Abstract
The genomic basis of primate phenotypic uniqueness remains obscure, despite increasing genome and transcriptome sequence data availability. Although factors such as segmental duplications and positive selection have received much attention as potential drivers of primate phenotypes, single-copy primate-specific genes are poorly characterized. To discover such genes genomewide, we screened a catalog of 38,037 human transcriptional units (TUs), compiled from EST and cDNA sequences in conjunction with the FANTOM3 transcriptome project. We identified 131 TUs from transcribed sequences residing within primate-specific insertions in 9-species sequence alignments and outside of segmental duplications. Exons of 120 (92%) of the TUs contained interspersed repeats, indicating that repeat insertions may have contributed to primate-specific gene genesis. Fifty-nine (46%) primate-specific TUs may encode proteins. Although primate-specific TU transcript lengths were comparable to known human gene mRNA lengths overall, 92 (70%) primate-specific TUs were single-exon. Thirty-two (24%) primate-specific TUs were localized to subtelomeric and pericentromeric regions. Forty (31%) of the TUs were nested in introns of known genes, indicating that primate-specific TUs may arise within older, protein-coding regions. Primate-specific TUs were preferentially expressed in reproductive organs and tissues (P < 0.011), consistent with the expectation that emergence of new, lineage-specific genes may accompany speciation or reproduction. Of the 33 primate-specific TUs with human Affymetrix microarray probe support, 21 were differentially expressed in human teratozoospermia. In addition to elucidating the likely functional relevance of primate-specific TUs to reproduction, we present a set of primate-specific genes for future functional studies, and we implicate nonduplicated pericentromeric and subtelomeric regions in gene genesis.
Keywords: EST, gene birth, genomics, reproduction
Whole-genome sequencing provides a foundation for comparative genomics. The basis of human-specific, and primate-specific, phenotypic distinctions (1) is under intense study. The completion of the chimpanzee (2), rhesus (3), and more recently other nonhuman primate genomes such as the orangutan and the marmoset is facilitating phylogenomic analyses directed at uncovering the genetic underpinnings of human uniqueness.
Explanations for lineage- and species-specific phenotypes include protein and regulatory sequence evolution relevant to human-specific traits. Although human/chimpanzee DNA sequence divergence (2), affecting ≈3 × 107 sites, has likely contributed to phenotypic differences, the high sequence correspondence has concurrently illustrated the closeness of the 2 species. In 1975, King and Wilson (4) suggested that protein sequence divergence between the 2 species is insufficient to account for the paradoxically substantial organismal differences. Human-specific phenotypes may result from such regulatory differences, but also from genome structure changes (5) and species-specific insertions/deletions (indels), which in human and chimpanzee total 3% of the genome (1). Use of the rhesus as a sufficiently close primate outgroup of humans and chimpanzees has helped identify adaptively evolved brain-expressed genes in the human lineage (6).
The “less-is-more” hypothesis implicates loss-of-function in human-specific phenotypes, portraying the human as a degenerate ape (6). An example is the human-specific, Alu-mediated inactivation of the CMAH gene, and hence of N-glycolylneuraminic acid synthesis, implying differences in pathogen susceptibility between humans and nonhuman primates. Human-specific Siglec protein expression explains differences relative to chimpanzees in T cell mediated diseases, including AIDS (5). Rearrangements, gene genesis, and duplications enhance the accretion of evolutionary novelties in primate genomes (7).
We define a transcript as a mature, capped, polyadenylated, and (whether biologically applicable) spliced RNA. This definition includes mRNAs, as well as non-protein-coding, mRNA-like long noncoding RNAs (lncRNAs) (ref. 8 and references therein). The concept of a transcriptional unit (TU), a genome segment transcribed in a consistent fashion and defined by a collection of cDNAs and/or ESTs, which all map to the same locus with at least partial exon overlap and represent transcription in the same orientation, helps resolve the discrepancy between the numbers of known genes and observed RNAs (9). Although microRNAs and small nucleolar RNAs are generally well-conserved, longer ncRNAs (some of which have regulatory functions) are not (10). Clearly, a search for functional primate-specific genes should include ncRNAs.
We hypothesized that TUs of recent evolutionary origin exist in primates. To identify such TUs, we performed multispecies detection for every exon of every human TU. We identified TUs consisting entirely of exons that localize to primate/nonprimate indel regions in multispecies BLASTZ alignments. We defined these regions as present in at least some of the primates we considered, but absent in all of the nonprimates. Their presence is the derived, not ancestral, state. We characterized conservation, structure, and expression of primate-specific TUs, providing a foundation for functional studies. We also demonstrated that primate-specific TUs may function in human reproduction.
