Abstract
The spread of high-level pyrimethamine resistance in Africa threatens to curtail the therapeutic lifetime of antifolate antimalarials. We studied the possible evolutionary pathways in the evolution of pyrimethamine resistance using an approach in which all possible mutational intermediates were created by site-directed mutagenesis and assayed for their level of drug resistance. The coding sequence for dihydrofolate reductase (DHFR) from the malaria parasite Plasmodium falciparum was mutagenized, and tests were carried out in Escherichia coli under conditions in which the endogenous bacterial enzyme was selectively inhibited. We studied 4 key amino acid replacements implicated in pyrimethamine resistance: N51I, C59R, S108N, and I164L. Using empirical estimates of the mutational spectrum in P. falciparum and probabilities of fixation based on the relative levels of resistance, we found that the predicted favored pathways of drug resistance are consistent with those reported in previous kinetic studies, as well as DHFR polymorphisms observed in natural populations. We found that 3 pathways account for nearly 90% of the simulated realizations of the evolution of pyrimethamine resistance. The most frequent pathway (S108N and then C59R, N51I, and I164L) accounts for more than half of the simulated realizations. Our results also suggest an explanation for why I164L is detected in Southeast Asia and South America, but not at significant frequencies in Africa.
Keywords: adaptive landscape, drug resistance, evolution
Understanding the molecular evolution of drug resistance has potential clinical implications for the development of therapeutic protocols to forestall resistance, the rational design of modified drugs that target resistant proteins, and the deployment of more effective drugs less likely to promote resistance. More generally, understanding the acquisition of drug resistance can reveal broader features of the multidimensional fitness landscape that organisms traverse as they evolve, and the constraints that this landscape imposes on their evolution.
The malaria parasite offers promising opportunities for such studies, as the useful therapeutic lifetime of most first-line malaria drugs has been compromised by the evolution of parasite resistance. Notable examples include chloroquine (1), atovaquone (2), and pyrimethamine (3, 4). Both chloroquine resistance and pyrimethamine resistance are associated with multiple amino acid replacements in the target protein (5, 6). Mutations in other parasite genes also may contribute quantitatively to increase the resistance phenotype or enhance parasite fitness (7–9).
In this study, we focused on the evolution of resistance to pyrimethamine, a drug that competitively inhibits the parasite enzyme dihydrofolate reductase (DHFR; EC 1.5.1.3). DHFR is part of a bifunctional enzyme required for the synthesis of tetrahydrofolate, an essential precursor of purines and several amino acids (10, 11). Although pyrimethamine and other antifolates are currently the first-line treatment for malaria in many parts of Africa (12–14), high-level resistance has been reported elsewhere (4, 13, 15). The genetic basis of pyrimethamine resistance in Plasmodium falciparum is associated with a small number of amino acid replacements in the parasite DHFR (3, 4, 16–18). Each of these replacements has been observed in clinical isolates of resistant strains (14, 19–21). Resistant mutants in DHFR generally are associated with some combination of 4 amino acid changes—in particular, Asn-51 to Ile (N51I), Cys-59 to Arg (C59R), Ser-108 to Asn (S108N), and Ile-164 to Leu (I164L). These 4 replacements affect the enzyme-binding pocket and reduce the binding affinity for pyrimethamine (22). Some of the mutants also compromise the enzymatic efficiency of endogenous DHFR (6). Resistant mutants of DHFR appear to have arisen in Southeast Asia and then spread to Africa (23). No other point mutations in the DHFR gene are commonly associated with drug resistance, although a few rare variants associated with heightened resistance have been reported (24).
In view of the continuing clinical importance of antifolates in treating malaria in Africa, the pathways by which resistance may have evolved are of immediate interest. While protein evolution proceeds largely via the sequential replacement of individual amino acids, various processes, trade-offs, and interactions can constrain the temporal order in which these replacements occur. Factors that may constrain the realized evolutionary pathways or trajectories include:
Mutation bias, which in the malaria parasite is a strong bias toward AT and thus favors AT-rich codons over others (25)
Adaptive conflict, because some amino acid replacements that increase drug resistance also may impair the endogenous function of the protein (6)
Epistasis resulting from nonadditive interactions between mutant sites in the same gene, because the effect of any amino acid replacement may depend on the sequence context in which it occurs (26)
Epistasis resulting from nonadditive interactions between mutations in different genes, because the evolution of any enzyme in a metabolic pathway alters the selection pressures acting on other enzymes in the same metabolic pathway or other pathways (27).
