Abstract
Cryo-electron microscopy in conjunction with advanced image analysis was used to analyze the structure of the 26S proteasome and to elucidate its variable features. We have been able to outline the boundaries of the ATPase module in the “base” part of the regulatory complex that can vary in its position and orientation relative to the 20S core particle. This variation is consistent with the “wobbling” model that was previously proposed to explain the role of the regulatory complex in opening the gate in the α-rings of the core particle. In addition, a variable mass near the mouth of the ATPase ring has been identified as Rpn10, a multiubiquitin receptor, by correlating the electron microscopy data with quantitative mass spectrometry.
Keywords: ATPase, cryo-electron microscopy, mass spectrometry, protein degradation, AAA-ATPase
Cellular protein levels are regulated through protein synthesis and degradation. Given its destructive potential, intracellular protein degradation must be subject to rigorous spatial and temporal control. In eukaryotic cells, most proteins in the cytosol and the nucleus are degraded by the ubiquitin–proteasome system, and malfunctions of this system have been implicated in a wide variety of diseases (1–4). Unlike constitutively active proteases, the proteasome has the capacity to degrade almost any protein, yet it acts with exquisite specificity. The key stratagem is self-compartmentalization: the active sites of the proteolytic machine are sequestered from the cellular environment in the interior of the barrel-shaped 20S proteasome (5). Proteins destined for degradation are marked by ubiquitin, a degradation signal that is recognized by the regulatory 19S complexes (RPs) that associate with the core 20S proteasome or core particle (CP) to form the holoenzyme called the 26S proteasome. The 26S complex is a multimeric assembly with a mass of approximately 2.5 MDa (6).
The 20S core complex, which is highly conserved from archaea to higher eukaryotes, was amenable to structure determination by X-ray crystallography, and the resulting structures have revealed the salient features of this prototypical self-compartmentalizing protease (7–9). In contrast, the 26S holocomplex with 1 or 2 RPs attached to the barrel-shaped core has so far resisted all crystallization attempts. For protein complexes refractory to crystallization, cryo-electron microscopy (cryo-EM) of “single particles” is an alternative approach. The amounts of material required for cryo-EM are minute and, moreover, some degree of heterogeneity is tolerable. Although high resolution is often elusive, the medium resolution (1–2 nm) structures afforded by cryo-EM provide a platform for hybrid approaches which, in turn, can provide useful insights into the structure and mechanism of macromolecular assemblies (10).
Results and Discussion
MS Analysis of Purified 26S Proteasomes.
In the case of the 26S proteasome, progress in elucidating its structure has been hampered by the complexity of the system, its variability, and its fragility. 26S proteasome preparations from Drosophila melanogaster embryos (11, 12) appear biochemically and structurally homogeneous and have a well-defined complement of subunits. For an assessment of the subunit stoichiometry of the purified complex, we performed a quantitative mass spectrometry analysis. All canonical subunits (α1–7, β1–7, Rpt1–6, Rpn1–13, and the deubiquitinating enzyme UCH 37) were found to be present in equimolar amounts (Fig. 1), with the notable exception of Rpn10 (see below). Furthermore, we detected 2 additional ATPases, albeit in small amounts. Apparently they are paralogs of Rpt3 and Rpt6, and are exchangeable. Such a scenario is not without precedent; some of the CP β-type subunits are also exchangeable and can be up-regulated under certain physiological conditions (13). At the current resolution, the microheterogeneity caused by such exchanges can be neglected.
Fig. 1.
Subunit composition and relative abundance of the 26S proteasome subunits analyzed by quantitative mass spectrometry. Shown are the measured summed peptide intensities normalized by the protein molecular mass. In a second step the resulting intensities were standardized relative to the subunits of the 20S core particle (standard deviation is 0.24). All canonical subunits of the 26S proteasome were found to be present in equimolar amounts. In contrast, paralogs of Rpt3 and Rpt6 (indicated by asterisk) were detected in far substoichiometric quantities (<1:100), suggesting that these subunits are exchangeable. A subunit of the 26S base complex, Rpn10, was determined with a relative intensity of 1:4, indicative of a transient interaction with the 26S proteasome.
