Abstract
Click chemistry provides extremely selective and orthogonal reactions that proceed with high efficiency and under a variety of mild conditions, the most common example being the copper(I)-catalyzed reaction of azides with alkynes1,2. While the versatility of click reactions has been broadly exploited3–5, a major limitation is the intrinsic toxicity of the synthetic schemes and the inability to translate these approaches to biological applications. This manuscript introduces a robust synthetic strategy where macromolecular precursors react via a copper-free click chemistry6, allowing for the direct encapsulation of cells within click hydrogels for the first time. Subsequently, an orthogonal thiol-ene photocoupling chemistry is introduced that enables patterning of biological functionalities within the gel in real-time and with micron-scale resolution. This material system allows one to tailor independently the biophysical and biochemical properties of the cell culture microenvironments in situ. This synthetic approach uniquely allows for the direct fabrication of biologically functionalized gels with ideal structures that can be photopatterned and all in the presence of cells.
An emerging paradigm in organic synthesis is a focus on highly selective and orthogonal reactions that proceed with high efficiency and under a variety of mild conditions. A growing number of these reactions are grouped under the term “click chemistry,” which have been used to produce a catalog of functional synthetic molecules and subsequent materials1,4. Characteristics of modular click reactions include: (a) high yields with fast kinetics, (b) regiospecificity and stereospecificity, (c) insensitivity to oxygen or water, and (d) mild, solventless reaction conditions or in water.
While the versatility of click reactions has been broadly exploited in many fields including drug discovery7,8, material science9–11, and bioconjugation3,12,13, a major limitation is the intrinsic toxicity of the synthetic schemes and the inability to translate these approaches to biological applications. Though the 1,3-dipolar Huisgen cycloaddition between azides and alkynes2 is often seen as the quintessential click reaction, the crucial copper catalyst precludes its usage with biological systems14,15. This drawback, however, was recently circumvented via the development of novel cyclooctyne moieties whose ring strain and electron-withdrawing fluorine substituents give rise to an activated alkyne. This molecule has been shown to react quickly with azides in the absence of a metal catalyst, enabling the usage of traditional click chemistry in living systems6,16. Specifically, azide-labeled cell-surface glycans were reacted with fluorescently-labeled cyclooctynes in vivo to enable the visualization of dynamic subcellular development within zebrafish embryos17. Though this chemistry has been exploited in the labeling of biomolecules, it has not yet been utilized for biomaterial formation.
More recently, the radical-mediated addition of a thiol to an alkene known as the thiol-ene reaction has gained attention as an emerging click reaction18. In addition to being bio-orthogonal and biocompatible, the reaction is advantageous in that it is readily initiated with light, ultimately affording spatial and temporal control over where the reaction occurs.19 This reaction has been utilized to create 2D surface gradients of biomolecules20 as well complex materials21.
In alignment with the evolution of click chemistry, the combined utilization of multiple orthogonal reactions presents the opportunity to fabricate multifunctional and tunable materials without sacrificing synthetic simplicity or efficiency. While materials with highly defined structures have applications in microelectronics, membrane technology, and fuel cells, one increasingly important area of research is in developing biomaterial platforms that allow researchers to culture and study cells in 3D22. Though initial material development has proven successful at permitting cell growth, a growing topic of interest is the development of bioactive materials that promote and detect specific cell function via spatially-presented biochemical and biomechanical cues23. Ultimately, a platform offering such versatility would be of particular note to those interested in well-defined niches for 3D cell culture, understanding the role of biomechanical versus biochemical signals on cell function, as well as regenerating tissue structures24. Appropriately developed click chemistry can provide this versatility, enabling the fabrication of increasingly complex 3D culture constructs using just a few simple reactions.