In principle, the evolutionary pathways of drug resistance can be reconstructed by selection for resistance in cultured isolates. Experimental systems in which the parasite enzyme is studied in either transgenic bacteria (17) or yeast (28) have been developed, and both approaches have been successful in identifying amino acid sites important in clinical drug resistance, as well as novel resistance sites not observed in nature. Kinetic studies of putative intermediates found as polymorphisms in natural populations also have proven informative in inferring the stepwise evolution of drug resistance (6). The results suggest 3 likely pathways by which pyrimethamine resistance may have evolved (6).
In the present experiments, we adopted a third approach—genetically engineering and analyzing the phenotypes of all possible combinations of the amino acid replacements implicated in drug resistance. In previous experiments carried out to examine possible pathways for the evolution of various functions in various proteins (29–32), epistasis among the mutant sites substantially reduced the number of selectively accessible mutational pathways. Mutational bias and adaptive conflicts were not explicitly addressed, however. In the present analysis, we addressed these limitations in two ways. First, we assumed the mutational biases implied by changes in genome sequence in the evolutionary lineage of P. falciparum. Second, in the experimental studies, we used the transgenic bacterial system in which each bacterial strain contained an alternatively mutated DHFR coding sequence from P. falciparum. The endogenous bacterial enzyme was inhibited with an antifolate to which the parasite enzyme is naturally resistant. Thus, the transgenic mutant genotypes have an adaptive conflict that trades-off resistance to inhibition by pyrimethamine against efficient conversion of dihydrofolate into tetrahydofolate, which impinges on growth rate in the absence of pyrimethamine. Our results suggest that this trade-off may provide a hypothesis to help explain why the quadruple DHFR mutant so widespread in Southeast Asia has not yet spread in Africa.
Overall, our results confirm and also extend previous hypotheses on the evolutionary pathways of pyrimethamine resistance. Our inferences take earlier results a step further in the ability to estimate a probability for the realization of each possible pathway of the evolution of pyrimethamine resistance. The most frequent pathway accounts for more than half of all realizations in simulated evolutionary replays, and the top 3 pathways account for 87.5% of the realizations. We discuss our results in the context of clinical resistance of P. falciparum to antifolates, protein folding and function, and, more generally, protein evolution.
Results
By means of site-directed mutagenesis of a bacterial plasmid containing the coding sequence of P. falciparum DHFR, we created all 16 possible combinations of the mutants Asn51Ile (N51I), Cys59Arg (C59R), Ser108Asn (S108N), and Ile164Leu (I164L). The sequences of the mutagenized DHFR genes were confirmed by direct sequencing.
Drug-Resistance Assays.
Assays for pyrimethamine resistance were carried out by means of a bacterial complementation system (17). In this system, the endogenous bacterial enzyme was chemically inhibited by 5 μM trimethoprim, a concentration sufficient to prevent bacterial growth. The P. falcarium DHFR enzyme is insensitive to trimethoprim and thus can complement the inhibited bacterial enzyme. Therefore, the bacterial system requires sufficient DHFR enzyme activity to allow the cells to grow, which in the presence of pyrimethamine represents an adaptive conflict between drug resistance and catalytic activity.
The level of pyrimethamine resistance was estimated as the concentration of drug that inhibited cell growth by 50%, a metric referred to as IC50. Bacterial strains carrying each of the DHFR alleles were assayed across a range of pyrimethamine concentrations, and growth rate as a function of pyrimethamine concentration was estimated by polynomial regression. To determine the empirical distribution of IC50 estimates, the IC50 value for each of the 16 DHFR alleles was estimated in 4–8 independent biological replicates, each with 4 technical replicates.
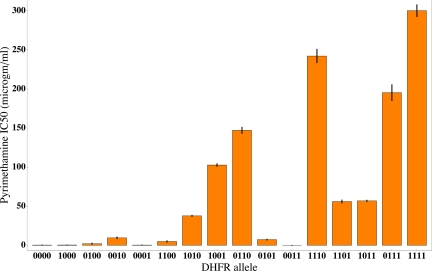
The results are shown in a bar graph in Fig. 1 and in tabular form in Table S1. The DHFR alleles indicated along the horizontal axis are given in the form of a vector of 0's and 1's corresponding, from left-to-right, to residues 51, 59, 108, and 164. Each 0 indicates a nonmutant codon, and each 1 indicates a mutant codon. Thus, 0000 represents the nonmutant sequence NCSI, and 1111 represents the quadruple mutant IRNL. The error bars are equal to the SD of each of the distributions of estimated IC50 values. Larger IC50 values can be estimated with greater precision than smaller ones. The coefficient of variation is about 30% of the mean for IC50 values <10 μg/mL and about 5% of the mean for values >10 μg/mL.