Drosophila 26S preparations, when applied to carbon film and negatively stained, appear rather homogenous, and double-capped complexes are abundant (12). However, suspension in a thin aqueous film before cryofixation causes partial disassembly even when great care is taken to avoid depletion of ATP, which is required for stability; this may be the result of multiple interactions with the water–air interface. Chemical fixation using glutaraldehyde or other cross-linkers is effective in reducing disintegration, but visual inspection of averages derived from such preparations indicates that some structural alterations do occur; therefore, we have not used any of these stabilizing measures.
Determination of the 26S Proteasome Density by Cryo-EM.
The intrinsic heterogeneity of 26S preparations and the fragility of the complexes give rise to a disturbing degree of variation in cryo-EM sample preparations. As a consequence, it is necessary to collect a large number of particle images, to be able to classify the particles into sufficiently populated classes representing the distinct states present in the sample. A recently developed automated image acquisition procedure (14) has enabled us to collect large data sets of 26S proteasomes, despite the relatively low abundance of double-capped holocomplexes.
A first 3D reconstruction (Fig. 2) was obtained by using a standard angular refinement procedure as implemented in the XMIPP software package (15). To minimize any model bias, we used a plain cylinder model as an initial reference to assign optimal projection directions and in-plane transformations to all experimental particles, and this model was iteratively refined until no further improvements were observed. The quality of the resulting reconstruction (left 2 columns in Fig. 2) can be assessed by using the low-pass filtered (2-nm cut-off) crystal structure of the 20S CP as an internal standard as well as for optimizing the isosurface threshold. The included total volume corresponds to a molecular mass of 2.5 MDa, which is in good agreement with the proteomics analysis (11). Moreover, the fit of the 20S CP into the cryo-EM density map, found after an extensive correlation search, is excellent [cross-correlation function (CCC) = 0.69]. Therefore, we conclude that the overall average structure with a nominal resolution of 2.5 nm is accurate. Nevertheless, given the dynamics of this molecular machine, we expect that significant local variations from the average structure do exist. Therefore, we calculated a 3D variance map (16) (right column in Fig. 2). Whereas no significant differences are observed in the CP, a number of conspicuous variances are seen in the RPs. One particularly prominent variance hotspot is visible in the RP near the interface between the base and the lid (see below).
Fig. 2.
Seven different views (from top to bottom) of the 26S proteasome reconstruction rotated around the pseudo-7-fold axis of the 20S CP. The left column shows isosurface representations of the entire 3D reconstruction of the 26S proteasome complex; the second from the left column shows isosurfaces of the reconstruction cut open along the pseudo-7-fold axis of the core particle to display its inner organization. The view direction corresponds to the C2-axis of the 20S CP. The isosurface threshold was set to include a protein mass of 720 kDa for the 20S CP. The superimposed crystal structure of the 20S CP from yeast was low-pass filtered to a resolution of 2.0 nm (red mesh) and fitted by an extensive-search correlation algorithm, demonstrating that the crystal structure and the cryo-EM structure are in excellent agreement. The second from the right column shows central slices in the x–y plane of the density distribution (mass is white) of the reconstruction. The right column shows a mesh representation of the reconstruction with an overlay isosurface in red highlighting the main variances.
Structural Analysis of the ATPase Density.
