) differed both from those of initial cytosolic fractions (Fig. 1, lane 3) and from control cytosolic fractions that were subjected to second subcellular fractionations (Fig. 1, lane 2) (the latter of contained 50–75 kDa proteins identified as contaminating cytokeratins). From the six excised lane 1 zones, MS/LS analyses identified proteins listed in Table 1.
Figure 1.
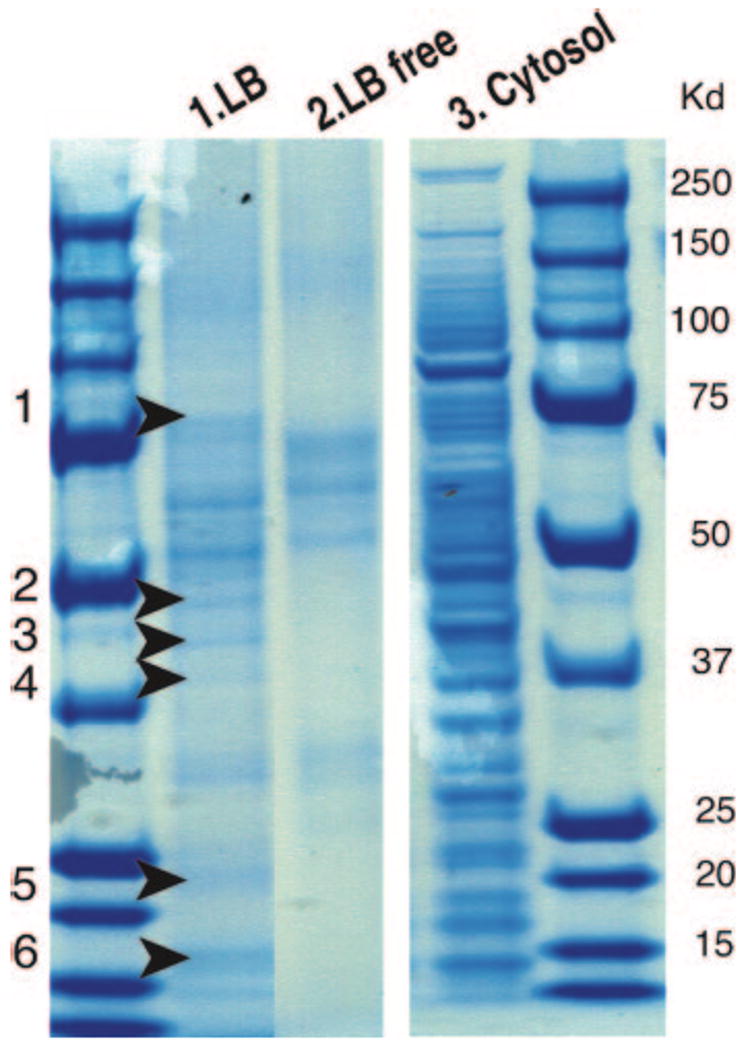
Protein profiles of LB and cytosolic fractions isolated from U937 cells. Samples were resolved on SDS-PAGE gels and protein stained, as described in Materials and Methods. After two sequential subcellular fractionations, lane 1 contained LB proteins in uppermost buoyant fractions of doubly isolated LBs whereas lane 2 contained proteins from uppermost subcellular fractions isolated from second control fractionations in which initial cytosolic fractions were subjected to a second ultracentrifugation. Lane 3 contained proteins from LB-depleted cytosolic fractions after the first subcellular fractionation. Six zones (arrows) from lane 1 were subjected to proteomic analyses of LB-associated proteins. Lane 1 LB proteins were reproducible in three different LB purifications.
U937 cell LB-associated proteins included many associated with LBs in other cells, such as the LB proteins, adipophilin, TIP47, CGI-49, and CGI-58, various enzymes involved in cholesterol, triglyceride, and retinyl lipid synthesis, and ER chaperone/folding proteins (heat shock protein 70, BiP, calnexin, and protein disulfide isomerase P5) (Table 1). Two cytoskeletal proteins, tubulin-α and actin, previously associated with LBs, were identified (Table 1). Several proteins potentially involved in vesicular trafficking were identified, including Ras-related Rab family member proteins (Table 1), previously associated with LBs, as well as three (VAT-1, transmembrane traffic protein, and GTP binding protein SAR1a) not known to be associated with LBs. In addition, other proteins (Table 1, see Miscellaneous) were identified, including some not yet recognized to be associated with LBs, S100 calcium binding protein A9, hypothetical proteins FLJ21820 and MGC10084, and HSPC028.
Novel findings for LB-associated proteins were several ribosomal component proteins and proteins involved in regulation of ribosomal protein translation. Ribosomal proteins were 60 S acidic ribosomal protein, 60 S ribosomal proteins L11 and L23, and 40 S ribosomal protein S20; translational control proteins were eukaryotic elongation factors 1-α, 1-γ, and 4A-1 (Table 1).
Several proteins mediating glycosylation and often associated with the ER were identified (Table 1), including two not previously associated with LBs: oligosaccharyl transferase subunit and dehydrodolichyl diphosphate synthase. These latter proteins are membrane-associated, and of the varied proteins identified in U937 LB fractions, more than half were recognized as membrane-spanning proteins or contained predicted membrane-inserting domains (Table 1).
Ultrastructural studies of lipid bodies in situ
Findings of ribosomal and ER-related proteins associated with isolated U937 LBs and the identification of many membrane-inserting proteins with LBs, that nominally possess only a delimiting phospholipid monolayer (4) and lack heretofore identified internal membranes, led us to examine the ultrastructure of LBs. As we found with isolated eosinophil LBs (24), the inherent buoyancy of isolated U937 LBs precluded their sedimentation into pellets amenable for transmission EM. Thus, we used EM of intact U937 cells, neutrophils, and eosinophils to examine the ultrastructure of LBs.