Fig. 1.
Mean IC50 values for pyrimethamine among the 16 possible combinations of mutant amino acid sites in DHFR. The error bars are the SDs of the estimated sampling distributions of IC50.
Fig. 1 shows that 2 of the 4 possible single mutants (1000 and 0001) are about equally sensitive to pyrimethamine as the nonmutant allele 0000. Thus, the first step in the evolutionary pathway toward pyrimethamine resistance is likely either 0010 or 0100. Beyond that, the favored evolutionary pathways are not clear, although the quadruple mutant is obviously much more resistant than any of the triple mutants.
While the average IC50 of the single, double, and triple mutants steadily increases, from 3.06 μg/mL to 49.9 μg/mL to 137.6 μg/mL, respectively, the complexity of the drug-resistance landscape is evident from Fig. 1. Compare, for example, the values among the double mutants, whose IC50 values range from 0 to almost 150 μg/mL. A notable example of epistatic interaction among the mutant sites is allele 0011, which is extremely sensitive to pyrimethamine, whereas its single-mutant constituents 0010 and 0001 have IC50 values of 9.56 μg/mL and 0.29 μg/mL, respectively. The growth of strains containing 0011 is impaired even in the absence of pyrimethamine. In particular, the growth rate of 0011 relative to that of 0000 in the absence of the drug is 0.169 ± 0.002, whereas the relative growth rates of 0010 and 0001 are 1.103 ± 0.009 and 0.968 ± 0.004, respectively. In other words, the combination of 2 mutants, each with a virtually normal growth rate in the absence of drug, results in a double mutant with severely impaired growth. The inability of 0011 to grow in pyrimethamine is a good illustration of the adaptive conflict between resistance to inhibition and residual enzyme activity.
The IC50 values in Fig. 1 are in good agreement with another widely used metric of drug resistance, the minimal inhibitory concentration (MIC), which refers to the smallest concentration of the drug that can completely inhibit growth (Table S1). Although the solubility of pyrimethamine limits the discrimination among the MIC values of the most highly resistant alleles, the concordance between the estimated IC50 and MIC values is very high. Because it is the relative ranking of the DHFR alleles that matters most in inferring the possible evolutionary pathways, this is the relevant comparison, and the linear association exhibits r2 = 0.81. The linear association between the raw resistance scores of IC50 and MIC has r2 = 0.68, but, as noted earlier, this comparison is less relevant. The close correspondence between the rank ordering of alleles according to IC50 and MIC seems to exclude the possibility that the analysis of the evolutionary pathways to pyrimethamine resistance is unduly influenced by the choice of resistance assay.
Evolutionary Model.
To analyze the data in Fig. 1, we used an evolutionary model in which selection acts to increase pyrimethamine resistance. In this model, selection pressure is assumed to be strong relative to mutation pressure, implying that the time to fixation or loss of a newly arising mutation is much shorter than the time between the occurrence of new mutations. In addition, the population size is assumed to be sufficiently large so that random genetic drift has a negligible effect on the probability of fixation. In the model, the selectively driven fixation or loss of each newly arising mutant occurs before the next mutation occurs, and so an evolutionary trajectory comprises a succession of new mutant alleles becoming fixed, each of which increases the level of drug resistance (30). In the mutational process, we allow all possible single-mutant neighbors to occur, including reversions of previously fixed mutations (33). Selection is based only on the relative values of IC50, and differences in growth rate in the absence of drug are not explicitly taken into account. The rationale for this is that IC50 values differ among alleles by a factor of more than 1,000, whereas for most alleles, the growth rates in the absence of drug differ by no more than a factor of 2 (Table S2). Thus, the presence of any significant drug pressure swamps selection attributable to intrinsic differences in growth rate; however, as discussed later, the case of the quadruple mutant is especially interesting in light of potential compensatory mutations.
The evolutionary model also incorporates the mutational bias of the malaria parasite. The genome of P. falciparum comprises approximately 82% A–T nucleotide pairs (34), reflecting a strong mutational bias toward A or T (25). We estimated the mutation matrix (Table S3) using a genome-wide collection of 1,105 SNPs) in intergenic regions, culled from publicly available sequencing reads from P. falciparum and its sister species P. reichenowi. Methods of analysis are described in Table S3.
Analysis of Evolutionary Pathways.