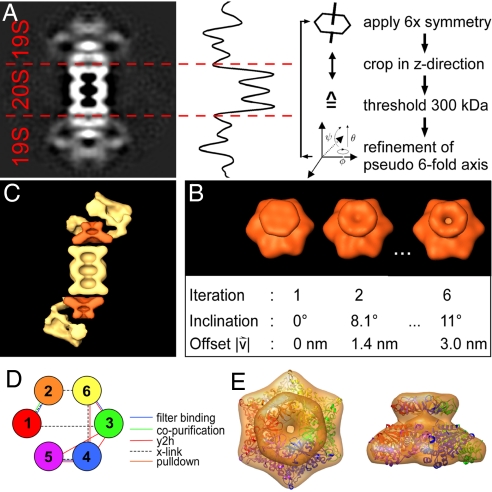
Little is currently known about the subunit topology of the RPs, beyond the assignment of Rpt1–6, Rpn1–2, and Rpn10 to the proximal (base) part and Rpn3–12 to the distal (lid) part (17). Interaction patterns between subunits, as derived from yeast 2-hybrid screens (18) or mass spectrometry (19), are valuable data per se but cannot in a straightforward manner be integrated into EM density maps. Therefore, we have made an attempt to determine the boundaries of a key component of the RP, namely the AAA-ATPase, within our reconstruction. Our procedure is based on the assumption that the ATPase subunits Rpt1–6 form a hexameric complex like most other AAA-ATPases. In fact, slices through a (nonsymmetrized) earlier tomographic reconstruction as well as the present single-particle reconstruction show 6 distinct centers of mass at the base of the RP mass (see Fig. S1), consistent with a (pseudo)-6-fold symmetry. Therefore, we have in a first step applied 6-fold symmetry to the entire complex around its long axis (Fig. 3A). The 2 RPs were then separated from the CP by identifying the depicted minima of a projection plot (see Fig. 3A) of the reconstruction. By assuming a protein density of 1.3 g/cm3 and a molecular mass of 300 kDa for the ATPase hexamer, a subvolume was extracted from the reconstructed volume. Finally, the exact position and orientation of the ATPase subvolume was refined by an exhaustive 6D (3 translational and 3 rotational parameters) search procedure. The search was iterated until no significant changes of the position and orientation parameters of the ATPase subvolume were found (Fig. 3B).
Fig. 3.
Segmentation strategy for the pseudo-6-fold ATPase within the 26S proteasome. In a first step (A) we segmented the 2 flanking 19S cap complexes from the 20S CP. The reconstruction volume of the 26S holocomplex was symmetrized by applying 6-fold symmetry along the pseudo-7-fold axis of the 20S CP. The central x–y slice of the resulting volume was extracted (Left). From this 2D image we calculated a 1D projection resulting in a line-profile (Center). The 2 flanking 19S cap complexes were cropped separately from the 20S by identifying the first 2 minima (in the line-profile) next to the core complex. This cut is indicated by red dashed lines and was applied to the entire symmetrized reconstruction volume, resulting in 2 subvolumes comprising one 19S cap complex each. Isosurfaces of the 19S subvolumes were calculated by setting the threshold such that a volume corresponding to 300 kDa was included. This segmented density was then fitted with an extensive 6D search (3 translational and 3 rotational parameters) into the original nonsymmetrized reconstruction volume; the shift and rotational parameters of this fitting procedure were determined and applied to the original volumes of the segmented 19S cap complexes This procedure was repeated until no significant changes in the alignment parameters were found. (B) An isosurface representation of 1 of the 6-fold-symmetrized 19S subvolumes (Upper) after the 1st, 2nd, and 6th iteration and the corresponding (Lower) alignment parameters. The inclination angles and the offset values are given relative to the pseudo-7-fold axis of the 20S core particle. (C) In a cut-open view of the 26S complex, the inclination of the ATPase toward the pseudo-7-fold axis of the 20S complex and the offset from the center are clearly visible. (D) The determined order of Rpt subunits (circles) allows placement of the coiled-coils on Rpt2, -3, and -5 and violates only 2 of 16 restraints (lines). The violated restraints are the yeast 2-hybrid interactions between Rpt4 and Rpt6 as well between Rpt3 and Rpt5. (E) The comparative model of the AAA-ATPase fits excellently into the 6-fold-symmetrized EM map. We did not attempt to model the linkers between N-domain (top) and AAA-domain.