U937 cell lipid bodies in situ
LBs within U937 cells appeared to be composed of amorphous lightly dense core material (Fig. 2A,B) and frequently showed an electron-dense rim devoid of a bilayer membrane (Fig. 2A, arrowheads), features typical of LBs in other leukocytes (40, 41). U937 LBs were frequently closely associated with the ER. LBs were partially (Fig. 2B, C) or completely (Fig. 2D) surrounded by ER cisternae, with some clear attachments between them (Fig. 2B, C, arrows). Cytoskeletal filaments were inserted onto LBs (Fig. 2E, F, arrowheads) or encapsulated these structures (Fig. 2F, arrows).
Figure 2.
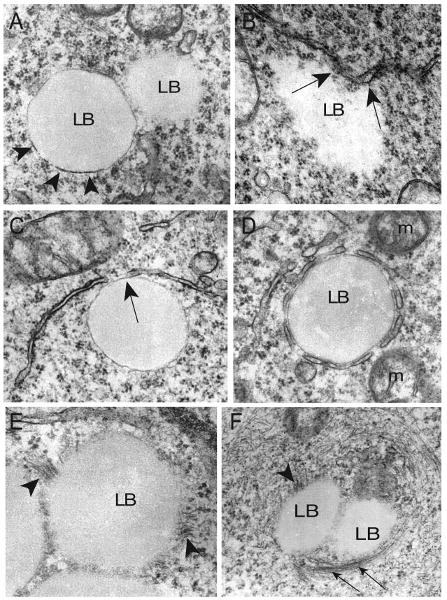
Morphology of lipid bodies (LBs) and their intracellular interactions in U937 cells. A) LBs showed a light-dense core with an electron-dense rim (arrowheads) or an ill-defined periphery. B–D) LB-endoplasmic reticulum (ER) interactions. ER partially (B, C) or entirely (D) surrounded LBs, with areas of clear attachment (B, C, arrows). E, F) Cytoskeleton filaments inserted onto (arrowheads) or encapsulated (arrows) LBs. Cultured cells were processed for transmission electron microscopy (TEM) as described (39). m, mitochondrion. Scale bar, 500 nm (A–C), 480 nm (D, F), 360 nm (E).
Ribosomes are associated with lipid bodies
Associations between LBs and ribosomes were evident in U937 cells. Free ribosomes (Fig. 3A, arrowheads) or rough ER profiles (Fig. 3A) were lining or attached to borders of LBs. Moreover, ribosomes or ribosome subunit-like particles were present within the lipid-rich cores of LBs (Fig. 3A, B, arrows). To further ascertain associations of ribosomes with leukocyte LBs, we studied human blood-derived granulocytes, neutrophils, and eosinophils by EM. These granulocytes were chosen for three reasons: 1) they are native leukocytes (in contrast to the U937 cell line), 2) they can contain prominent LBs, and 3) they exhibit differential LB osmiophilic staining when examined by EM. Eosinophil LBs characteristically are very electron-dense whereas neutrophil LBs are more electron lucent (40), like those in U937 cells.
Figure 3.
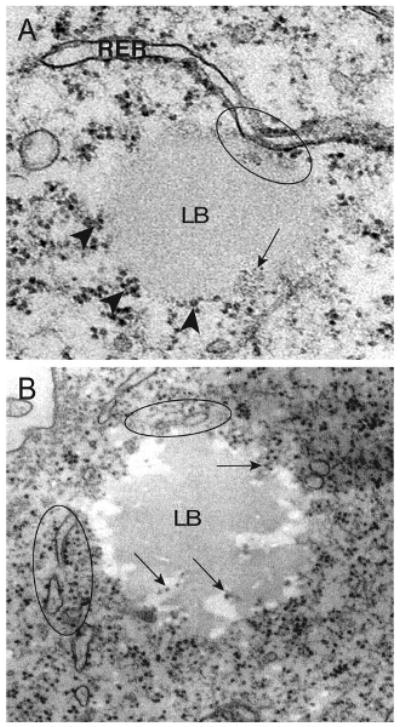
Ribosomes are associated with lipid bodies (LB) within U937 cells. A, B) Clusters of ribosomes were attached to the LB periphery (A, arrowheads) or distributed inside LBs (A, B, arrows). Note that profiles of the rough endoplasmic reticulum (RER) (circles) interact with or surround LBs. Scale bar, 400 nm (A), 500 nm (B).
Clear associations between LBs and ribosomes were observed in both neutrophils and eosinophils. Free ribosomes were lining or attached to LB borders, often with heavy clusters of ribosomes (Fig. 4A). In addition, ribosome-like particles were present within LBs (Fig. 4Aii). In both granulocytes and U937 cells, some LBs devoid of associated ribosomes were observed concomitantly with heavily ribosome-associated LBs (Fig. 4Ab, B). To provide additional evidence that LBs interact with ribosomes, eosinophils were stimulated with eotaxin or RANTES and examined by EM. Eosinophils activated by eotaxin or RANTES in vitro, like those recruited during inflammatory processes in vivo, exhibit morphological changes associated with activation, including the induction of new LB formation (23). Stimulated eosinophils contained large LBs with ribosomes not only attached to the circumferential surfaces of LBs, but also spread within their core contents (Fig. 4C). Ribosomes were also associated with ER cisternae wrapping around LBs (Fig. 4Cb, arrows). Thus, EM studies of U937 cells and two human leukocytes provided evidence of intimate associations of ribosomes around and within LBs, in support of proteomic findings of ribosomal subunit proteins in isolated U937 LBs.
Figure 4.