We used computer simulation to explore the evolutionary implications of the results in Fig. 1. A flow chart of the computer algorithm is provided in Fig. S1. In brief, replicate evolutionary landscapes were defined by random sampling of IC50 values for each of the 16 possible alleles from normal distributions with the allele-specific genotypic means and SDs given in Fig. 1. The possible evolutionary paths along each landscape were then explored by randomly choosing single-step mutations, including possible reverse mutations, according to the mutation model given in Table S3. Each new mutant allele was discarded if its IC50 value was smaller than that of the prevailing allele; otherwise, the mutant was fixed or lost according to the difference in the ranks of the IC50 values defining the evolutionary landscape. This procedure reflects the principle that the probability of fixation of a new mutation is proportional to its selective advantage, while the use of ranks renders the multitude of simulated fitness landscapes commensurate. Evolution on each simulated landscape was continued until either fixation of the allele with the maximum IC50 or fixation of a different allele at a submaximal fitness peak, whose single-mutant neighbors all had lower IC50 values, occurred.
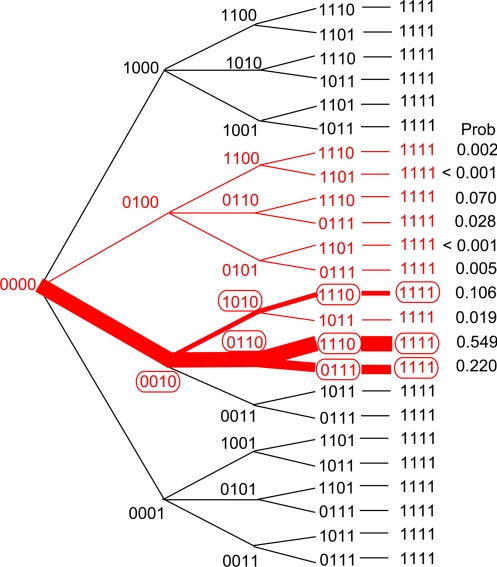
Simulation of 10,000 landscapes with 1,000 independent excursions in each landscape identified 10 evolutionary pathways that were traversed at significant (although very uneven) frequencies. These evolutionary pathways are shown in red in Fig. 2, where vectors of 0's and 1's again depict the amino acid residues 51, 59, 108, and 164. Numerous other pathways were realized, each at a negligible frequency, including all pathways with mutational reversions. These rare pathways mainly reflected the random sampling of extreme values from the distributions in Fig. 1, resulting from the large number of simulated landscapes.
Fig. 2.
Major inferred pathways for the evolution of pyrimethamine resistance. The top 10 pathways are shown in red, along with their estimated probabilities.
To estimate the relative probabilities of the prominent pathways more precisely, we chose 100 of the landscapes at random and explored each with 1 million independent evolutionary excursions. The results confirm the prominence of the 10 previously mentioned pathways and yield the estimates of the relative probability of each pathway shown in Fig. 2 in the column headed “Prob.” Only those evolutionary pathways traced in red constitute the top 10 in the simulations, with the more likely pathways traced in thicker lines.
The relative frequencies of the 10 major pathways were virtually identical when 10,000 landscapes were traversed 1,000 times each and when 100 landscapes were traversed 1 million times each. This consistency reflects the relatively small SDs among the IC50 distributions (Fig. 1). The probabilities given in Fig. 2 are determined primarily by the differences in IC50 values, and are affected only slightly by the magnitude of the mutation bias.
Just 3 pathways in Fig. 2 account for 87.5% of the pathways traversed at significant frequencies in the simulations. All 3 of these pathways feature S108N as the first step, and the 2 most likely pathways include C59R as the second step. Similarly, all 3 of these most likely pathways have I164L incorporated either last or next to last.
Comparison With DHFR Polymorphisms.
The results in Fig. 2 are consistent with the combinations of amino acid replacements observed at significant frequencies in worldwide surveys of P. falciparum (Table 1; refs. in Table S4). The common polymorphisms, indicated by red ovals in Fig. 2, coincide with the intermediates predicted in the most likely pathways.
Table 1.