The resulting structure has a hexagonal base, measuring 12 nm across the flats, and on top of it a “mouth” with a diameter of 7.9 nm; the total height is 7.2 nm. The segmented structure is highly reminiscent of the structure of the PAN complex, the archetypal activator of the 20S proteasome that is found in some archaea (20). Likewise reassuring is the fact that in our Drosophila data set there is a subpopulation of approximately 900 particles that upon averaging is almost congruent with the 20S CP plus the segmented ATPase, probably representing (dis-) assembly intermediates of the 26S holocomplex (Fig. S2). There is an additional rather elongate mass embracing the ATPase module and making contact to α-subunits of the CP; we tentatively assigned this mass to subunit Rpn1 and/or Rpn2 (Fig. S3). At the current resolution it is not possible to delineate the boundaries of Rpn1 or Rpn2 throughout the map. Our assignment is based on the reported physical interactions of Rpn1 and Rpn2 with the AAA-ATPase (21) and with the α-ring of the 20S CP (22), as well as on their predicted domain architecture; Rpn1 and Rpn2 likely consist of relatively small HEAT repeats (38–50 residues each) enabling the formation of extended structures (23).
The most intriguing finding is that the (pseudo-) 6-fold symmetry axis of both ATPase modules does not coincide with the (pseudo-) 7-fold symmetry axis of the CP; both ATP modules are shifted by ≈3.0 nm with respect to the axis of the CP. This finding indicates that the assembly mechanism of 20S/RP is markedly different from the complex of CP and 11S activator; the 11S particle is 7-fold symmetric and its symmetry axis coincides with the one of the CP in the 20S/11S holocomplex (24). For the 20S/AAA-ATPase the symmetry mismatch does not allow the ATPase subunits to interact with the subunits of the α-ring in an equivalent manner. In fact, only 3 of the ATPase subunits contain a motif at the C terminus that can insert into pockets in the α-subunits and induce gate opening (25). In addition to the different symmetry of the AAA-ATPases compared with the 11S activator, the requirement of ATP for the assembly of the 19S holocomplex in contrast to the 20S/11S complex makes it plausible that the architectures of these holocomplexes are not similar.
Interestingly, the axis of one of the 2 ATPase modules in the double-capped complexes is always inclined with respect to the 7-fold axis of the core complex, breaking the 2-fold symmetry of the holocomplex (Fig. 3C). We assume that the different inclinations at both cylinder ends reflect 2 different functional states of the RP. Our experimental observation is in excellent agreement with the “wobbling model” as proposed in ref. 26, which is based on theoretical considerations. In this model, an inclination of the ATPase relative to the 20S CP is implicated in the mechanism of opening the 20S CP gate.
Each subunit consists of a non-ATPase N-domain and an AAA-fold (27); each N-domain is further predicted to consist of a 25- to 60-residue N-terminal region, a coiled-coil, and an intermediate segment. The N-domain of the Archaeoglobus fulgidus ortholog PAN (PAN-N) could be crystallized as a homohexamer (41). We built comparative models of the N-domains and the AAA-domains of Rpt1–6 by using PAN-N and the AAA-folds of FtsH (PDB code 2ce7) and p97 (1e32) as structural templates. Next, we determined the configuration, that is, the order of Rpt1–6 in the hexamer by using these models as well as published biochemical interactions as determined by yeast 2-hybrid assays, chemical cross-linking, filter binding, copurification, and different pulldown experiments (Table S1). We built all possible arrangements of the ATPases and scored them on the basis of the proteomics data (see Materials and Methods). None of the models satisfied all experimental restraints, yet 12 of 120 possible configurations violated only 2 restraints of 16. Among these models, only 1 configuration, Rpt 1/2/6/3/4/5, allowed an invariant proline in Rpt2, -3, and -5 (corresponding to Pro-62 of PAN-N), to adopt a cis isoform, which is required for coiled-coil formation. Notably, this configuration differs from the previously proposed order Rpt1/2/6/4/5/3 (28), which violates 3 restraints. The difference between our configuration and the previously determined one can be attributed to interactions reported in the meantime. In addition, the narrow pore of the N-domain ring (≈11 Å) can be spanned by the cross-linkers that were used (28), which was presumably not anticipated (Fig. 3D). After establishing the order of the ATPases we rebuilt homology models for N- and AAA-hexamers and fitted them into the EM map (Fig. 3E). Using the EM map, we can thus obtain a model for the structure of N-domain and AAA-domain. An ambiguity remains because a (anti-clockwise) rotation of the N-domain by 60° results in a conformation that would also be compatible with the linker length.