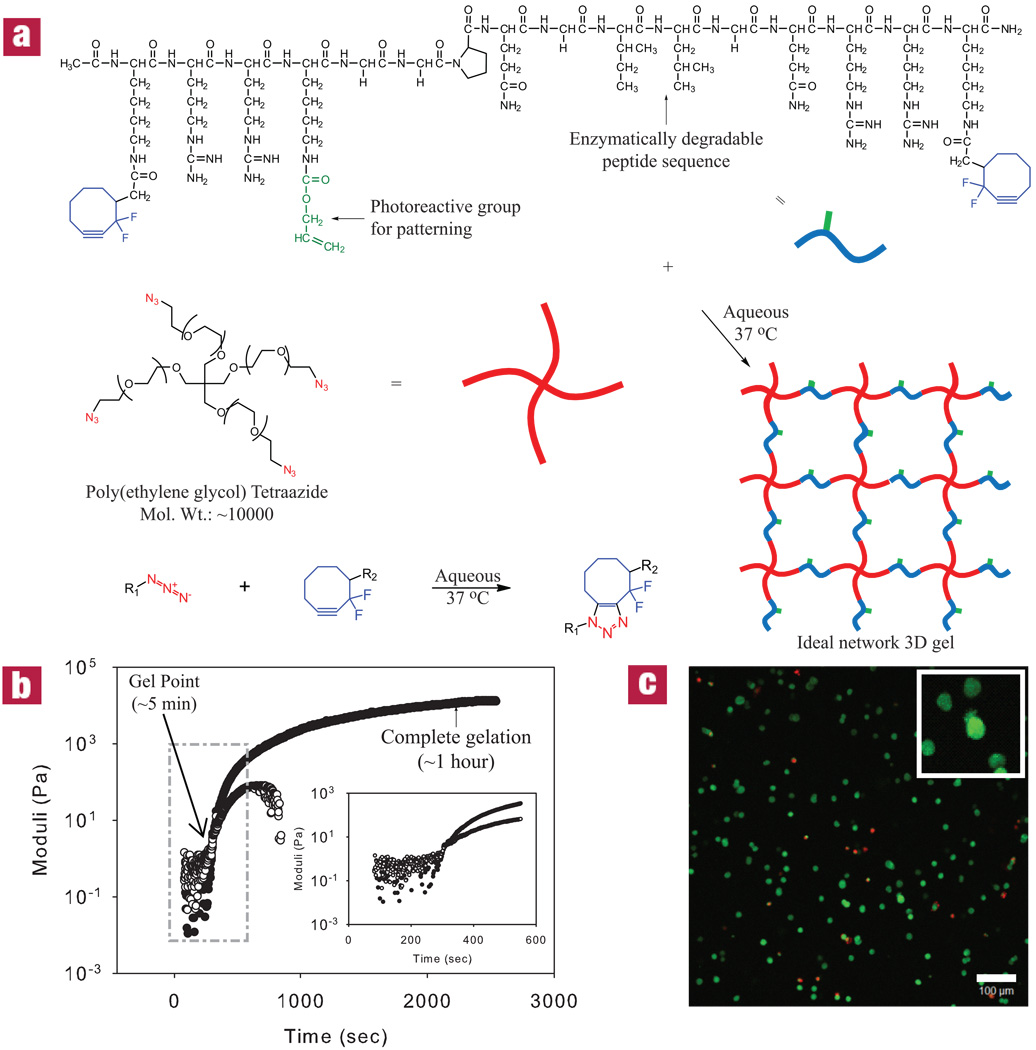
Here, a hydrogel platform is introduced that utilizes two orthogonal click chemistries; one for hydrogel formation and another for biochemical patterning within the preformed material. The modular aspect of these reactions allows for independent control of the network structure and chemistry, and facile incorporation of biological epitopes. Network formation is accomplished using a recently developed Cu-free variant to the traditional click reaction, the Huisgen cycloaddition, between an azide (-N3) and an alkyne (-C≡C-) to form a triazole6. This method employs a di-fluorinated cyclooctyne moiety (DIFO3), whose ring strain and electron-withdrawing fluorine substituents promote the [3+2] dipolar cycloaddition with azides without the use of a catalyst25 (Fig. 1a). This reaction has been carried out under physiological conditions in the presence of living cells with no reported toxicity17. Beyond this bioconjugation approach for cell labeling, multifunctional macromolecular monomers were synthesized to create ideal network structures with minimal defects and local heterogeneties. Specifically, multifunctional azides and activated alkynes were reacted in a one-to-one fashion to yield end-linked polymer gels, under reaction conditions that enable cell encapsulation and result in gels with initially uniform material properties.
Figure 1. Cytocompatible click hydrogel formation reaction and kinetics.
(a) Click-functionalized macromolecular precursors undergo the [3+2] Huisgen cycloaddition to form a 3D ideal network hydrogel via a step-growth polymerization mechanism. (b) Rheology can be used to monitor dynamic network formation and indicates gelation within minutes and complete reaction occurring in less than one hour at 37 °C for a 13.5 wt% monomer solution. G’ is shown as closed circles, while G’’ are open circles. (c) A Live/Dead stain at 24 hours of 3T3s encapsulated within this material indicates a predominantly viable population (live cells are shown in green, while dead cells are red). Image represents a 200 µm confocal projection. Scale bar = 100 µm.
A four-arm poly(ethylene glycol) (PEG) tetraazide was reacted with bis(DIFO3) di-functionalized polypeptide in an aqueous environment at 37 °C (schematic shown in Fig. 1a). The choice of PEG allows one to tailor readily the biophysical properties of the gel, while eliminating non-specific interactions that often result when proteins adsorb to materials. Biological functionality can be readily introduced into the hydrogel backbone by the choice of the crosslinking peptide sequence. Here, a matrix metalloproteinase (MMP)-cleavable (GPQG↓ILGQ) is selected, so that cells can actively remodel their surroundings via secreted enzymes26. Cells encapsulated in hydrogels containing an enzymatically-degradable crosslinker sequence spread and migrate throughout the material with dramatically increased viability compared with non-degradable alternatives27,28.