Common polymorphic DHFR alleles in P. falciparum
| Allele | Location |
|---|---|
| S108N (0010) | Indonesia, Philippines, Cameroon, Burkina Faso, Bangladesh, Senegal, Sudan |
| N51I/S108N (1010) | Kenya, Burkina Faso, Cameroon, Sudan, Ethiopia, India |
| C59R/S108N (0110) | Laos, Indonesia, Thailand, Philippines, Kenya, Cameroon, Bangladesh, Sudan, Iran, India |
| N51I/C59R/S108N (1110) | Senegal, Sudan, Ethiopia, Kenya, Zambia, Benin, Cameroon, Ivory Coast, Gabon, Guinea, Mali, Uganda, Thailand, Bangladesh |
| C59R/S108N/I164L (0111) | Bangladesh, Iran, India |
| N51I/C59R/S108N/ I164L (1111) | Bangladesh, India, Thailand |
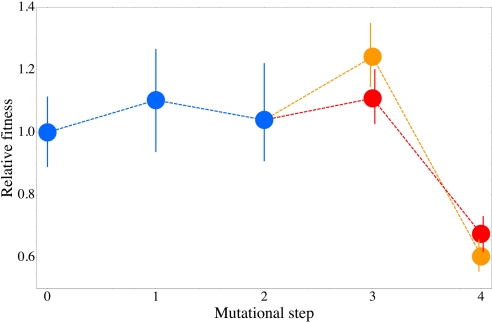
Note that according to Table 1, the quadruple mutant has not yet been found at significant frequencies in Africa. The absence of this allele in Africa is all the more surprising in light of the compelling evidence that pyrimethamine-resistant alleles in Africa derived from those present in Southeast Asia, where the quadruple mutant is quite common (23). Our results also may have relevance to this issue. In the absence of pyrimethamine, the growth rates of the strains bearing 0010 (S108N), 0110 (S108N + C59R), or either of the triple mutants are not manifestly impaired relative to the previous step or even the nonmutant allele (Fig. 3; Table S2). These results are consistent with previous inferences based on comparisons of the Michaelis constant (Km) and catalytic turnover rate (kcat) of the wild-type and mutant enzymes (35). However, when the fourth mutation (either N51I or I164L) is added, creating the quadruple mutant, a strongly deleterious interaction between N51I and I164L occurs, with the result that the fourth mutation decreases the growth rate in the absence of pyrimethamine by 30%–40% (Fig. S1), even though it increases the IC50 value by 25%–50% (Fig. 1). Thus, in the absence of compensatory mutation (see Discussion), the potential fitness cost of incorporating the last mutation in the pathway is substantial.
Fig. 3.
Relative growth rates of each intermediate in the 2 major evolutionary pathways. The growth rate of each strain after each step in the pathway was measured relative to the strain carrying the immediately preceding allele. The final steps in the pathway 0110–1110–1111 are shown in orange, and those in the pathway 0110–0111–1111 are shown in red. The error bars are 95% confidence intervals.
Discussion
Previous in vitro studies of the mutant DHFR proteins and their susceptibility to inhibition by pyrimethamine have identified 3 hypothetical pathways for the stepwise evolution of resistance (6). These are precisely the 3 most likely pathways identified in Fig. 2. Our data take the analysis a step further by providing estimates of the relative likelihoods of these pathways. The order of amino acid replacements S108N, C59R, N51I, I164L is favored by a factor of about 2 over the order S108N, C59R, I164L, N51I, which in turn is favored by another factor of about 2 over the order S108N, N51I, C59R, I164L. Together, these 3 pathways account for 87.5% of the excursions over the evolutionary landscape that occur with significant frequency based on the data in Fig. 1.
Pyrimethamine acts by competing with dihydrofolate for access to the binding pocket of DHFR. Because endogenous DHFR activity is essential for viability, the evolution of resistance occurs through increased substrate specificity (36). The key kinetic parameters are the Michaelis constant (Km), the pyrimethamine dissociation constant (Ki), and the catalytic turnover rate (kcat) (6). The competitive binding dynamics are biphasic, which implies that the rate of product production is largely independent of catalytic capacity (kcat/Km) if the relative preference of the enzyme for substrate (Ki/Km) is below some threshold. Consistent with this picture, the pathways with the highest probabilities in Fig. 2 all improve the substrate specificity at the expense of catalytic capacity. According to table 3 in ref. (6), relative to the nonmutant DHFR, the quadruple mutant shows an approximate 500-fold increase in Ki/Km at the expense of about a 6-fold decrease in kcat/KM. Interestingly, among the single mutants, S108N yields the largest increase in IC50 relative to the nonmutant allele and is the most clinically important (14, 21), even though C59R yields larger increases in both Ki/Km and kcat/Km (6). This finding implies that protein attributes beyond kinetic constants alone also contribute to differences in IC50. Obvious candidates include protein folding, stability, potential for aggregation, rate of degradation, and so forth, and the role of natural selection acting through such factors has been discussed previously (37). While DHFR is an established model system for the study of protein folding (38), such data are not yet available for the pyrimethamine-resistant DHFR alleles.