Results
We started with 42,887 human TUs from experimental (cDNA and EST) data. We excluded TUs that mapped to no, or multiple, locations in the hg18 assembly by the University of Californa, Santa Cruz (UCSC) BLAST-Like Alignment Tool (11). The remaining 38,037 TUs were subjected to our BLASTZ-based liftOver (12) assessment of primate-specificity. We assessed the conservation of each human TU vs. 9 genomes (for list, see line 1, Dataset S1) by running liftOver for each human exon (Dataset S1 and Dataset S2). We identified 131 TUs such that all exons of each TU did not map to any nonprimate genomes. These TUs are operationally defined here as primate-specific, although they might not be present in all primates.
We developed a computational pipeline for summarizing expression of any TU. The pipeline extracts tissue information from the GenBank entry of every cDNA and EST of the TU. An expression profile was defined as relevant to primate-specific phenotypes if it indicated brain or reproductive expression. Brain-expressed genes originating in the primate lineage may underlie neural reorganization and complex behaviors (13). Because reproductive genes tend to evolve rapidly and may contribute to speciation (14), reproductive genes unique to primates may have had a role in species-specific phenotypes.
Conservation of Human TUs in 9 Nonhuman Genomes.
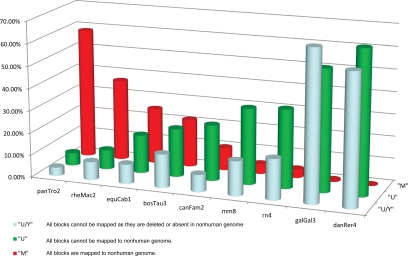
Comparative mapping was performed for each exon of each human TU against each nonhuman genome (Dataset S1 and Dataset S2). A human TU defined as mapped or unmapped has all exons either fully mapped or fully unmapped to a nonhuman genome, respectively. The percentage of mapped TUs decreases as evolutionary distance from human increases (Fig. 1), validating the assessment of TU conservation by exon liftOver.
Fig. 1.
Percentages of 24,785 cDNA-supported human TUs that could be mapped to 9 nonhuman genomes. Only the 24,785 human TUs supported by full-length cDNA sequences were considered, to preclude problems related to unsequenced but conserved regions of EST-represented transcripts. A TU is considered M if all its blocks mapped to the nonhuman genome and U if all its blocks did not map. A block is considered mapped if at least 95% of its bases are mapped. “Y” in “U/Y” indicates that the unmapped block is deleted or absent in the nonhuman genome although its flanking sequences are present in the BLASTZ alignment. In this case, the figure shows the percentage of the total TUs such that all blocks of each TU are not found in the nonhuman genome. We computed the number of such blocks and aggregated them at the TU level (Dataset S1). Totals per nonhuman species are <100% because TUs with some but not all human blocks mappable to the nonhuman genome (partially-mappable TUs) were not considered in this analysis.
To confirm whether the 131 TUs were primate-specific according to a different method, we masked repeats in TU reference transcripts and performed a BLASTN search against chimpanzee, orangutan, macaque, mouse lemur, bushbaby, and treeshrew genomes (treeshrew is the closest nonprimate outgroup to primates). The orangutan, lemur, and treeshrew genomes were not accessible by means of UCSC liftOver, but BLASTN allowed us to check for presence of the primate-specific TU exons in those genomes. Of the 108 TUs amenable to this analysis (Dataset S3), 18 (17%) seem to be conserved in treeshrew; this percentage might reflect TUs conserved beyond primates but identified as primate-specific false-positives by liftOver. Also, 27% of 90 TUs lacking BLASTN treeshrew conservation were present in prosimians. These 24 primate-specific TUs may have originated before the prosimian split, with the other 66 arising afterward.
A Subset of Primate-Specific TUs May Encode Proteins.
We screened the longest positive-strand ORF of every TU for domain and protein matches with CDD search and BLASTP, respectively (see Materials and Methods). The analysis revealed 7 TUs with 100% identity, and 11 with 26–98% identity, to protein database entries. Fifteen additional TUs contained previously undescribed nonrepetitive ORFs devoid of domain and database hits, potentially a primate-specific fraction of the human protein sequence space. In summary, 59 (45%) of the 131 primate-specific TUs were deemed to possess protein-coding capacity (Dataset S4).
We analyzed the 59 TUs for ORF conservation. Following a precedent (47), we used conservation of start and stop codons as proxy for ORF conservation, because Ka/Ks ratio calculations might fail due to lack of suitably distant outgroups. Forty-four TUs had intact start and stop codons conserved in human, chimpanzee, and rhesus (Dataset S4, columns AN–AQ). ORF boundary conservation is preliminary evidence for the protein-coding nature of the TUs.
Genomic Properties of Primate-Specific TUs.