Hydrogels were formed using a 13.5 wt% total macromer solution containing a 1:1 ratio of alkyne to azide functionalities. Ultimately, this gel composition affords a high water content, elasticity similar to many tissue matrices, and the ability to image cells in 3D. Dynamic time sweep rheological experiments were conducted to monitor network evolution during this step polymerization (Fig. 1b). The crossover point, an estimate of gelation at which the elastic modulus (G’) is equal to the storage modulus (G’’), occurs in less than 5 min (290 ± 30 sec). Furthermore, the data indicates a final G’ value of 12.0 ± 0.6 kPa at t ~ 1 hour, signifying a structurally robust network that maintains its 3D shape with loading. The step-growth mechanism was confirmed by the statistical gelation model for step-growth networks developed by Flory and Stockmayer (Supplementary Fig. S1). In addition, dynamic magic-angle spinning (MAS) 1H NMR was carried out to further examine the reaction kinetics of network formation (Supplementary Fig. S2). Under normal solution-phase NMR conditions, the NMR spectral lines would quickly become extremely broad, yielding useless spectra as the polymer network begins to form due to dipolar relaxation in the motionally restricted (semi-solid) phase being formed. With the sample oriented at the magic-angle (ca. 54.736 degrees), rotating at a frequency that exceeds the static dipolar linewidth, this dipolar line-broadening can be eliminated, yielding high-resolution 1H NMR spectra throughout the polymerization reaction. Characteristic peaks associated with the alkyne DIFO3 functionality were found to completely disappear upon reaction with azides within 1 hour with a second-order rate constant of 8.9 × 10−5 M−1 s−1. Both the rheological and the MAS NMR data suggest that the formed hydrogel is nearly ideal, agreeing with previous work with click-based networks10. Ultimately, the time scale and mechanism of this reaction are such that it permits cell encapsulation with high viability comparable to traditional hydrogel systems (>90% at 24 hours post encapsulation, Fig. 1c & Supplementary Fig. S6).
Post network formation, a second click reaction allows facile modification of the cell’s niche through the conjugation of biomolecules at specific locations with the gel. Specifically, by including a photoreactive allyl ester within the crosslinking peptide sequence via the commercially available Fmoc-Lys(alloc)-OH amino acid (Fig. 1a), relevant biochemical cues can be covalently incorporated within the hydrogel using the bio-orthogonal thiol-ene coupling reaction. Originally designed as an orthogonal protecting group for lysine29, allyloxycarbonyl (alloc) contains a vinyl group capable of undergoing a thiol-ene photocoupling reaction with any thiol-containing compound, including cysteine11,30. The alloc protecting group is stable to Fmoc deprotection and peptide trifluoroacetic acid cleavage from resin, rendering it a suitable and versatile choice as the photoreactive component of our hydrogel crosslinker. Additionally, the electron-rich alloc allyl ester is not susceptible to Michael-type addition, eliminating the possibility for non-specific chemical immobilization31.