The fitness cost of the quadruple mutation in the absence of pyrimethamine (Fig. 3) may help explain why the quadruple mutant has not yet spread in Africa. An intriguing link between the evolution of drug resistance and the intensity of malaria transmission has been noted: Drug resistance may evolve more readily in areas of low transmission (7). A possible mechanism for this is that some drug-resistance mutants may have deleterious effects on fitness in the absence of the drug, which may be tolerated because of compensatory mutations occurring elsewhere in the genome (8). In areas of low transmission, inbreeding is relatively high and genetic recombination is restricted, and so the deleterious drug-resistance determinant and the compensatory mutations can remain genetically associated. In areas of high transmission, however, frequent mixed infectious result in genetic recombination, which breaks down the association between the mutant genes.
Consistent with the hypothesis of compensatory mutation, a copy-number polymorphism in the gene encoding GTP-cyclohydrolase I, the first gene in the folate biosynthetic pathway, has recently been shown to be associated with the DHFR quadruple mutant in Thailand (9). Elsewhere in the genome, a selective sweep associated with pyrimethamine resistance has been reported across a region of chromosome 13 (39). It is an interesting, testable hypothesis that this region of chromosome 13 may include 1 or more mutations that also alleviate the deleterious fitness effects of DHFR resistance alleles.
While previous studies of mutationally accessible pathways to high-fitness sequences (29–32, 40) have explored the evolution of novel functions, pathways were not penalized if they simultaneously caused the organism to lose other functionality. In this sense, our experiment provides the first glimpse of the constraint imposed on evolutionary pathways by the adaptive conflict between inhibitor resistance and maintenance of sufficient endogenous activity. Nevertheless, levels of constraint on pathways in DHFR are comparable to those found in previous work, in which constraint was a reflection of epistasis in a single trait, and many (but not all) evolutionary pathways were rendered inaccessible to selection.
Of great interest in the exploration of evolutionary landscapes is the possible existence of submaximal fitness peaks at which a population may become stranded because each 1-step mutation has lower fitness. The DHFR landscape features 1 such submaximal peak. The sequence 1001 (N51I + I164L) has an IC50 of about 100 μg/mL and is surrounded in the landscape by 1101 (N51I + C59R + I164L), 1011 (N51I + S108N + I164L), 1000 (N51I), and 0001 (I164L), which have IC50 values of approximately 56, 57, 0.4, and 0.3 μg/mL, respectively (Fig. 1). Thus, 1001 is accessible through either 1000 or 0001, but neither pathway is realized sufficiently frequently to be among the top 10 (Fig. 2), mainly because among the single mutants, the mutation yielding S108N (G–C to A–T) is strongly favored to be fixed. Interestingly, the low fitness of the quadruple mutant in the absence of pyrimethamine (Fig. 3) is not observed in 1001 (N51I + I164L). Evidently, the deleterious interaction between N51I and I164L occurs only on a background of C59R + S108N.
The observed polymorphisms in natural populations of P. falciparum noted in Table 1 are consistent with the 3 major pathways identified in our analysis. This agreement was not necessarily expected. Drug treatment in a laboratory setting is carefully controlled and reproducible, whereas that in the field is variable depending on drug dosage, frequency, potency, pharmacokinetics, compliance, and other factors. Moreover, our results were obtained from studies in the E. coli system, not in P. falciparum itself, and some significant disparities might have been anticipated in view of the vast evolutionary distance between these organisms. Thus, it is reassuring to find that the inferred major pathways receive additional support from the naturally occurring polymorphisms, as well as from the kinetic studies discussed earlier. More generally, such support augers well for a similar use of model organisms in future studies of the evolution of drug resistance not only in the malaria parasite, but also in many other organisms.
Materials and Methods
The sequence coding for DHFR amino acids 1–239 was isolated from P. falciparum strain K1 by PCR and cloned into the vector pET17 (Novagen) without the T7 tag. The resulting plasmid was transformed into E. coli strain HMS174(DE3) (Novagen), and the construct was confirmed by sequencing. DHFR mutants were introduced by QuikChange site-directed mutagenesis (Stratagene) and reintroduced into strain HMS174(DE3). Residues Asn-51, Cys-59, Ser-108, and Ile-164 were changed to Ile, Arg, Asn, and Leu, respectively, individually and in all possible combinations, using oligonucleotides (Operon). All of the mutants were confirmed by DNA sequencing.