The majority of the 131 primate-specific human TUs were detectable in both chimpanzee and rhesus [human, chimp, rhesus (HCR)-specific TUs; Table 1]. The reference transcripts of the TUs totaled 125,924 nt. This number is an estimate of the combined length of primate-specific genomic sequence giving rise to previously undescribed genes. Including introns, primate-specific TUs covered 436,865 bp (0.013%) of the human genome.
Table 1.
HCR-, HC-, and H-specific TUs from the human FANTOM3 TU catalog
| TUs with full-length cDNA support | TUs with EST-only support | Total | |
|---|---|---|---|
| HCR-specific | 44 | 63 | 107 |
| HC-specific | 10 | 6 | 16 |
| H-specific | 3 | 5 | 8 |
| Total | 57 | 74 | 131 |
Primate-Specific TUs Have Transcript Lengths Comparable with the Average Length of Human cDNAs, and Few Exons.
We removed all introns <30 nt and concatenated their flanking UCSC “blocks” into a single exon, because short introns in those alignments are generally artifacts not due to splicing. After this concatenation, most (94; 71%) of the TUs turned out to possess only 1 exon. Primate-specific TUs are unlikely to be retrogenes or unprocessed pseudogenes, because they were single-copy, outside of segmental duplications, and largely devoid of nonself BLASTN hits (SI Appendix). Forty-six TUs were present in human, chimpanzee, and rhesus, but not in any nonprimates considered. Of these 46 TUs, 36 were single-exon and protein-coding (Dataset S5). Ninety-eight percent of the primate-specific TUs have 3 or fewer exons. In contrast, a typical human known gene has 9 exons (16). The remaining 10 of the 46 TUs were spliced (Dataset S6), and 3 were nested in introns of known genes in the antisense direction, contradicting the assumption that most nested genes are intronless (17).
Pericentromeric and subtelomeric regions contain segmental duplications (18). Although we filtered out TUs with ambiguous mappings, including segmental duplications, 4 (9%) of our 57 cDNA-supported TUs and 19 (26%) of the 74 EST TUs are subtelomeric; and 3 (5%) of the cDNA TUs and 6 (8%) EST TUs are in pericentromeric regions. Therefore, certain primate-specific TUs may have arisen at terminal or pericentromeric chromosomal regions outside of segmental duplications. An intriguing TU of this type in a single-copy subtelomeric region, BC015579, has a long ORF, good EST support, and 20% repeat coverage. Its expression profile is oncofetal, and transcripts are present in stem cells.
All 215,758 human cDNAs mappable to single hg18 locations had a mean length of 1,782 nt. The 57 cDNA-supported TUs had a mean transcript length of 1,551 nt, broadly comparable to those of all human TUs including known genes.
The lengths of the 131 primate-specific TU reference transcripts ranged from 176 bp, well above the lengths of known microRNAs and other small-RNA classes, to >4 kb. We tested the genomic interval containing every primate-specific TU for overlap with known human microRNAs and small nuclear/nucleolar RNAs. None of the 131 TUs had any overlap with known small RNAs. Therefore, it appears unlikely that any of these TUs are host genes of known small, including micro, RNAs. Consequently, primate-specific TUs appear more likely to encode lncRNAs or proteins.
Exons and Promoters of Many Primate-Specific TUs Overlap Interspersed Repeats.
Interspersed repeats contributed to exonic sequences for most of the primate-specific TUs. The main classes of exonic repeats were: LINE (63 TUs), LTR (60 TUs), and Alu (47 TUs; Dataset S7). Seventy (53%) TUs are repeat mosaics, combining exonic repeat classes. Some TUs exhibited complex, higher-order repeats, conceptually analogous to SVA sequences (19). Only 14 TUs were devoid of interspersed repeats.
We analyzed transcription start sites (putative promoters) of the 62 TUs supported by full-length cDNAs and/or possessing long (>100 aa) ORFs, because they were more likely to correspond to full-length genes. The most frequent repetitive element class at their promoters was ERV, at 16 of the 62 TUs.
We separately considered exonic repeat content of the 59 protein-coding TUs. Twenty-seven (46%) of those 59 TUs had exonic primate-specific Alu repeats. This exonic Alu presence is consistent with the known contributions of Alu repeats to exon sequences in genes (20–22). Exonic LINE/LTR repeats were enriched in the 59 protein-coding, relative to all 131 primate-specific, TUs (Table 2; χ2 = 5.305, P = 0.021). Fifteen of 17 brain-expressed and 23 of 24 reproductively expressed TUs had exonic LTR and/or LINE repeats. Hence, formation of primate-specific protein-coding TUs relevant to brain and reproductive changes in primate evolution could have been facilitated by insertions of LINE/LTR elements.
Table 2.