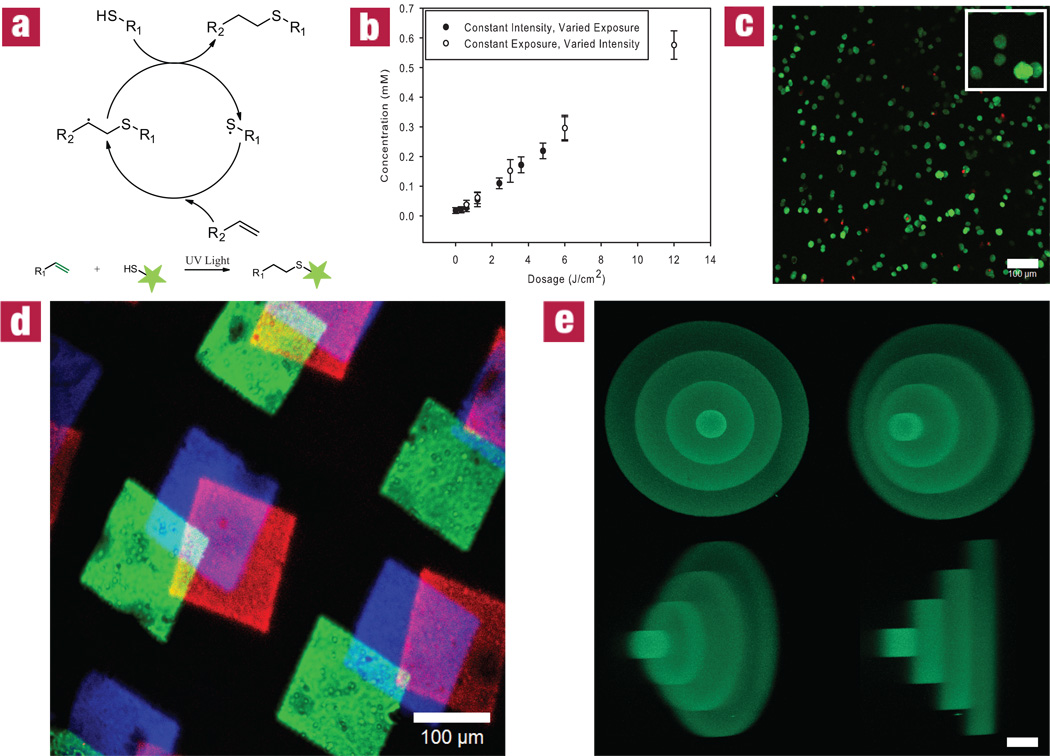
The thiol-ene reaction is a radical-mediated addition of a thiol to an alkene, involving the catalytic propagation of a thiyl radical through a vinyl functional group and the chain transfer from the resulting carbon radical to a thiol32 (Fig. 2a). Thiols can be deprotonated to thiyl radicals using photolytically-cleaved, hydrogen-abstracting initiator systems. Selectively exposing specific locations within the material to light affords spatial and temporal control of where this photocoupling reaction occurs in real time. The extent of patterning can ultimately be controlled by regulating the light intensity and exposure time (Fig. 2b) and utilizes cytocompatible wavelengths (365 nm) and intensities (~10 mW/cm2). Light exposure can be controlled using conventional photolithographic, single-photon, and multi-photon techniques, each affording a higher degree of reaction specificity than the last. This thiol-ene reaction is compatible with cells, as indicated by the high viability maintained throughout patterning (>90% at 24 hours post encapsulation, Fig. 2c & Supplementary Fig. S6). 2D patterns were transferred throughout the z-axis of a gel using stereolithography (Fig. 2d & Supplementary Fig. S8). We demonstrate that the reaction scheme is fully additive by incorporating three different peptides at varied positions within the gel (Fig. 2d & Supplementary Fig. S10). More complex 3D structures of arbitrary size and shape can be patterned within the gel by systematically scanning the focal point of a pulsed near IR laser where functionalization is desired. The latter technique affords micron-scale pattern resolution, as illustrated in Fig. 2e & Supplementary Fig. S9, and is performed in time scales similar to that required for 3D confocal imaging.
Figure 2. Cytocompatible, biochemical patterning within preformed click hydrogels.
(a) The thiol-ene reaction mechanism provides a means to quantitatively couple sulfhydryls (-SH) with vinyl functionalities (-C=C) in the presence of light. (b) Upon swelling into the material, relevant thiol-containing biomolecules are covalently affixed to the hydrogel network at varying concentrations by altering the dosage of exposed light (intensity and exposure time). (c) A Live/Dead stain at 24 hours after photolithographic patterning of 3T3s indicates a predominantly viable population (live cells are shown in green, while dead cells are red) and that the patterning process is cytocompatible. (d) The thiol-ene reaction is confined to user-defined regions in space using photomasks to introduce three different fluorescently-labeled peptide sequences within the gel, a process that can be repeated at desired times and spatial locations to introduce additional biochemical cues. (e) By controlling the focal point of the laser light in three-dimensions using a confocal microscope, micron-scale spatial patterning resolution is achieved. Values in (b) are reported as mean ± SD (n=5). The image in (c) represents a 200 µm confocal projection. The images in (d) and (e) represent confocal micrographs of fluorescently-tagged peptides patterned within the networks. Scale bar = 100 µm for (c), 100 µm for (d), and 50 µm for (e).