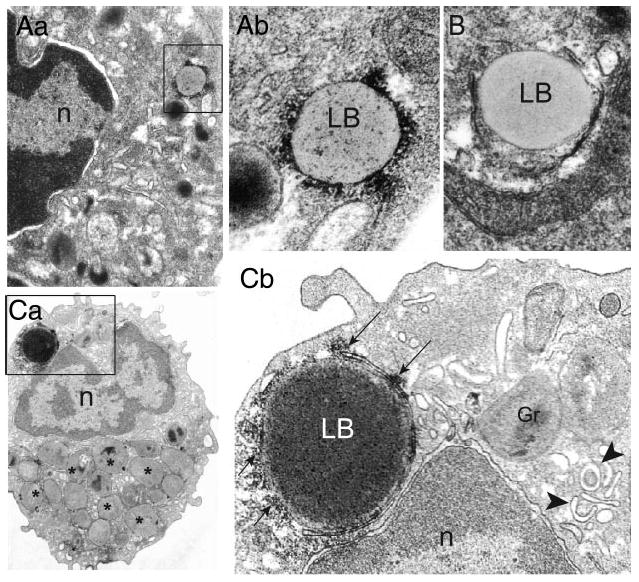
Lipid bodies (LBs) from human blood leukocytes are sites for ribosomes. Aa) In resting neutrophils, electron-lucent LBs (box) exhibited heavy ribosome clusters in both periphery and core. Ab) Higher magnification of the boxed area. B) An endoplasmic reticulum (ER) -encircled LB from the same sample does not show internal ribosomes. Ca) Eotaxin-stimulated eosinophil shows a large electron-dense LB (box) with clusters of attached ribosomes at its periphery (arrows in panel Cb). Note the presence of ribosomes in the LB core and the clear LB interaction with the surrounding rough ER. Panel Cb corresponds to the boxed area in panel Ca. Eosinophil specific granules with content losses (*) and eosinophil sombrero vesicles (EoSVs) (arrowheads), morphological concomitants of eosinophil activation (39), are indicated respectively in panels Ca and Cb. n, nucleus; gr, granule. Scale bar, 440 nm (Aa), 300 nm (Aa, B, Cb), 1 μM (Ca).
Internal structure of leukocyte lipid bodies
EM of U937 LBs at times demonstrated subtle variegated inhomogeneous densities within LB cores (e.g., Fig. 2D) and at times indicated membranous structures extending within LB cores (Fig. 5A). Such findings, while suggestive, did not clearly reveal internal membranes in LBs. In contrast, EM of LBs from eosinophils provided clear evidence of extensive networks of membranous structures within LBs in both resting (Fig. 5B) and agonist-stimulated eosinophils (Fig. 5C). The same complex membranous structures present within LBs (Fig. 5B, C) were also visualized outside of and adjacent to LBs (Fig. 5D, E, arrows). Typical ER cisternae were also revealed within LBs (Fig. 5Ba, Bb, arrowheads).
Figure 5.
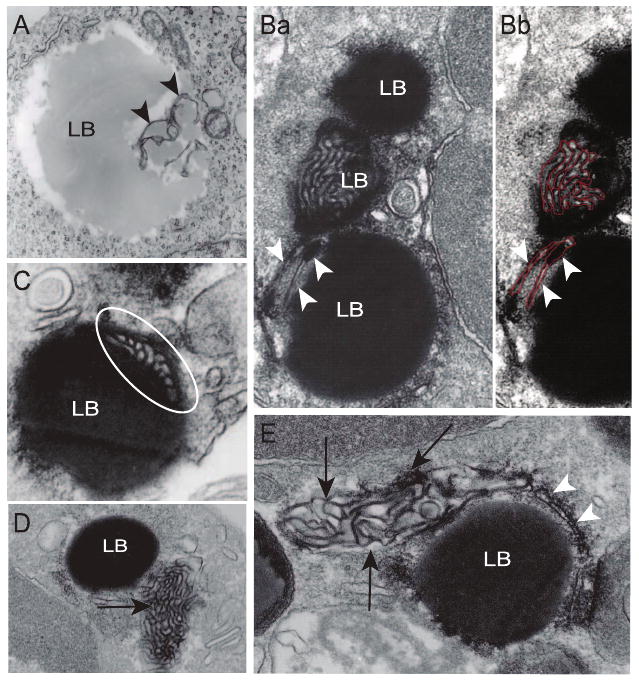
Internal structure of lipid bodies (LBs) in human U937 cells and blood leukocytes. A) A large and electron-lucent U937 LB shows internal membranous structures (arrowheads). Ba) An organized membranous network (highlighted in red in panel Bb) is clearly seen within an osmiophilic eosinophil LB. Note that typical ER cisternae (arrowheads) are seen as part of another LB. C) Part of an elaborate membranous structure (circle) is revealed within a LB (circle). D, E) The same membranous network (arrows) is observed in close apposition to other apparently homogenous LBs. Arrowheads in panel E indicate a classical ER cisternum wrapping around LB. Eosinophils were stimulated with RANTES (C, E) or eotaxin (D), or medium alone (B). Cells were processed for EM as described (39). Scale bars, 600 nm (A, C), 330 nm (B), 800 nm (D), 300 nm (E).
Discussion
LBs in diverse prokaryotic and eukaryotic cells are multifunctional organelles (1), although the origins and functions of LBs in leukocytes have not been fully considered in analyses of LBs (2). In leukocytes, LBs are organelles at which phospholipid-esterified substrate arachidonate, arachidonate-liberating phospholipases, and eicosanoid-synthesizing PGHSs and 5-LO are colocalized to provide for the focal formation of eicosanoids (20, 23, 42). In nonleukocytic cells, Chinese hamster ovary cells (10), squamous carcinoma cells (11), hepatoma cells (12, 13), and 3T3-L1 adipocytes (14), proteomic analyses of their LBs have revealed a diversity of LB-associated proteins. In the present study, we used human U937 monocytic cells for proteomic analyses of isolated LBs and combined these analyses with EM of both U937 cells and human blood leukocytes. Together, these investigations yielded insights into the protein compositions, organellar interrelationships, internal membranous structures, and ER origins of leukocyte LBs.