LTR/LINE repeats in exons of primate-specific TUs
| Putative protein-coding primate-specific TUs |
All primate-specific TUs |
|||||
|---|---|---|---|---|---|---|
| LTR repeats | LINE repeats | LTR/LINE repeats | LTR repeats | LINE repeats | LTR/LINE repeats | |
| H-specific | 2 | — | 2 | 4 | — | 4 |
| HC-specific | 5 | 7 | 9 | 9 | 10 | 15 |
| HCR-specific | 25 | 27 | 40 | 47 | 53 | 82 |
| Total | 32 | 34 | 51 | 60 | 63 | 101 |
Each instance of an LTR or LINE repeat in a TU was counted individually.
Four TUs not mappable to any nonhuman genomes were potentially human-specific. All had repeat-encoded ORFs (Dataset S8). Therefore, we did not pinpoint any unique human-specific proteins.
Primate-Specific TUs Are Preferentially Expressed in the Reproductive System, and Are Deregulated in a Reproductive Disorder.
We compared expression in brain/neuronal, as well as in reproductive, tissues in the 131 primate-specific TUs against all 42,887 FANTOM TUs. We scored reproductive expression for a TU if at least 1 transcript for that TU had reproductive organ/tissue expression (see Materials and Methods). Primate-specific TUs were preferentially expressed in reproductive organs and tissues at a 5% significance level (Table 3; χ2 statistic = 6.409, P value = 0.011). A similar test for brain and neuronal tissues showed no enrichment. We conclude that primate-specific TUs are enriched for reproductive system expression relative to all human TUs.
Table 3.
Enrichment of reproductive and neuronal expression in primate-specific TUs
| Type of TUs | Reproductive expression | Total | Brain/neuronal expression | Total |
|---|---|---|---|---|
| Primate-specific TUs | 56 | 131 | 27 | 131 |
| All FANTOM TUs | 14203 | 42887 | 11082 | 42887 |
| χ2 = 6.409, p-value = 0.011 | χ2 = 1.875, p-value = 0.171 | |||
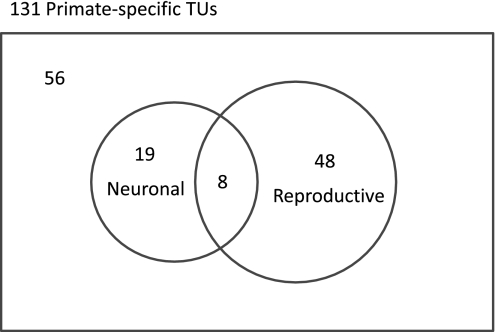
Forty-eight primate-specific TUs were expressed solely in reproductive tissues, 19 had solely neuronal expression, and 8 had reproductive and neuronal expression (Fig. 2). This expression profile underscores the significant enrichment for reproductive expression, and shows that 48 TUs likely function within the reproductive system, because they are not detectable anywhere else. Five reproductive tissues (F: placenta, ovary, uterus; M: testis, prostate) were detected, with male reproductive expression mainly in testis. Our result is consistent with the association of recent-origin genes with both reproductive expression (23) and reproduction-related behavior (24).
Fig. 2.
Exclusivity and overlap in neuronal and reproductive expression profiles of primate-specific TUs. Neuronal and reproductive expression profiles, as defined in Materials and Methods, are represented in a Venn diagram. Of the 131 TUs, there are 19 TUs with neuronal-only expression, 48 with reproductive-only expression, 8 with both neuronal and reproductive expression, and 56 with neither.
Thirty-three of our 131 TUs were represented by Affymetrix U133 probes (Dataset S9). Of these 33 TUs, 19 (58%) are putatively coding and 6 are subtelomeric. Using the National Center for Biotechnology Information (NCBI) GEO database, we searched for differential expression of the TUs in published microarray datasets. Twenty-one TUs demonstrated differential expression in spermatozoa from 14 men with consistent and severe teratozoospermia, a condition in which <4% of sperm cells are morphologically normal, relative to 17 normal fertile men (25). Of these 21 TUs, 10 had cDNA/EST support for expression in reproductive organs and tissues. These results are particularly interesting given the enrichment of expression in reproductive organs and tissues among primate-specific TUs, because if a TU has reproductive system expression and is also differentially expressed in a reproductive system disorder, then the TU likely has a function relevant to human reproduction. Summarily, the majority of Affymetrix-profiled primate-specific TUs are differentially expressed in a human reproductive disorder, teratozoospermia.
Primate-Specifc TUs Are Frequently Intercalated in the Introns of Known Protein-Coding Genes.