RPN1-RPN2 Are Not Localized in the Center of the ATPase.
Recently, structural and biochemical studies performed with different mixtures of purified Rpn1, Rpn2, and 20S CP but in the absence of the ATPase module led to the proposal that Rpn1/Rpn2 may form a heterodimer of 2 stacked concentric rings, which is physically attached to the 20S CP (22). For the complete 26S proteasome, this model further implies that the Rpn2/Rpn1 dimer is localized in the center of the AAA-ATPases and might provide a direct physical link between substrate recruitment and proteolysis. However, the internal volume of our AAA-ATPase hexamer model could accommodate a maximum of ≈75 kDa, whereas the putative Rpn1/Rpn2 dimer has a molecular mass of ≈220 kDa. Moreover our EM map shows that the cavity enclosed by the ATPase harbors no significant protein mass. Therefore, our data on the 26S holocomplex do not support the model proposed for 20S/Rpn2/Rpn1 (22).
Proposed Position of RPN10.
The variance map (Fig. 2) indicates that a substantial mass may be present or absent in 1 of the 2 RPs. To elaborate on this observation, we performed a 3D maximum-likelihood classification (ML3D) on these data (29). The question arises as to the identity of the variable mass. The fact that it is found only in ≈50% of all particles analyzed and that it is present in only 1 of the 2 RPs implies that it is present in a substoichiometric amount of approximately 1:4. The only subunit found in a similar stoichiometry is Rpn10 (see Fig. 1). Rpn10 has a molecular mass of 42.6 kDa, which is in good agreement with the mass (60 ± 25 kDa) derived from the volume of the additional mass in the class averages, when using the mass of the 20S CP for calibration. Although the C-terminal ubiquitin-interacting motif of Rpn10 (approximately 1/3 of the sequence) was shown to be disordered in solution (30), most of the density of Rpn10 should show up in a low-resolution map. We also tried to confirm the position of Rpn10 by assembly modeling (31). In a preliminary approach, we represented all proteins within the 26S proteasome as strings of spheres, each representing a single protein domain, and built models that comply with the EM data and reported physical protein–protein interactions (Fig. S4). Although this approach did not yield a unique solution, it is notable that the proposed position of Rpn10 coincides with 1 of 3 predominant locations of Rpn10 in the models.
Taken together, our observations provide strong circumstantial evidence, albeit not a rigorous proof, that the variable mass is Rpn10. Interestingly, there is a whole body of experimental data suggesting that Rpn10 is a receptor for multiubiquitin chains and can bind to the 26S proteasome reversibly (32). Its location close to the mouth of the ATPase module appears to be well placed to deliver substrates for the ATP-dependent processing steps. At present, no high-resolution structure of Rpn10 is available that could be used for docking into the EM density. Given the fact that the C-terminal half of Rpn10 was shown to be highly flexible in solution, it might be difficult to obtain such a structure (30). However, it might be possible to locate Rpn10 by using ubiquitinated substrate bound to it and to take snapshots of early steps of substrate uptake and processing by the RPs.
Materials and Methods
Purification and Mass Spectrometry.
26S proteasomes from Drosophila were prepared as described in refs. 11 and 12. Fractions used for structural studies were loaded on a native gradient (2–15%) polyacrylamide gel and subjected to electrophoresis, and proteasome bands were excised and analyzed by mass spectrometry using a hybrid linear ion trap–orbitrap instrument (Thermo Fisher Scientific Inc.) (33). Although not as accurate as isotope-based technologies (34), the “label-free” proteomics approach used in this study achieves good quantitation results for high-intensity signals (35). The measured intensities for the 26S proteasome sample were found to be in the high-range region (109 to 1010 counts per second) where accuracy of the measured signals is high. We determined the stoichiometry of the sample by normalizing the sum of all of the identified peptides by the molecular masses of the individual proteins. The resulting intensities were standardized relative to the subunits of the 20S CP; only small variations (standard deviation of 0.24) were found, indicating that the procedure yields accurate enough results.