Proteomic analyses of U937 LBs, doubly purified by subcellular fractionation to minimize contamination, identified many proteins found with LBs as well as other proteins not previously associated with LBs. U937 LBs contained four well-recognized LB-associated proteins: adipophilin, TIP47 (mannose-6-phosphate receptor binding protein), CGI-49, and CGI-58. PAT family proteins, including adipophilin and TIP47, pervade LB cores in other cells (18), including macrophages (16); their functions in lipid metabolism in other cells have been reviewed (9). CGI-58 may be involved in fatty acid transfer from triglycerides to phospholipids (10), and its genetic defect underlies the Chanarin-Dorman syndrome characterized by accumulations of triglyceride-rich LBs in multiple cells, including leukocytes (43). CGI-49 is of uncertain function.
Other proteins common to U937 LBs and LBs of other cells were many related to lipid metabolism (Table 1), including the synthesis, storage, utilization, and degradation of cholesterol esters, triglycerides, and retinyl esters. Two cytoskeletal proteins, previously localized to LBs, actin and tubulin-α, may have pertinence to the cytoskeletal insertions into U937 cells demonstrable by EM (Fig. 2E, F) as well as the recognized capacity for neutrophil LBs to exhibit rapid intracytoplasmic movement and kiss and run interactions with phagosomes (32).
The findings of Ras-related Rab proteins in prior proteomic studies of LBs (10, 11, 13–15, 44) were recapitulated with the identification of Rabs 1A, 1B, 7, 10, 11, 14, and 18 in isolated U937 LBs (Table 1). Other proteins, likely involved in vesicular trafficking, found with U937 LBs included some not previously associated with LBs: VAT-1 (synaptic vesicle membrane protein) homologue, SNAP29 (vesicle membrane fusion protein), transmembrane traffic protein, GTP binding protein SAR 1a, and Rap-1a. Human VAT-1 homologue is a member of medium-chain dehydrogenases/reductases family (45). Protein p22, a calcium binding protein, may mediate constitutive membrane traffic (46), have roles in microtubule and ER organization (47), and interact with microtubules and glyceraldehyde-3-phosphate dehydrogenase (48), the latter also identified in U937 LBs (Table 1). AMID, an apoptosis-inducing factor/homologous mitochondrion-associated inducer of death, which lacks a mitochondrial localization signal, was previously localized to the cytoplasm or the outer mitochondrial membrane, and induces caspase-independent apoptosis (49), is of uncertain function in LBs. Transmembrane protein, an integral membrane protein often localized to the lumen of the ER, may be involved in vesicular trafficking (50). TD54 may be involved in vesicle trafficking; and in yeast, two-hybrid screening binds perilipin (51). The functional roles of these candidate vesicular transport-mediating proteins vis-à-vis LBs remain largely unknown, although overexpression of Rab 18 has promoted LB association with the ER (44).
ER-associated proteins identified in U937 LBs included oligosaccharyl transferase subunit, dehydrodolichyl diphosphate synthase, and dolichyl-phosphate-glucosyl transferase as well as ER-associated chaperones or folding proteins. The latter included BiP, an intraluminal ER protein, calnexin, a transmembrane ER protein, and protein disulfide isomerase; these have been localized to LBs in other cell types (Table 1). EM studies demonstrated close associations of ER with LBs in U937 cells (Fig. 2B–D). The points of LB contacts with ER cisternae have been noted to be ribosome-rich, as we found, and have been suggested to be named as an ER subdomain, “lipid droplet-associated membrane” (44). While close associations of ER with LBs might contribute in part to the ER proteins identified in the proteomic analyses, LBs are considered to be derived from the ER and perhaps to have continuing interactions with it (3, 17).
Of the proteins identified with U937 LBs, several may have specific relevance for leukocyte LBs. Two proteins may be related to their myeloid origin. HSPC028, a basic leucine zipper protein, was first sequenced from CD34+ hematopoietic progenitor cells (52), and its function is uncertain. Myeloid S100A9 (MRP14) can bind and transport arachidonate, and may shuttle unsaturated fatty acids to membranes (53, 54). With the activities of leukocyte LBs in arachidonate-derived eicosanoid formation, SA100A9 might participate in local arachidonate metabolism or delivering neutrophil LB-derived arachidonate to activate phagosomal NADPH oxidase (32, 55). Another protein potentially involved in arachidonate metabolism in LBs is triglyceride lipase. Neutrophils store arachidonate in triglyceride pools in LBs (25) and transfer arachidonate from triglycerides into phospholipids (56). Triglyceride lipase could release arachidonate from triglycerides to enter LB phospholipids, from which regulated activation of PLA2 would provide arachidonate for eicosanoid synthesis. Likewise, LB-associated CG-58 might also mediate fatty acid transfer from triglycerides to phospholipids (10).
Novel findings from proteomic analyses were the identification of several ribosomal subunit proteins as well as translation initiation factors in isolated LBs. Although care was taken to minimize contamination of isolated buoyant LBs, it is possible that some ribosomes were durably associated with LBs throughout the isolation. EM analyses of U937 cells, neutrophils, and eosinophils, however, were also informative. Free ribosomes or rough ER profiles were attached to the borders of LBs (Fig. 3A), and ribosomes or particles resembling ribosomal subunits were present within U937 (Fig. 3A, B) and neutrophil (Fig. 4Ab) LBs. In agonist-stimulated eosinophils, which contained large LBs, ribosomes were both attached to the periphery of LBs and distributed within LB cores (Fig. 4Cb). Thus, EM of U937 cells, neutrophils, and eosinophils demonstrated ribosomes present around and within LBs, in support of proteomic findings of ribosomal subunit proteins in isolated U937 LBs. Recently, proteomic analyses of Drosophila and yeast LBs have likewise identified ribosomal proteins (57, 58), and proteomic analyses of LBs from hepatitis C virus core protein expressing hepatoma cell have identified ribosomal and RNA-interacting (DEAD box) proteins (13).