In eukaryotes, nested genes are rare and can be located within an intron of another gene (15, 16). It is noteworthy that 39 (31%) of our primate-specific TUs were nested. Nesting occurred in both sense and antisense orientations, was never associated with sense-antisense exon overlaps, and always represented the intercalation of a novel primate-specific TU with a clearly distinct known gene. Of these 39 TUs, 9 were spliced, with up to 6 exons of their own per TU. Therefore, primate-specific genes may arise over evolutionary time at loci harboring older, protein-coding genes.
Human- and Chimpanzee-Specific Protein-Coding TUs Absent in Rhesus and Nonprimates Are Rare.
Nine human TUs (Dataset S10) had exons that matched only the chimpanzee genome, consistent with gene origination after the last common ancestor of humans and Old World monkeys (≈25 Mya) but before the human-chimpanzee split (≈5–7 Mya). AK128008, expressed in the testes, lies on the 10q subtelomere, is supported by 1 cDNA and 4 ESTs, and has 2 L1 fragments making up only 18% of its exonic sequence. Its 3′ end contains a canonical polyA signal. AK128008 appears to be a previously undescribed protein-coding TU comprised of largely single-copy sequence and putatively specific to humans and chimpanzees.
Primate-Specific TUs Exhibit Few Chromosomal Distribution Trends.
Smaller chromosomes had fewer primate-specific TUs, which attests to an unbiased chromosomal distribution. Discordant trends may nevertheless exist. Chromosome 7 harbored the greatest number (12) of putative primate-specific TUs, perhaps because of its high gene density. Chromosome 20 had the greatest density (0.08 TUs/megabase) of primate-specific TUs. There are 2 primate-specific TUs on chromosome Y, more than is expected (χ2 = 16.31, P < 0.0001). The Y chromosome is covered by extensive tandem duplications of gene arrays whose structural variation is accompanied by a high mutation rate (26). Numerous Y-linked genes are dispensable, under relaxed constraints, and/or repeatedly lost and replaced in mammalian evolution (27).
Discussion
Rare primate-specific genes suggested by earlier reports (7, 13, 28, 29) motivated our genomewide search. From the FANTOM3 human TU catalog, we isolated 131 putative primate-specific TUs (Fig. 3). We made several observations:
Many (70%) primate-specific TUs have only 1 exon. Intronless genes can arise from retroposition, which intensified in primates 38–50 Myr ago (30). Alternatively, the “introns-late” model (31), implying intron accretion over evolutionary time, may explain why young TUs are unspliced or have fewer exons than older conserved genes.
Although novel genes may form in pericentromeric and subtelomeric segmental duplications, primate-specific TUs were generally not resident in segmental duplications. An underappreciated role for single-copy regions in primate gene genesis is suggested by data and another report of evolutionary innovations in nonduplicated, subtelomeric regions (32).
Alu repeats, a key marker of primate specificity, overlapped exons of 36% of our primate-specific TUs, but were limited to a minor portion of transcribed sequence. Only 5 TUs had 5′ and/or 3′ ends within Alu sequences, consistent with repeat-mediated recombination in gene genesis (33). Alu-mediated modification in primate evolution is perhaps more relevant to structural changes in existing genes than to the origination of new genes.
Repetitive element integration can cause loss of gene function. Yet, ERV repeats may be beneficial to their host genes, acting as promoters (34). We detected ERVs at transcription starts of several primate-specific genes. ERV insertions have disseminated binding sites of the p53 tumor suppressor transcription factor in an evolutionary lineage-specific manner (35). Initiation from promoters contributed by mobile elements is a property of the primate-specific TUs in our dataset. Recruitment of formerly dormant genomic fragments into new transcriptional frameworks is consistent with the exaptation paradigm (36). Such de novo gene origins may explain new organismal functions (37).
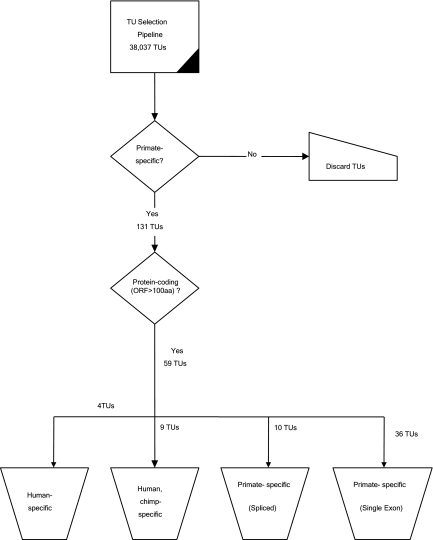
Fig. 3.
Extraction and classification of primate-specific TUs. The selection of 38,037 TUs were from the 42,887 FANTOM3 TUs by eliminating TUs that have multimapping or are not found in hg18. The exons of all TUs were individually mapped to nonhuman genomes. 131 primate-specific TUs whose exons are not detectable in nonprimates were found and TUs with ORFs at least 300 nt long qualified as protein-coding.