EM.
EM data were recorded in a fully automated manner using a microscope equipped with an energy filter that was operated in a 0-loss mode at an accelerating voltage of 300 kV. Magnification on the detector plane (CCD camera) was 82,500×, corresponding to a 0.36-nm pixel size in the object plane. The defocus was set to nominal values of 2 μm and 4 μm, respectively; at these settings, the first 0 of the contrast transfer function (CTF) is at 2.0 nm and 2.8 nm. Micrographs and their corresponding power spectra were visually inspected. Altogether, 5,134 micrographs of a total of 11,605 were selected for further processing. The contrast transfer functions were determined and outliers were set to a weighted average of successfully determined defocus values of the surrounding micrographs (for details see ref. 14). The selected electron micrographs were deconvoluted by a phase-flipping procedure described elsewhere (36). Particles showing the typical shape of the 26S proteasome holocomplex (20S core flanked by two 19S complexes) were selected in an interactive manner. Two particle stacks with a box size of 160 × 160 pixels were created, comprising 18,931 (2 μm underfocus) and 17,285 (4 μm underfocus) particles.
3D Variance Analysis and Classification.
The first 3D reconstruction was low-pass-filtered to a resolution of 6 nm and used to generate 3 seeds for subsequent ML3D classification that yielded 1 class with a strongly enhanced additional mass. This class was used to create 2 references to a subsequent (supervised) classification: an asymmetric reference with the additional mass and a C2-symmetrized reference without the additional mass. ML3D refinements of these references against the 2-μm and 4-μm defocus data sets served to separate the data into 2 classes, corresponding to 26S proteasomes with and without the additional mass and comprising 17,294 and 16,906 particles, respectively.
Both classes were refined separately by progressively reducing the angular sampling interval to 5° and finally 2°. Final reconstructions were obtained by using an ART with blobs algorithm (37) and the data from the 2-μm and 4-μm defocus subsets were combined by using an 3D Wiener filter (38), resulting in 2 models with a resolution of 20.7 Å (without additional mass) and of 21.3 Å (with additional mass) (Fig. 4 and Fig. S5).
Fig. 4.
Two subclasses of the 26S proteasome were identified. The top row shows isosurface representations of a subclass where an extra mass is visible (white arrows). The bottom row shows the second subclass where this extra mass is missing. From left to right, a 6-fold rotation around the pseudo-7-fold axis of the 20S particle was applied, resulting in 6 different views of the complex. The viewing direction corresponds to the C2-axis of the core particle. The isosurface threshold was set to include a protein mass of 720 kDa for the 20S CP.
AAA-ATPase Modeling.
First, we built comparative models of the N-domains and the AAA-domains of Rpt1–6 by using PAN-N and the AAA-folds of FtsH (PDB code 2ce7) and p97 (1e32) as structural templates using MODELLER (39). For the initial model we assumed the previously proposed order of the ATPases Rpt1/2/6/4/5/3 (28). Both rings were then fitted separately into the 6-fold-symmetrized EM map by using Chimera (40), whereby the rotation of the N-ring with respect to the AAA-ring was chosen such that domains of the same protein are adjacent.
We further elucidated the order of the Rpt1–6 subunits in the ring by assessing 5! AAA-ring configurations (the position of Rpt1 was kept fixed) against reported interactions found in the literature (Table S1). Specifically, each reported interaction between a subset of subunits was compiled into a conditional connectivity restraint (31) that penalizes deviation from an expected minimal distance between the corresponding Cα atoms by using a harmonic function. The excepted minimal distance between interacting subunits is a function of errors of the comparative model; in case the interaction restraint is derived from chemical cross-links, we added the length of the linker arm to the expected minimal distance. The 5! different AAA-ring configurations were sampled by rotating Rpt2–6 into all possible positions. For all models, the numbers of violated restraints as well as the score were determined. These configurations were then filtered for models that had Rpt2, -3, and -5 in the 2, 4, 6 positions of the ring. The comparative models were then rebuilt in the resulting unique conformation (1/2/6/3/4/5) and the rings were again fitted by using Chimera, yielding a correlation coefficient of 0.71.