That LBs in leukocytes may be sites of ribosomal function is consonant with prior ultrastructural studies of LBs in human mast cells: 1) 3H-uridine accumulated in these LBs; 2) RNA was localized within LBs by affinity-staining with an RNase-gold probe and by anti-ribosomal antibody (Ab) and antiuridine Ab immunogold labelings; 3) poly(A)mRNA was detected within LBs by in situ hybridization with a poly(U) probe; and 4) several human autoimmune sera to ribosomal component proteins immunolabeled LBs (59, 60). Moreover, our ongoing studies in a mast cell line transfected with a GFP-5-LO encoding plasmid are demonstrating translation and de novo synthesis of GFP-5-LO protein within LBs (unpublished results). Thus, ribosomal localization at and within LBs in leukocytes may be linked to compartmentalized protein synthesis at LBs.
Finally, a new finding from our ultrastructural studies was the identification of internal membranous structures within LBs, especially notable within eosinophils. Robenek et al., whose freeze fracture EM studies demonstrated concentric lamellae within LBs when conventional transmission EM revealed apparently homogenous lipid staining, have suggested that vagaries in chemical fixation and/or lipid solvents have prevented recognition of internal structures within LBs in most cells (17). Upon closer examination, EM images of LBs, however, such as those from U937 cells (Fig. 2D), often demonstrated inhomogeneities within the neutral lipid-rich cores of LBs. In eosinophils, our prior transmission EM at times revealed honeycomb patterns within eosinophil LBs (see Fig. 3 in ref. 22); and EM immunogold staining for 5-LO, an enzyme that associates with membranes for its activation and catalysis, was distributed throughout LBs (22).
One contemporary model of LB biogenesis has neutral lipids accumulating between the cytoplasmic and luminal leaflets of ER membranes, followed by the budding off of LBs surrounded by a phospholipid monolayer derived from the cytoplasmic leaflets of ER membranes (17). This model, which has not been corroborated by EM images of developing LBs, accords with the recognized phospholipid monolayer that surrounds LBs (4), but fails to account for a solely noncircumferential topology of membrane-associated proteins in LBs. More than half of the proteins identified in the U937 LB proteome were membrane-spanning proteins or contained predicted membrane insertion domains (Table 1). With this model, these proteins could insert only in the circumferential membrane of LBs. LTC4 synthase, localized fully within eosinophil LBs (22), is a transmembrane-spanning protein, like the luminal ER protein, calnexin, also commonly found in LBs (Table 1). The freeze fracture immunogold EM studies of Robenek et al. demonstrated that caveolin-1 was located on the luminal leaflets of ER membranes and distributed within LBs (17). The same group likewise localized TIP47 and adipophilin within LBs, but noted how TIP47 and adipophilin, as polar proteins, and membrane-associated caveolin-1 could partition within LB cores that were assumed to contained only membrane-devoid neutral lipids, has been an enigma (16, 17).
The freeze fracture EM studies that demonstrated lamellae, often in concentric circles, in LBs in smooth muscle and macrophages (16–18) would be compatible with internalized ER-derived membranes. Our findings that ribosomes, which are associated with cytoplasmic surfaces of rough ER membranes, and several ER luminal proteins were also present within LBs suggest a new model for LB formation (Fig. 6) by which LBs form by incorporating multiple loops of ER membranes (both cytoplasmic and luminal leaflets of membranes within the developing LB). Such a model would be in accord with the freeze fracture EM studies and our EM demonstrations of membranes within leukocyte LBs. Such a model by incorporating cytoplasmic membranes of the ER to which ribosomes are attached would provide a means for ribosomes to become incorporated with LBs. Moreover, by providing for the inclusion of luminal regions of the ER within LBs, this model would reconcile numerous heretofore unresolved observations that include how luminal ER proteins, such as caveolin-1 and calnexin, may be localized within LBs. Both PGHS-1 and -2 are integral membrane proteins and contain C-terminal KDEL-like sequences that target PGHS to the luminal surfaces of the ER and to nuclear envelope membranes, as confirmed by immunogold EM (61). In studies of LBs, immunogold EM has localized PGHSs throughout LBs in multiple leukocytes (27, 28). Loops or sheets of ER-derived membranes within LBs would also provide the means for membrane-associated or membrane-spanning proteins to be localized within LBs and not solely in the circumferential membrane monolayer. Accumulations of neutral lipids would develop among the membranes within LBs. Although such a model of LB organization might be germane principally to LBs in leukocytes, it is also likely that LBs in other cells share a common organization. As noted earlier, PGHSs have been localized to LBs in diverse nonleukocytic cells (30, 31). Both stanniocalcin and its membrane-associated receptor are present on LBs in ovarian steroidogenic cells and adipocytes (62), but how a membrane receptor could be associated with LBs had been uncertain. The presence of ER-derived membranes within LBs in steroidogenic cells and adipocytes would provide a means for receptors and other membrane-associated proteins to be localized within LBs.
Figure 6.