Estimates of IC50 values (Table S1) were obtained as follows. Overnight cultures grown in rich LB medium, containing 50 μg/mL of carbamicillin to maintain the DHFR plasmid, were diluted, and the cells were allowed to grow into the log phase. Aliquots of these cultures were diluted to ≈107 cells/mL in a series of concentrations of pyrimethamine in LB containing 5 μM trimethoprim, and then dispensed into the wells of 96-well microtiter plates. The plates were incubated in the dark at 37 °C for 19–20 h, and the resulting cell concentrations were estimated spectrophotometrically from the optical density at 600 nm. Preliminary experiments identified the interval in which the IC50 was likely to be found, and final estimates were based on concentrations bracketing this interval. Growth rate as a function of pyrimethamine concentration was estimated by polynomial regression, and the IC50 value was estimated from this curve. To minimize the experimental error as much a possible, the overall IC50 value for each of the 16 DHFR alleles was estimated from the IC50 values observed in 4–8 independent biological replicates and 4 technical replicates of each biological replicate.
For the MIC assays (Table S1), bacterial strains were streaked onto fresh agar medium consisting of LB broth and trimethoprim plus various concentrations of pyrimethamine in increments of 25 μg/mL, after which the plates were incubated at 37 °C for 18 h. For each DHFR allele, the MIC was estimated with a minimum of 3 biological replicates.
Excursions through evolutionary landscapes were simulated by means of an algorithm implemented in PERL. A flow chart of this algorithm is shown in Fig. S1. Mean pathway probabilities and their 95% confidence intervals were estimated using scripts written in R version 2.2.1 (R Foundation for Statistical Computing). Ad hoc analysis and computer graphics were carried out in Mathematica (Wolfram Research).
Supplementary Material
Acknowledgments.
We thank Kalsang Namgyal for excellent technical support in the experiments, Sarah Volkman, Yongyuth Yuthavong, and Prapon Wilairat for their thoughtful advice, and David Houle and Michael Ferdig for their review and suggestions regarding the manuscript. This work was supported by National Institues of Health grant R01GM079536 (to D.L.H.). T.C. is supported by grant MRG5080418 from the Thailand Research Fund, Commission on Higher Education. S.K. is an International Research Scholar of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905922106/DCSupplemental.
References
- 1.Wellems TE. Plasmodium chloroquine resistance and the search for a replacement antimalarial drug. Science. 2002;198:124–126. doi: 10.1126/science.1078167. [DOI] [PubMed] [Google Scholar]
- 2.Looareesuwan S, et al. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am J Trop Med Hyg. 1996;54:62–66. doi: 10.4269/ajtmh.1996.54.62. [DOI] [PubMed] [Google Scholar]
- 3.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase–thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snewin VA, England SM, Sims PFG, Hyde JE. Characterisation of the dihydrofolate reductase–thymidylate synthetase gene from human malaria parasites highly resistant to pyrimethamine. Gene. 1989;76:41–52. doi: 10.1016/0378-1119(89)90006-1. [DOI] [PubMed] [Google Scholar]
- 5.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc Natl Acad Sci USA. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hastings IM. Malaria control and the evolution of drug resistance: An intriguing link. Trends Parasitol. 2003;19:70–73. doi: 10.1016/s1471-4922(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 8.Nzila A, et al. Why has the dihydrofolate reductase 164 mutation not consistently been found in Africa yet? Trans R Soc Trop Med Hyg. 2006;99:341–346. doi: 10.1016/j.trstmh.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Nair S, et al. Adaptive copy number evolution in malaria parasites. PLoS Genet. 2008;4:e1000243. doi: 10.1371/journal.pgen.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel OG, Mberu EK, Nzila AM, Macreadie IG. Sulfa drugs strike more than once. Trends Parasitol. 2004;20:1–3. doi: 10.1016/j.pt.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Kompis IM, Islam K, Then RL. DNA and RNA synthesis: Antifolates. Chem Rev. 2005;105:593–620. doi: 10.1021/cr0301144. [DOI] [PubMed] [Google Scholar]