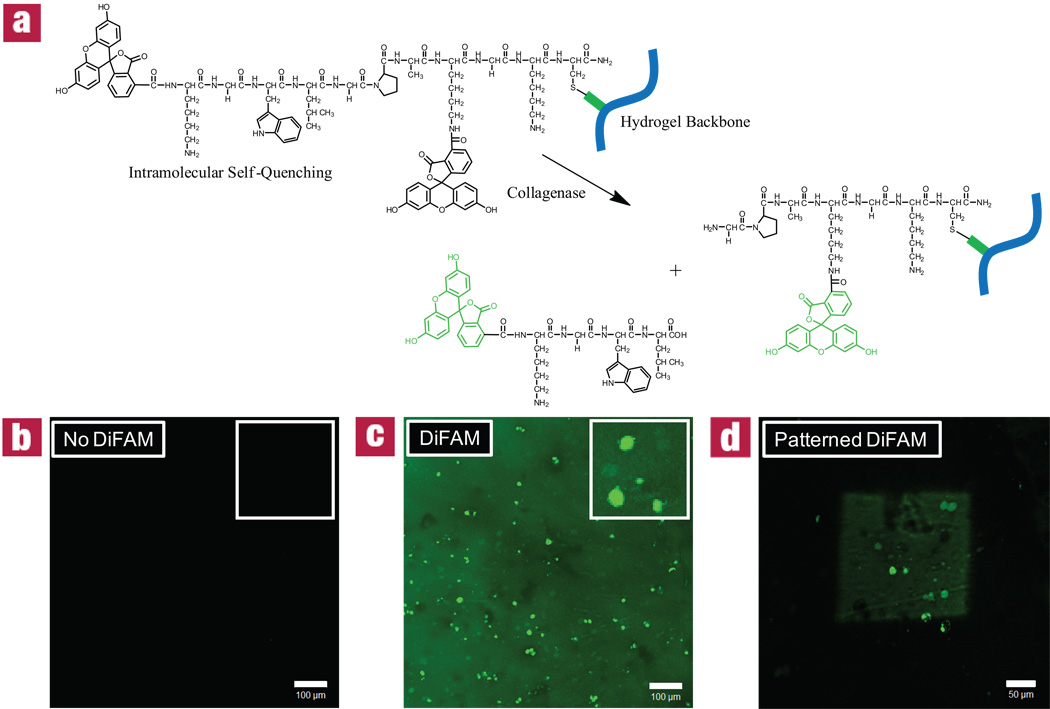
The thiol-ene reaction allows any thiol-containing compound to be pendantly attached at user-defined locations within the hydrogel. While adding thiol functionality to small molecules is fairly straightforward, cysteine-containing peptides require no additional synthetic modifications to be patterned within a gel. To illustrate the versatility that is afforded by this type of patterning scheme, a detection molecule was covalently incorporated as a pendant functionality that increases its fluorescence when exposed to cellular protease activity within the network. Specifically, a di-fluorescein collagenase-sensitive peptide sequence (DiFAM) was selectively patterned into the gels. This peptide, FAM-KGWL↓GPAK(FAM)GKC-NH2, exhibits intramolecular self quenching until it is enzymatically cleaved (Fig. 3a). While the gel fluoresces slightly where the quenched molecule has been patterned, the probe is found to fluoresce with much higher intensity in regions of collagenase activity immediately surrounding the cells (Fig. 3b–d). This DiFAM probe serves as a proof of concept that these materials are able to report real-time information concerning local encapsulated cell behavior, and that these detection assays can be confined to user-defined regions within the gel.
Figure 3. Visualizing 3T3 collagenase activity via patterned detection peptide within 3D click hydrogels.
3T3s were encapsulated into hydrogel networks at 3 × 106 cells/mL. After 24 hours, a di-fluorescein collagenase-sensitive peptide sequence (DiFAM) which exhibits intramolecular self-quenching until enzymatically cleaved (a) was swollen into networks at 0.5 mgs/mL and exposed to 365 nm collimated light at 10 mW/cm2 for 10 minutes through a variety of photomasks: (a) full mask; (b) no mask; (c) full mask with 200 µm square opening. Here, patterned regions gently fluoresce while areas of high collagenase activity (near cell surface) fluoresce with greater intensity. Images represent 200 µm confocal projections at 3 days. Scale bars = 100 µm for (b) and (c) and 50 µm for (d).
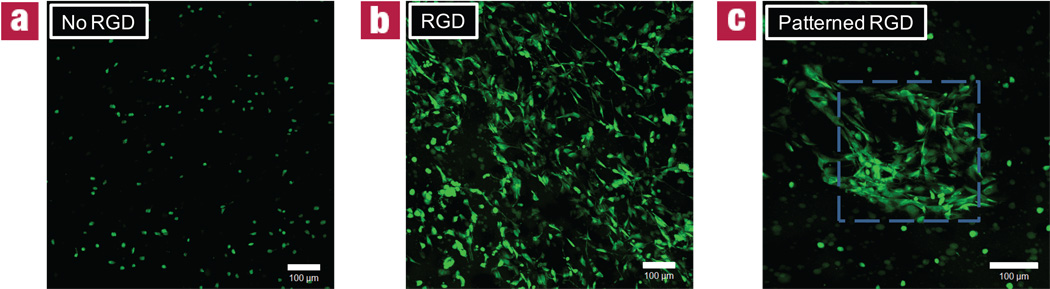
Just as this system allows for the patterning of reporter probes, biochemical functionalities that direct cell behavior can be incorporated within these materials in a location-specific manner. Incorporation of the RGD sequence, a fibronectin motif, has been extensively used to promote cell adhesion uniformly throughout biomaterials33,34, as well as in patterned channels35–37. Here, a cysteine-containing, fluorescently-labeled RGD sequence, AF488-AhxRGDSC-NH2, was selectively affixed within a cell-impregnated hydrogel. The fibronectin motif induces localized morphological and migratory changes within the patterned regions (Fig. 4). Where the RGD is present, cells are able to attach to and locally degrade the surrounding network, giving rise to a spread morphology. However, when this functionality is absent, cells maintain a rounded morphology. Fig. 4c illustrates that these induced differences in cell behavior can be selectively confined to patterned regions within a single gel.