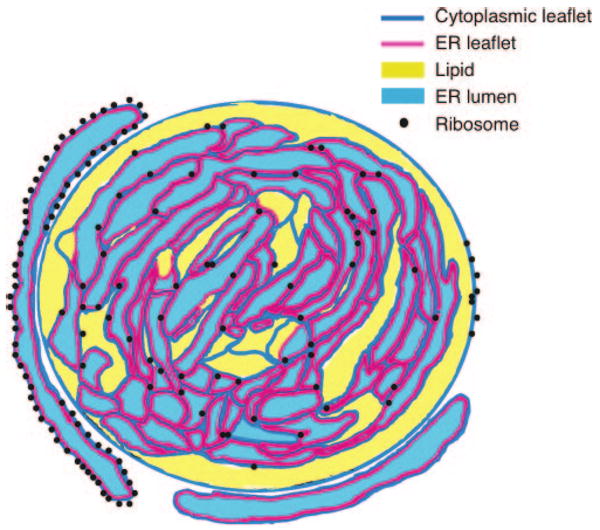
Model of the structure of lipid bodies. LBs contain enfolded ER membranes, accounting for the localization of luminal ER proteins within LBs and providing internal membranes into which membrane-associated proteins may insert. The circumferential border of LBs is the phospholipid monolayer of the cytoplasmic leaflet of the ER. Neutral lipids accumulate within LBs and may obscure their internal membranes. Ribosomes associated with the ER are at the periphery of LBs as well as within them.
In summary, the evolving understanding of LBs as organelles extends beyond their conventional roles in neutral lipid metabolism and their more recently indicated involvement in vesicular trafficking. Based on proteomic and ultrastructural studies of human leukocyte LBs, we propose a novel model for LB biogenesis and identify LBs as organelles containing ER-derived membranous domains and proteins as well as ribosomes, and with functional capabilities that may include local protein synthesis.
Acknowledgments
This work was supported by National Institutes of Health grants AI33372, AI20241, AI22571, and HL70270. R.C.N.M. was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil). We thank Dr. Steven Gygi and Ross Tomaino in the Taplin BioMassSpectrum Facility, Harvard Medical School for assisting in MS/LS analyses.
References
- 1.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 2.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 3.Martin S, Parton RG. Caveolin, cholesterol, and lipid bodies. Semin Cell Dev Biol. 2005;16:163–174. doi: 10.1016/j.semcdb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 5.Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- 6.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 7.Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- 8.Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3–12 coats nascent lipid droplets. J Biol Chem. 2003;278:37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 9.Londos C, Sztalryd C, Tansey JT, Kimmel AR. Role of PAT proteins in lipid metabolism. Biochimie (Paris) 2005;87:45–49. doi: 10.1016/j.biochi.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 11.Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. Association of stomatin with lipid bodies. J Biol Chem. 2004;279:23699–23709. doi: 10.1074/jbc.M310546200. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem (Tokyo) 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- 14.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 15.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets: Effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem. 2005;280:42405–42419. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 16.Robenek H, Lorkowski S, Schnoor M, Troyer D. Spatial integration of TIP47 and adipophilin in macrophage lipid bodies. J Biol Chem. 2005;280:5789–5794. doi: 10.1074/jbc.M407194200. [DOI] [PubMed] [Google Scholar]
- 17.Robenek MJ, Severs NJ, Schlattmann K, Plenz G, Zimmer KP, Troyer D, Robenek H. Lipids partition caveolin-1 from ER membranes into lipid droplets: updating the model of lipid droplet biogenesis. FASEB J. 2004;18:866–868. doi: 10.1096/fj.03-0782fje. [DOI] [PubMed] [Google Scholar]
- 18.Robenek H, Robenek MJ, Troyer D. PAT family proteins pervade lipid droplet cores. J Lipid Res. 2005;46:1331–1338. doi: 10.1194/jlr.M400323-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Pacheco P, Bozza FA, Gomes RN, Bozza M, Weller PF, Castro-Faria-Neto HC, Bozza PT. Lipopolysaccharide-induced leukocyte lipid body formation in vivo: innate immunity elicited intracellular loci involved in eicosanoid metabolism. J Immunol. 2002;169:6498–6506. doi: 10.4049/jimmunol.169.11.6498. [DOI] [PubMed] [Google Scholar]
- 20.D'Avila H, Melo RCN, Parreira GG, Werneck-Barroso E, Castro-Faria-Neto HC, Bozza PT. Mycobacterium bovis Bacillus Calmette-Guerin induces TLR2-mediated formation of lipid bodies: Intracellular domains for eicosanoid synthesis in vivo. J Immunol. 2006;176:3087–3097. doi: 10.4049/jimmunol.176.5.3087. [DOI] [PubMed] [Google Scholar]
- 21.Bozza PT, Payne JL, Goulet JL, Weller PF. Mechanisms of platelet-activating factor-induced lipid body formation: requisite roles for 5-lipoxygenase and de novo protein synthesis in the compartmentalization of neutrophil lipids. J Exp Med. 1996;183:1515–1525. doi: 10.1084/jem.183.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bozza PT, Yu W, Penrose JF, Morgan ES, Dvorak AM, Weller PF. Eosinophil lipid bodies: specific, inducible intracellular sites for enhanced eicosanoid formation. J Exp Med. 1997;186:909–920. doi: 10.1084/jem.186.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandeira-Melo C, Phoofolo M, Weller PF. Extranuclear lipid bodies, elicited by CCR3-mediated signaling pathways, are the sites of chemokine-enhanced leukotriene C4 production in eosinophils and basophils. J Biol Chem. 2001;276:22779–22787. doi: 10.1074/jbc.M101436200. [DOI] [PubMed] [Google Scholar]
- 24.Weller PF, Monahan-Earley RA, Dvorak HF, Dvorak AM. Cytoplasmic lipid bodies of human eosinophils. Subcellular isolation and analysis of arachidonate incorporation. Am J Pathol. 1991;138:141–148. [PMC free article] [PubMed] [Google Scholar]