Many primate-specific TUs were expressed in brain and neuronal, as well as reproductive, tissues, as expected of young genes contributing to neuromediated phenotypic distinctions and, perhaps concurrently, speciation. However, only reproductive, not neuronal, expression was enriched. This expression profile is reminiscent of frequent male germ-line function in recent-origin genes (38), not limited to mammals (39). Also, the majority of primate-specific TUs represented on commercial Affymetrix microarrays were differentially expressed in human teratozoospermia, a reproductive disorder. Reproductive proteins can evolve rapidly, and undergo adaptive evolution after speciation events (40); these proteins include zona pellucida egg coat components that participate in egg-sperm binding and are implicated in reproductive isolation. Our results suggest that primate-specific, including lncRNA, genes are important in reproductive function and disease. We speculate that some of these lncRNAs may function by regulating, or interacting with, reproductive proteins, because functional protein-lncRNA interactions have been demonstrated for mammalian lncRNAs.
Most of our TUs were found in the genomes of all 3 primates analyzed. Thus, a limited set of novel TUs may affect organismal functions common to these primates, whereas phenotypic changes postdating the last common ancestor of humans and Old World Monkeys could have been mediated by means other than gene birth.
Biological functions of lncRNAs remain largely unelucidated. Nevertheless, specific lncRNAs regulate transcription factors (refs. 41–43 and references therein), epigenetically modulate gene expression, control nucleocytoplasmic protein translocation, and have diverse other roles (44). We present initial evidence for function of primate-specific, particularly including lncRNA, genes, which comprised 55% of our TUs (Dataset S4). A reproductive function for primate-specific TUs differentially expressed in teratozoospermia is suggested by their expression profile.
Another avenue of future investigation concerns sequence divergence that may have enabled emergence of primate-specific TUs. It might be instructive to consider the evolutionary fate of specific genomic sites required for exonification of single-copy sequences, in particular polyA signals, splice donors, and splice acceptors at orthologous loci, to explain the emergence of primate-specific TUs and to evaluate the interplay between generation of transcribed new genes on one hand, and subsequent selective pressures on these genes on the other.
Candidate primate-specific TUs might be conserved in nonprimates, but may have undergone accelerated sequence evolution, making their conservation unrecognizable in distant lineages and obscuring their shared origins from ancestral nonprimate genes. We do not believe exceptional divergence to be a major complication, because lowering the liftOver minMatch threshold did not improve detection of nonprimate alignments (see Materials and Methods). Accordingly, primate-specific TUs are consistent with de novo insertions, not sequence divergence.
Updating the catalog of primate-specific TUs is feasible as more cDNAs and ESTs become available. While our manuscript was in preparation, Toll-Riera et al. (45) identified primate orphan genes whose properties including transposon content paralleled those of our TUs. However, none of those genes overlapped the genomic location of our TUs (Dataset S14). Therefore, computational pipelines for primate-specific gene discovery might possess a high false negative rate, meaning that the global primate-specific TU catalog remains far from saturated. ENCODE highlighted transcribed sequences that are not conserved but are supported by tiling-array expression and transcription factor binding data (46). Our approach is capable of searching for primate-specificity in ENCODE “racefrags” and “transfrags,” and should aid identification of primate-specific transcribed regions when ENCODE methods are applied to the remainder of the human genome and the entire genomes of nonhuman primates, as well as identification of primate-specific exons in genes conserved beyond primates.
Materials and Methods
We examined human TU multispecies alignments using solely publicly available resources (SI Appendix).
Selection of Human TUs.
TUs were divided into 26,258 with a representative cDNA (cDNA TUs) and 16,629 without (EST TUs). Each TU, originally mapped to the hg17 assembly, was remapped to hg18. TUs (n = 24,794), each with a uniquely mapped cDNA, were used. Single-EST TUs (n = 3,704) matched the hg18 EST database. Of the remaining 12,925, 9,539 matched hg18-mapped ESTs; for each TU, we chose the EST with the longest genomic span as representative. Remaining TUs were discarded. As a result, 24,794 cDNA and 13,243 EST (38,037 total) TUs were selected.
Multispecies Comparisons of Human TU Structures.
Chromosomal mappings for UCSC “blocks” (1 or more blocks represent 1 exon) for the 38,037 TUs were extracted from hg18 all_mrna and all_est databases. UCSC liftOver converted the coordinates of human blocks to nonhuman genomes using chain files of precomputed whole-genome BLASTZ alignments (ftp://hgdownload.cse.ucsc.edu/goldenPath/hg18/). A chain is made up of nonoverlapping matched blocks for a pair of genomes (e.g., human and 1 nonhuman genome).
Interpretation of UCSC LiftOver Results Within a Computational Framework of Defining Primate Specificity.