Figure 4. Effect of patterned RGD on 3T3 population within 3D click hydrogels.
3T3s were encapsulated into hydrogel networks at 3 × 106 cells/mL. After 24 hours, thiol-functionalized RGD, a fibronectin motif known to promote cell attachment, was swollen into networks at 3 mgs/mL and exposed to 365 nm collimated light at 10 mW/cm2 for 10 minutes through a variety of photomasks: (a) full mask; (b) no mask; (c) full mask with 250 µm square opening (illustrated by the dashed lines). 3T3s were stained at day 10 with CellTracker orange and imaged using confocal microscopy. Here, cells only adopt a spread morphology in user-defined regions of RGD. Images represent 200 µm confocal projections. Scale bars = 100 µm.
As presented, this work utilizes two novel bio-orthogonal click chemistry schemes to combine and exploit features of previously mutually exclusive technologies. Namely, the enzymatically-degradable hydrogel platform provides an ideal network into which biomacromolecules can be photopatterned that detect, as well as promote, specific cellular functions. The material chemistry affords a simplified synthetic microenvironment that captures critical aspects of extracellular matrices, allowing for the direct observation of cellular processes in 3D, including migration, proliferation, and morphological changes. The ability to then spatially tune the material properties provides an additional tool to manipulate cell function. Since reactive monomer components can be easily exchanged, the material is readily tailorable with multiple functionalities for 3D cell studies.
Methods
Synthesis of Click-Functionalized Macromolecular Precursors
Synthesis of PEG-tetraazide
4-arm poly(ethylene glycol) tetraazide was synthesized following a published synthetic route11. In short, methanesulfonyl chloride (5x) was added to 4-arm PEG (Mn ~ 10,000 Da) (Jenkem) and subsequently reacted with sodium azide (5x). Additional detail is available in Supplementary Fig. S11.
Synthesis of Bis(cyclooctyne)-functionalized peptide crosslinker
The enzymatically-degradable, allyl ester-containing peptide Ac-KRRK(alloc)GGPQGILGQRRK-NH2 was synthesized (ABI 433A peptide synthesizer) via standard Fmoc solid-phase methodology and HBTU/HOBt activation. Resin was treated with trifluoroacetic acid/triisopropylsilane/water (95:2.5:2.5) for 2 hours and precipitated (3x) using ice-cold diethyl ether (Supplementary Fig. S3). DIFO3 was coupled to the ▯-amino groups of the terminal lysines via standard HATU coupling chemistry. Peptides were purified using semi-preparative RP-HPLC (Waters Delta Prep 4000) using a 70 minute linear gradient (5 to 95%) of acetonitrile and 0.1% trifluoroacetic acid (Sigma). Peptide purity was confirmed by an analytical RP-HPLC and MALDI-TOF-MS (Applied Biosystems DE Voyager) using α-cyano-4-hydroxycinnamic acid matrix (Sigma): calculated ([M+H]+ 2329.7); observed ([M+H]+ 2329.1) (Supplementary Fig. S4)
Synthesis of self-quenched collagenase-sensitive detection peptide (DiFAM)
H-KGWLGPAK(Dde)GKC-NH2 (0.25 mmol) was synthesized via standard Fmoc solid-phase methodology and HBTU/HOBt activation. Carboxyfluorescein (1 mmol, Novabiochem) was coupled to the N-terminus via standard HATU coupling chemistry. The Dde protecting group was removed using 2% hydrazine in DMF, and a second carboxyfluorescein (1 mmol) was coupled to the ▯-amino groups of the deprotected lysine using standard HATU coupling chemistry. Resin was treated with trifluoroacetic acid/triisopropylsilane/water (95:2.5:2.5) for 2 hours and precipitated (3x) using ice-cold diethyl ether. Peptides were purified using semi-preparative RP-HPLC (Waters Delta Prep 4000) using a 70 minute linear gradient (5 to 95%) of acetonitrile and 0.1% trifluoroacetic acid (Sigma). DiFAM peptide purity was confirmed by an analytical RP-HPLC and MALDI-TOF-MS (Applied Biosystems DE Voyager) using α-cyano-4-hydroxycinnamic acid matrix (Sigma): calculated ([M+H]+ 1860.04); observed ([M+H]+ 1861.94) (Supplementary Fig. S5)
Synthesis of fluorescently-labeled adhesive ligand
H-AhxRGDSC-NH2 (0.25 mmol) was synthesized via standard Fmoc solid-phase methodology and HBTU/HOBt activation. Alexa Fluor® 488 carboxylic acid, 2,3,5,6-tetrafluorophenyl ester (1 mg, Invitrogen) was dissolved in NMP with a catalytic amount of DIEA and stirred with resin overnight at room temperature. Resin was treated with trifluoroacetic acid/triisopropylsilane/water/dithiothreitol (94.5:2.5:2.5:0.5) for 1 hour and precipitated (3x) using ice-cold diethyl ether. This product, denoted AF488-AhxRGDSC-NH2, was used with no further purification.