- 12.Sibley CH, et al. Pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: What next? Trends Parasitol. 2001;17:582–588. doi: 10.1016/s1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- 13.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 14.Kublin JG, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparium malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 15.Roper C, et al. Antifolate antimalarial resistance in southeast Africa: A population-based analysis. Lancet. 2003;361:1174–1181. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- 16.Ferlan JT, Mookherjee S, Okezie IN, Fulgence L, Sibley CH. Mutagenesis of dihydrofolate reductase from Plasmodium falciparum: Analysis in Saccharomyces cerevisiae of triple mutant alleles resistant to pyrimethamine and WR99210. Mol Biochem Parasitol. 2001;113:139–150. doi: 10.1016/s0166-6851(01)00207-9. [DOI] [PubMed] [Google Scholar]
- 17.Chusacultanachai S, Thiensathit P, Tarnchompoo B, Sirawaraporn W, Yuthavong Y. Novel antifolate resistant mutations of Plasmodium falciparum dihydrofolate reductase selected in Escherichia coli. Mol Biochem Parasitol. 2002;120:61–72. doi: 10.1016/s0166-6851(01)00440-6. [DOI] [PubMed] [Google Scholar]
- 18.Plowe CV. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. 2009;103(Suppl 1):S11–S14. doi: 10.1016/j.trstmh.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, et al. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol Biochem Parasitol. 1997;89:161–177. doi: 10.1016/s0166-6851(97)00114-x. [DOI] [PubMed] [Google Scholar]
- 20.Sims PFG, Wang P, Hyde JE. On the efficacy of antifolate antimalarial combinations in Africa. Parasitol Today. 1999;15:132–134. doi: 10.1016/s0169-4758(97)01203-9. [DOI] [PubMed] [Google Scholar]
- 21.Nzila AM, et al. Toward an understanding of the mechanism of pyrimethamine-sulfadoxine resistance in Plasmodium falciparum: Genotyping of dihydrofolate reductase and dihydropteroate synthase in Kenyan parasites. Antimicrob Agents Chemother. 2000;44:991–996. doi: 10.1128/aac.44.4.991-996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuthavong Y, et al. Malarial (Plasmodium falciparum) dihydrofolate reductase–thymidylate synthase: Structural basis for antifolate resistance and development of effective inhibitors. Parasitology. 2005;130:249–259. doi: 10.1017/s003118200400664x. [DOI] [PubMed] [Google Scholar]
- 23.Roper C, et al. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 24.Bates SJ, et al. Rare, highly pyrimethamine-resistant alleles of the Plasmodium falciparum dihydrofolate reductase gene from 5 Africa sites. J Infect Dis. 2004;190:1783–1792. doi: 10.1086/425078. [DOI] [PubMed] [Google Scholar]
- 25.DePristo MA, Zilversmit MM, Hartl DL. On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene. 2006;378:19–30. doi: 10.1016/j.gene.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 26.Weinreich DM, Chao L. Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution. 2005;59:1175–1182. [PubMed] [Google Scholar]
- 27.Keightley PD, Kacser H. Dominance, pleiotropy and metabolic structure. Genetics. 1987;117:319–329. doi: 10.1093/genetics/117.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sibley CH, Macreadie IG. Novel approaches to tackling malarial drug resistance using yeast. IUBMB Life. 2001;52:285–289. doi: 10.1080/152165401317291138. [DOI] [PubMed] [Google Scholar]
- 29.Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science. 2005;310:499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- 30.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 31.Reetz MT, Wang L-W, Bocola M. Directed evolution of enantioselective enzymes: Iterative cycles of CASTing for probing protein-sequence space. Angewadte Chemie. 2006;118:1258–1263. doi: 10.1002/anie.200502746. [DOI] [PubMed] [Google Scholar]
- 32.Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2007;312:97–100. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- 33.DePristo MA, Hartl DL, Weinreich DM. Mutational reversions during adaptive protein evolution. Mol Biol Evol. 2007;24:1608–1610. doi: 10.1093/molbev/msm118. [DOI] [PubMed] [Google Scholar]
- 34.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandefur CI, Wooden JM, Quaye IK, Sirawaraporn W, Sibley CH. Pyrimethamine-resistant dihydrofolate reductase enzymes of Plasmodium falciparum are not enzymatically compromised in vitro. Mol Biochem Parasitol. 2007;154:1–5. doi: 10.1016/j.molbiopara.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rastelli G, et al. Interaction of pyrimethamine, cycloguanil, WR99210 and their analogues with Plasmodium falciparum dihydrofolate reductase: Structural basis of antifolate resistance. Bioorg Med Chem. 2000;8:1117–1128. doi: 10.1016/s0968-0896(00)00022-5. [DOI] [PubMed] [Google Scholar]
- 37.DePristo MA, Weinreich DM, Hartl DL. Missense meandering through sequence space. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 39.Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 40.Poelwijk FJ, Kiviet DJ, Sans SJ. Evolutionary potential of a duplicated repressor–operator pair: Simulating pathways using mutation. PLoS Comput Biol. 2006;2:e58. doi: 10.1371/journal.pcbi.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.