Extraction of BLASTZ alignments by liftOver for a human query generates a liftOver output. Output types reflecting divergent regions alignable at lower liftOver thresholds, and/or human regions with multiple or ambiguous nonhuman matches, were not considered. The “#Deleted in new” liftOver output indicated that the human exon is absent, although flanking sequences on one or both sides of it are present, in the nonhuman genome, and was used to infer presence or absence of human-TU orthologs in that genome.
Nine cDNA TUs with >99 blocks were excluded as artefacts. For the remaining 24,785 cDNA TUs, 212,185 blocks were found, and 54,399 blocks were found for the 13,243 EST TUs.
The liftOver program was executed with block-level mappings of the TUs as queries, against each of the 9 genomes, with a minMatch = 0.95. A human block is “mapped” if 95% of its bases map to a segment in the nonhuman genome's chain. If a block cannot be mapped by liftOver at the given threshold, it is written into the “unmapped” file with a message of 1 of 4 types: E1, E2, E3, E4 (SI Appendix).
To focus on completely primate-specific insertions, we selected only blocks from the unmapped file deleted in a nonprimate genome (message E1). A TU is mapped (M) if all its blocks are mapped, partially mapped if only some of its blocks are mapped, and unmapped (U) if all of its blocks are unmapped.
Primate-Specific Characteristics.
The information in the liftOver mapped and unmapped files was used to define putative HCR-, human-chimpanzee (HC)-, and human (H)-specific TUs (Dataset S11). These 3 categories of TUs are collectively referred to as primate-specific TUs.
Validation of the Primate-Specific TU Dataset by LiftOver at a Relaxed Stringency Level Confirms That High Stringency Did Not Artificially Inflate the Number of Primate-Specific Tus.
A 0.95 liftOver minMatch parameter might identify TUs as primate-specific even if their exons possess nonprimate conservation with <95% sequence identity. To validate our 131 TUs, we performed liftOver with a relaxed 0.70 minMatch stringency to generate an alternate dataset of primate-specific TUs (Dataset S12 and Dataset S13). Whereas the less stringent minMatch = 0.70 increased the number of mapped exons, it resulted in more one-to-many matches in human-to-primate liftOver, and thus, in more E4 (Duplicated-in-new) outputs that we classify as unmapped exons. There was minimal reclassification of primate-specific TUs from the minMatch = 0.95 dataset into TUs detectable in nonprimates under minMatch = 0.70.
Under the relaxed minMatch stringency, there was no change to the number of “U/Y” TUs, consistent with indel, not extreme-divergence, origin for these TUs (0.95 v 0.70 Comparison, Dataset S12 and Dataset S13). We also performed a liftOver of the exons of all minMatch 0.70 primate-specific human TUs against mouse and rat at minMatch = 0.10. No hits were found, because the E1 liftOver message generally locates indels, instead of aligned-but-diverged regions.
Chromosomal Location Analysis and Expression Profiling of TUs.
Each TU's mapping was evaluated for pericentromeric or subtelomeric localization. The expression profile of each TU was checked by 4 procedures (SI Appendix). To test for protein-coding potential, we checked for the presence of an ORF within the representative cDNA or EST of each TU using the NCBI ORF Finder. A BLASTP analysis of the longest ORF was done vs. NCBI NR protein database to search for protein homologs.
We also investigated the 131 putative primate-specific TUs for differential gene expression in public Affymetrix U133 data. We manually reviewed each TU in the UCSC Genome Browser, verified probesets that represent the TU, and searched the NCBI GEO database for their expression datasets.
Supplementary Material
Acknowledgments.
We thank Distinguished Prof. Morris Goodman (Wayne State University) for communicating the manuscript; Assoc. Prof. Guillaume Bourque (National University of Singapore, Singapore) for critiques of the draft; and Maureen Osak, undergraduate student. (Hillsdale College, Hillsdale, MI) for assistance with data curation. This work was supported by the Genome Institute of Singapore Competitive Intramural Grant 06-114101, the Wayne State University Start-up Fund 23111R (to L.L.), and National Science Foundation HOMIND Award BCS 0827546 (to L.L. as Co-Investigator).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904569106/DCSupplemental.