- 25.Triggiani M, Oriente A, Seeds MC, Bass DA, Marone G, Chilton FH. Migration of human inflammatory cells into the lung results in the remodeling of arachidonic acid into a triglyceride pool. J Exp Med. 1995;182:1181–1190. doi: 10.1084/jem.182.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W, Bozza PT, Tzizik DM, Gray JP, Cassara J, Dvorak AM, Weller PF. Co-compartmentalization of MAP kinases and cytosolic phospholipase A2 at cytoplasmic arachidonate-rich lipid bodies. Am J Pathol. 1998;152:759–769. [PMC free article] [PubMed] [Google Scholar]
- 27.Dvorak AM, Morgan E, Schleimer RP, Ryeom SW, Lichtenstein LM, Weller PF. Ultrastructural immunogold localization of prostaglandin endoperoxide synthase (cyclooxygenase) to non-membrane-bound cytoplasmic lipid bodies in human lung mast cells, alveolar macrophages, type II pneumocytes, and neutrophils. J Histochem Cytochem. 1992;40:759–769. doi: 10.1177/40.6.1316915. [DOI] [PubMed] [Google Scholar]
- 28.Dvorak AM, Morgan ES, Tzizik DM, Weller PF. Prostaglandin endoperoxide synthase (cyclooxygenase): ultrastructural localization to nonmembrane-bound cytoplasmic lipid bodies in human eosinophils and 3T3 fibroblasts. Int Arch Allergy Immunol. 1994;105:245–250. doi: 10.1159/000236764. [DOI] [PubMed] [Google Scholar]
- 29.Bozza PT, Payne JL, Morham SG, Langenbach R, Smithies O, Weller PF. Leukocyte lipid body formation and eicosanoid generation: cyclooxygenase-independent inhibition by aspirin. Proc Natl Acad Sci U S A. 1996;93:11091–11096. doi: 10.1073/pnas.93.20.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meadows JW, Pitzer B, Brockman DE, Myatt L. Expression and localizatio of adipophilin and perilipin in human fetal membranes: association with lipid bodies and enzymes involved in prostaglandin synthesis. J Clin Endocrinol Metab. 2005;90:2344–2350. doi: 10.1210/jc.2004-1199. [DOI] [PubMed] [Google Scholar]
- 31.Arend A, Masso R, Masso M, Selstam G. Electron microscope immunocytochemical localization of cyclooxygenase-1 and -2 in pseudopregnant rat corpus luteum during luteolysis. Prostaglandins Other Lipid Mediat. 2004;74:1–10. doi: 10.1016/j.prostaglandins.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Van Manen HJ, Kraan YM, Roos D, Otto C. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proc Natl Acad Sci U S A. 2005;102:10159–10164. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beil WJ, Weller PF, Peppercorn MA, Galli SJ, Dvorak AM. Ultrastructural immunogold localization of subcellular sites of TNF-alpha in colonic Crohn's disease. J Leukoc Biol. 1995;58:284–298. doi: 10.1002/jlb.58.3.284. [DOI] [PubMed] [Google Scholar]
- 34.Dvorak AM, Morgan ES, Weller PF. Ultrastructural immunolocalization of basic fibroblast growth factor to lipid bodies and secretory granules in human mast cells. Histochem J. 2001;33:397–402. doi: 10.1023/a:1013771827069. [DOI] [PubMed] [Google Scholar]
- 35.Yu W, Cassara J, Weller PF. Phosphatidylinositide 3-kinase localizes to cytoplasmic lipid bodies in human polymorphonuclear leukocytes and other myeloid-derived cells. Blood. 2000;95:1078–1085. [PubMed] [Google Scholar]
- 36.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 37.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 38.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 39.Melo RCN, Perez SAC, Spencer LA, Dvorak AM, Weller PF. Intragranular vesiculotubular compartments are involved in piecemeal degranulation by activated human eosinophils. Traffic. 2005;6:866–879. doi: 10.1111/j.1600-0854.2005.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weller PF, Ackerman SJ, Nicholson-Weller A, Dvorak AM. Cytoplasmic lipid bodies of human neutrophilic leukocytes. Am J Pathol. 1989;135:947–959. [PMC free article] [PubMed] [Google Scholar]
- 41.Melo RCN, D'Avila H, Fabrino DL, Almeida PE, Bozza PT. Macrophage lipid body induction by Chagas disease in vivo: putative intracellular domains for eicosanoid formation during infection. Tissue Cell. 2003;35:59–67. doi: 10.1016/s0040-8166(02)00105-2. [DOI] [PubMed] [Google Scholar]
- 42.Bandeira-Melo C, Bozza PT, Weller PF. The cellular biology of eosinophil eicosanoid formation and function. J Allergy Clin Immunol. 2002;109:393–400. doi: 10.1067/mai.2002.121529. [DOI] [PubMed] [Google Scholar]
- 43.Lefevre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, Lakhdar H, Wollenberg A, Verret JL, Weissenbach J, et al. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet. 2001;69:1002–1012. doi: 10.1086/324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118:2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 45.Hayess K, Kraft R, Sachsinger J, Janke J, Beckmann G, Rohde K, Jandrig B, Benndorf R. Mammalian protein homologous to VAT-1 of Torpedo californica: isolation from Ehrlich ascites tumor cells, biochemical characterization, and organization of its gene. J Cell Biochem. 1998;69:304–315. [PubMed] [Google Scholar]
- 46.Barroso MR, Bernd KK, DeWitt ND, Chang A, Mills K, Sztul ES. A novel Ca2+-binding protein, p22, is required for constitutive membrane traffic. J Biol Chem. 1996;271:10183–10187. doi: 10.1074/jbc.271.17.10183. [DOI] [PubMed] [Google Scholar]