Rheological Experiments
Dynamic frequency, time, and strain sweep rheology experiments were performed on a TA Ares rheometer with parallel plate geometry (20 mm diameter) at 25 °C. Initial gel network formation of a 13.5 wt% solution was monitored by observing G’ and G’’ at a constant frequency of 100 rad/s as a function of time. Gel properties were monitored via frequency sweep measurements at fixed strain amplitude (10%) to measure the hydrogel storage, G’, and loss, G’’, moduli.
3T3 Fibroblast Cell Culture
General Cell Culture
NIH 3T3s were cultured in high-glucose Dulbecco’s modified eagle’s medium (Gibco) containing 10% fetal bovine serum (Invitrogen), 2% penicillin/streptomycin (Gibco), 0.4% fungizone (Gibco), and 0.2% gentamicin (Gibco) in 5% CO2 at 37 °C.
Cell Encapsulation
3T3s were suspended at 3 × 106 cells/mL in a 13.5 total wt% monomer solution in media and allowed to react for one hour to form a cell-laden hydrogel sheet.
Biochemical Patterning
Hydrogels were swollen in phenol red-free media (pH = 7.4) containing 0.05 wt% Irgacure 2959 (Ciba) and 3 mg/mL patterning agent AF488-AhxRGDSC-NH2 for one hour. Using conventional photolithographic techniques, gels were exposed to collimated UV light (365 nm wavelength at 10 mW/cm2) through a patterned photomask for 10 minutes. Under these photopatterning conditions, 1–2% of the alloc functional groups are consumed to yield ~0.1 mM of conjugated peptide, implying that multiple signals can be incorporated at biologically relevant concentrations. Alternatively, two-photon techniques were exploited for complex patterning by placing gels on a 710 LSM NLO confocal microscope stage (Carl Zeiss) and selectively exposing to pulsing focused 720 nm laser light through a 20×/0.8 Plan-Apochromat objective (Carl Zeiss), with x-y control afforded by Region of Interest scanning and z-control by focal depth. Z-planes were scanned at 1 µm increments with a laser power of 400 mW/µm2 and a scan speed of 2.4 µs/µm2. For both the two-photon and photolithographic approaches, photocoupling of the patterning agent to the hydrogel network occurs only within areas exposed to light. After patterning is complete, the gel is washed for approximately two hours with fresh media to remove any unbound material, yielding the final patterned hydrogel (Supplementary Fig. S7&Supplementary Fig.S9). The process of swelling in the patterning agent, photopatterning, and washing can be repeated for multiple cues within the same gel (Supplementary Fig. S10). For gels that are of reasonable thickness for 3D cell culture (~200 microns to 1 mm), characteristic diffusion times of the unreacted peptides from the gel are on the order of a few minutes to a few hours.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. C. Bertozzi and J. Baskin for their initial donation of DIFO3, Dr. R. Shoemaker for his assistance with MAS NMR, Dr. A. Kloxin and B. Fairbanks for their useful discussions on photopatterning, A. Aimetti for communication on peptide work, and Dr. C. Kloxin for his insightful critiques in the preparation of this manuscript. Thanks are also given to HHMI and the Janelia Farm Research Campus for support in the usage of their two-photon confocal microscope. Fellowship assistance to C.A.D. was awarded by the US Department of Education’s Graduate Assistantships in Areas of National Need program and the National Institutes of Health (T32 GM-065103). Correspondence and requests for materials should be addressed to K.S.A.
Footnotes
Competing financial interests
The authors declare that they have no competing financial interests.