References
- 1.Varki A, Altheide TK. Comparing the human and chimpanzee genomes: Searching for needles in a haystack. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 2.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 3.Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 4.King M, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 5.Kehrer-Sawatzki H, Cooper DN. Understanding the recent evolution of the human genome: Insights from human-chimpanzee genome comparisons. Hum Mutat. 2006;28:99–130. doi: 10.1002/humu.20420. [DOI] [PubMed] [Google Scholar]
- 6.Olson M, Varki A. Sequencing the chimpanzee genome: Insights into human evolution and disease. Nat Rev Genet. 2003;1:20–28. doi: 10.1038/nrg981. [DOI] [PubMed] [Google Scholar]
- 7.Nahon J. Birth of ‘human-specific’ genes during primate evolution. Genetica. 2003;118:193–208. doi: 10.1023/a:1024157714736. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R, et al. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA. 2009;15:85–96. doi: 10.1261/rna.1127009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frith M, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 10.Pang K, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Kent W. BLAT - The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmieder S, et al. Primate-specific spliced PMCHL RNAs are non-protein coding in human and macaque tissues. BMC Evol Biol. 2008;8:330. doi: 10.1186/1471-2148-8-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner L, Hoekstra HE. Causes and consequences of the evolution of reproductive proteins. Int J Dev Biol. 2008;52:769–780. doi: 10.1387/ijdb.082577lt. [DOI] [PubMed] [Google Scholar]
- 15.Levinson B, Kenwrick S, Lakich D, Hammonds G, Jr, Gitschier J. A transcribed gene in an intron of the human factor VIII gene. Genomics. 1990;7:1–11. doi: 10.1016/0888-7543(90)90512-s. [DOI] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 17.Yu P, Ma D, Xu M. Nested genes in the human genome. Genomics. 2005;86:414–422. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Bailey J, Yavor AM, Viggiano L, Misceo D, Horvath JE. Human-specific duplication and mosaic transcripts: Recent paralogous structure of chromosome 22. Am J Hum Genet. 2002;70:83–100. doi: 10.1086/338458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills R, Bennet A, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Makalowski W, Mitchell GA, Labuda D. Alu sequences in the coding regions of mRNA: A source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 21.Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 22.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pannetier M, Renault L, Jolivet G, Cotinot C, Pailhoux E. Ovarian-specific expression of a new gene regulated by the goat PIS region and transcribed by a FOXL2 bidirectional promoter. Genomics. 2005;85:715–726. doi: 10.1016/j.ygeno.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Dai H, et al. The evolution of courtship behaviors through the origination of a new gene in Drosophila. Proc Natl Acad Sci USA. 2008;105:7478–7483. doi: 10.1073/pnas.0800693105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platts A, Dix DJ, Chemes HE, Thompson KE. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum Mol Genet. 2007;16:763–773. doi: 10.1093/hmg/ddm012. [DOI] [PubMed] [Google Scholar]
- 26.Repping S, et al. High mutation rates have driven extensive structural polymorphism amony human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 27.Delbridge M, Graves JA. Mammalian Y chromosome evolution and the male-specific functions of Y chromosome-borne genes. Rev Reprod. 1999;4:101–109. doi: 10.1530/ror.0.0040101. [DOI] [PubMed] [Google Scholar]
- 28.Wu Q, Tommerup N, Ming Wang S, Hansen L. A novel primate specific gene, CEI, is located in the homeobox gene IRXA2 promoter in Homo sapiens. Gene. 2006;371:167–173. doi: 10.1016/j.gene.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 29.Takamatsu K, et al. Identification of two novel primate-specific genes in DSCR. DNA Res. 2002;9:89–97. doi: 10.1093/dnares/9.3.89. [DOI] [PubMed] [Google Scholar]
- 30.Marques A, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:1970–1979. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koonin E. The origin of introns and their role in eukaryogenesis: A compromise solution to the introns-early versus introns-late debate? Bio Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard K, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:1599–1611. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, et al. Repetitive element-mediated recombination as a mechanism for new gene origination in Drosophila. PLoS Genet. 2008;4:e3. doi: 10.1371/journal.pgen.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannert N, Kurth R. Retroelements and the human genome: New perspectives on an old relation. PNAS. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T, et al. Species-specific endogenous retroviruses shape the trancriptional network of human tumor suppressor protein p53. PNAS. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brosius J, Gould SJ. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc Natl Acad Sci USA. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long M, Betrán E, Thornton K, Wang W. The origin of new genes: Glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 38.Emerson J, Kaessmann H, Betrán E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 39.Vibranovski M, Zhang Y, Long M. Out of the X chromosomal gene movement in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner L, Hoekstra HE. Adaptive evolution of fertilization proteins within a genus: Variation in ZP2 and ZP3 in deer mice (Peromyscus) Mol Biol Evol. 2006;23:1656–1669. doi: 10.1093/molbev/msl035. [DOI] [PubMed] [Google Scholar]
- 41.Lanz R, McKenna NJ, Onate SA, Albrecht U, Wong J. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 42.Willingham A, et al. A strategy for probing the function of noncodingRNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 44.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toll-Riera M, et al. Origin of primate orphan genes: A comparative genomics approach. Mol Biol Evol. 2008;26:603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- 46.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollard K, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.