- 47.Andrade J, Zhao H, Titus B, Timm Pearce S, Barroso M. The EF-hand Ca2+-binding protein p22 plays a role in microtubule and endoplasmic reticulum organization and dynamics with distinct Ca2+-binding requirements. Mol Biol Cell. 2004;15:481–496. doi: 10.1091/mbc.E03-07-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrade J, Pearce ST, Zhao H, Barroso M. Interactions among p22, glyceraldehyde-3-phosphate dehydrogenase and microtubules. Biochem J. 2004;384:327–336. doi: 10.1042/BJ20040622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall KR, Gong M, Wodke L, Lamb JH, Jones DJ, Farmer PB, Scrutton NS, Munro AW. The human apoptosis-inducing protein AMID is an oxidoreductase with a modified flavin cofactor and DNA binding activity. J Biol Chem. 2005;280:30735–30740. doi: 10.1074/jbc.M414018200. [DOI] [PubMed] [Google Scholar]
- 50.Blum R, Feick P, Puype M, Vandekerckhove J, Klengel R, Nastainczyk W, Schulz I. Tmp21 and p24A, two type I proteins enriched in pancreatic microsomal membranes, are members of a protein family involved in vesicular trafficking. J Biol Chem. 1996;271:17183–17189. doi: 10.1074/jbc.271.29.17183. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi T, Omatsu N, Omukae A, Osumi T. Analysis of interaction partners for perilipin and ADRP on lipid droplets. Mol Cell Biochem. 2006;284:167–173. doi: 10.1007/s11010-005-9045-y. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Q, Ye M, Wu X, Ren S, Zhao M, Zhao C, Fu G, Shen Y, Fan H, Lu G, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ haematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–1560. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roulin K, Hagens G, Hotz R, Saurat JH, Veerkamp JH, Siegenthaler G. The fatty acid-binding heterocomplex FA-p34 formed by S100A8 and S100A9 is the major fatty acid carrier in neutrophils and translocates from the cytosol to the membrane upon stimulation. Exp Cell Res. 1999;247:410–421. doi: 10.1006/excr.1998.4382. [DOI] [PubMed] [Google Scholar]
- 54.Kerkhoff C, Klempt M, Kaever V, Sorg C. The two calcium-binding proteins, S100A8 and S100A9, are involved in the metabolism of arachidonic acid in human neutrophils. J Biol Chem. 1999;274:32672–32679. doi: 10.1074/jbc.274.46.32672. [DOI] [PubMed] [Google Scholar]
- 55.Berthier S, Paclet MH, Lerouge S, Roux F, Vergnaud S, Coleman AW, Morel F. Changing the conformation state of cytochrome b558 initiates NADPH oxidase activation: MRP8/MRP14 regulation. J Biol Chem. 2003;278:25499–25508. doi: 10.1074/jbc.M209755200. [DOI] [PubMed] [Google Scholar]
- 56.Chilton FH, Murphy RC. Remodeling of arachidonate-containing phosphoglycerides within the human neutrophil. J Biol Chem. 1986;261:7771–7777. [PubMed] [Google Scholar]
- 57.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dvorak AM, Morgan ES, Weller PF. RNA is closely associated with human mast cell lipid bodies. Histol Histopathol. 2003;18:943–968. doi: 10.14670/HH-18.943. [DOI] [PubMed] [Google Scholar]
- 60.Dvorak AM. Mast cell secretory granules and lipid bodies contain the necessary machinery important for the in situ synthesis of proteins. Chem Immunol Allergy. 2005;85:252–315. doi: 10.1159/000086520. [DOI] [PubMed] [Google Scholar]
- 61.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 62.Paciga M, McCudden CR, Londos C, DiMattia GE, Wagner GF. Targeting of big stanniocalcin and its receptor to lipid storage droplets of ovarian steroidogenic cells. J Biol Chem. 2003;278:49549–49554. doi: 10.1074/jbc.M307302200. [DOI] [PubMed] [Google Scholar]
- 63.Wolins NE, Rubin B, Brasaemle DL. TIP47 associates with lipid droplets. J Biol Chem. 2001;276:5101–5108. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- 64.Than NG, Sumegi B, Bellyei S, Berki T, Szekeres G, Janaky T, Szigeti A, Bohn H, Than GN. Lipid droplet and milk lipid globule membrane associated placental protein 17b (PP17b) is involved in apoptotic and differentiation processes of human epithelial cervical carcinoma cells. Eur J Biochem. 2003;270:1176–1188. doi: 10.1046/j.1432-1033.2003.03475.x. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi T, Omatsu N, Matsushita S, Osumi T. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J Biol Chem. 2004;279:30490–30497. doi: 10.1074/jbc.M403920200. [DOI] [PubMed] [Google Scholar]
- 66.Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3–L1 adipocytes. J Biol Chem. 2004;279:42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 67.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 68.Smirnova E, Goldberg EB, Makarova KS, Lin L, Brown WJ, Jackson CL. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2006;7:106–113. doi: 10.1038/sj.embor.7400559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohashi M, Mizushima N, Kabeya Y, Yoshimori T. Localization of mammalian NAD(P)H steroid dehydrogenase-like protein on lipid droplets. J Biol Chem. 2003;278:36819–36829. doi: 10.1074/jbc.M301408200. [DOI] [PubMed] [Google Scholar]
- 70.Caldas H, Herman GE. NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum Mol Genet. 2003;12:2981–2991. doi: 10.1093/hmg/ddg321. [DOI] [PubMed] [Google Scholar]
- 71.Fong TH, Wu CH, Liao EW, Chang CY, Pai MH, Chiou RJ, Lee AW. Association of globular beta-actin with intracellular lipid droplets in rat adrenocortical cells and adipocytes. Biochem Biophys Res Commun. 2001;289:1168–1174. doi: 10.1006/bbrc.2001.6080. [DOI] [PubMed] [Google Scholar]
- 72.Wu CC, Howell KE, Neville MC, Yates JR, 3rd, McManaman JL. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21:3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 73.Prattes S, Horl G, Hammer A, Blaschitz A, Graier WF, Sattler W, Zechner R, Steyrer E. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J Cell Sci. 2000;113:2977–2989. doi: 10.1242/jcs.113.17.2977. [DOI] [PubMed] [Google Scholar]