References
- 1.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angewandte Chemie-International Edition. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Huisgen R. 1.3-Dipolare Cycloadditionen-Ruckschau Und Ausblick. Angewandte Chemie-International Edition. 1963;75:604–637. [Google Scholar]
- 3.Moses JE, Moorhouse AD. The growing applications of click chemistry. Chemical Society Reviews. 2007;36:1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- 4.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discovery Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 5.Hawker CJ, Wooley KL. The convergence of synthetic organic and polymer chemistries. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]
- 6.Baskin JM, et al. Copper-free click chemistry for dynamic in vivo imaging. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasan R, Li J, Ng SL, Kalesh KA, Yao SQ. Methods of using click chemistry in the discovery of enzyme inhibitors. Nature Protocols. 2007;2:2655–2664. doi: 10.1038/nprot.2007.323. [DOI] [PubMed] [Google Scholar]
- 8.Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nature Biotechnology. 2005;23:1303–1307. doi: 10.1038/nbt1149. [DOI] [PubMed] [Google Scholar]
- 9.Franc G, Kakkar A. Dendrimer design using Cu-I-catalyzed alkyne-azide "click-chemistry". Chemical Communications. 2008:5267–5276. doi: 10.1039/b809870k. [DOI] [PubMed] [Google Scholar]
- 10.Malkoch M, et al. Synthesis of well-defined hydrogel networks using Click chemistry. Chemical Communications. 2006:2774–2776. doi: 10.1039/b603438a. [DOI] [PubMed] [Google Scholar]
- 11.Polizzotti BD, Fairbanks BD, Anseth KS. Three-dimensional biochemical patterning of click-based composite hydrogels via thiolene photopolymerization. Biomacromolecules. 2008;9:1084–1087. doi: 10.1021/bm7012636. [DOI] [PubMed] [Google Scholar]
- 12.Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. Journal of Organic Chemistry. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 13.Franke R, Doll C, Eichler J. Peptide ligation through click chemistry for the generation of assembled and scaffolded peptides. Tetrahedron Letters. 2005;46:4479–4482. [Google Scholar]
- 14.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 15.Chen JK. Fish 'n clicks. Nature Chemical Biology. 2008;4:391–392. doi: 10.1038/nchembio0708-391. [DOI] [PubMed] [Google Scholar]
- 16.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 17.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dondoni A. The Emergence of Thiol-Ene Coupling as a Click Process for Materials and Bioorganic Chemistry. Angewandte Chemie-International Edition. 2008;47:8995–8997. doi: 10.1002/anie.200802516. [DOI] [PubMed] [Google Scholar]
- 19.Hoyle CE, Lee TY, Roper T. Thiol-enes: Chemistry of the past with promise for the future. Journal of Polymer Science Part a-Polymer Chemistry. 2004;42:5301–5338. [Google Scholar]
- 20.Khire VS, Benoit DSW, Anseth KS, Bowman CN. Ultrathin gradient films using thiol-ene polymerizations. Journal of Polymer Science Part a-Polymer Chemistry. 2006;44:7027–7039. [Google Scholar]
- 21.Killops KL, Campos LM, Hawker CJ. Robust, efficient, and orthogonal synthesis of dendrimers via thiol-ene "Click" chemistry. Journal of the American Chemical Society. 2008;130:5062–5064. doi: 10.1021/ja8006325. [DOI] [PubMed] [Google Scholar]
- 22.Langer R, Vacanti JP. Tissue Engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 23.Cushing MC, Anseth KS. Hydrogel cell cultures. Science. 2007;316:1133–1134. doi: 10.1126/science.1140171. [DOI] [PubMed] [Google Scholar]
- 24.Anseth KS. A biologist looks to 'click chemistry' for better three-dimensional tissue models. Nature. 2008;453:5. [Google Scholar]
- 25.Codelli JA, Baskin JM, Agard NJ, Bertozzi CR. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J Am Chem Soc. 2008;130:11486–11493. doi: 10.1021/ja803086r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Lutolf MP, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: Engineering cell-invasion characteristics. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature Biotechnology. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 29.Kunz H, Unverzagt C. The Allyloxycarbonyl (Aloc) Moiety-Conversion of an Unsuitable into a Valuable Amino Protecting Group for Peptide-Synthesis. Angewandte Chemie-International Edition in English. 1984;23:436–437. [Google Scholar]
- 30.Lee TY, Smith Z, Reddy SK, Cramer NB, Bowman CN. Thiol-allyl ether-methacrylate ternary systems. Polymerization mechanism. Macromolecules. 2007;40:1466–1472. [Google Scholar]
- 31.Mather BD, Viswanathan K, Miller KM, Long TE. Michael addition reactions in macromolecular design for emerging technologies. Progress in Polymer Science. 2006;31:487–531. [Google Scholar]
- 32.Cramer NB, Reddy SK, O'Brien AK, Bowman CN. Thiol-ene photopolymerization mechanism and rate limiting step changes for various vinyl functional group chemistries. Macromolecules. 2003;36:7964–7969. [Google Scholar]
- 33.Ruoslahti E, Pierschbacher MD. New Perspectives in Cell-Adhesion-Rgd and Integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 34.Burdick JA, Anseth KS. Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 35.Hahn MS, Miller JS, West JL. Three-dimensional biochemical and biomechanical patterning of hydrogels for guiding cell behavior. Advanced Materials. 2006;18:2679–2684. [Google Scholar]
- 36.Lee SH, Moon JJ, West JL. Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y, Shoichet MS. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nature Materials. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.