Supplementary Material
Acknowledgments.
We thank Kornelius Zeth and Andrei Lupas for insightful discussions and for providing us with the atomic models of PAN-N. This work was supported by the 3-D Repertoire grant, the 3-D EM Network of Excellence grant, and the High throughput-3DEM grant, all within the Research Framework Program 6 (FP6) of the European Commission. A.S. was supported by the National Institutes of Health (R01 GM54762, R01 GM083960, and U54 RR022220), National Science Foundation (IIS-0705196), Hewlett-Packard, IBM, Intel, and NetApp. S.H.W.S. and J.M.C. were supported by the National Institutes of Health (HL070472) and by the Spanish Comision Interministerial de Ciencia y Tecnologia (BFU2004-00217), the Ministerio de Educación y Ciencia (CSD2006-00023, BIO2007-67150-C03-1 and -3), and Comunidad Autonoma de Madrid S-GEN-0166-2006). We acknowledge the Barcelona Supercomputing Center (Centro Nacional de Supercomputación) for providing computing resources. K.L. has been funded by the Clore Foundation Predoctoral Fellowship. F.F. is grateful for a long-term fellowship from the Human Frontier Science Program Organization.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905081106/DCSupplemental.
References
- 1.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. The early history of the ubiquitin field. Protein Sci. 2006;15:647–654. doi: 10.1110/ps.052012306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- 4.Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–711. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 6.Voges D, Zwickl P, Baumeister W. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 7.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 8.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 angstrom resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 9.Unno M, et al. The structure of the mammalian 20S proteasome at 2.75 angstrom resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 10.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 11.Hölzl H, et al. The regulatory complex of Drosophila melanogaster 26S proteasomes: Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol. 2000;150:119–129. doi: 10.1083/jcb.150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walz J, et al. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 13.Niedermann G. Immunological functions of the proteasome. Curr Top Microbiol Immunol. 2002;268:91–29. doi: 10.1007/978-3-642-59414-4_5. [DOI] [PubMed] [Google Scholar]
- 14.Nickell S, et al. Automated cryoelectron microscopy of “single particles” applied to the 26S proteasome. FEBS Lett. 2007;581:2751–2756. doi: 10.1016/j.febslet.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Scheres SHW, et al. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penczek PA, Yang C, Frank J, Spahn CMT. Estimation of variance in single-particle reconstruction using the bootstrap technique. J Struct Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 18.Davy A, et al. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2001;2:821–828. doi: 10.1093/embo-reports/kve184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: Insights from MS of intact complexes. PLoS Biol. 2006;4:1314–1323. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 21.Fu HY, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J Struct Biol. 2004;146:425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 Å structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Smith DM, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeki Y, Tanaka K. Unlocking the proteasome door. Mol Cell. 2007;27:865–867. doi: 10.1016/j.molcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Lupas A, Baumeister W, Hofmann K. A repetitive sequence in subunits of the 26S proteasome and 20S cyclosome. Trends Biochem Sci. 1997;22:195–196. doi: 10.1016/s0968-0004(97)01058-x. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch Biochem Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- 29.Scheres SHW, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 30.Wang QH, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 32.Kiss P, et al. Zn2+-induced reversible dissociation of subunit Rpn10/p54 of the Drosophila 26S proteasome. Biochem J. 2005;391:301–310. doi: 10.1042/BJ20050523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen JV, et al. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 35.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 36.Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct. 2002;31:303–319. doi: 10.1146/annurev.biophys.31.082901.134202. [DOI] [PubMed] [Google Scholar]
- 37.Marabini R, Herman GT, Carazo JM. 3D reconstruction in electron microscopy using ART with smooth spherically symmetric volume elements. Ultramicroscopy. 1998;72:53–65. doi: 10.1016/s0304-3991(97)00127-7. [DOI] [PubMed] [Google Scholar]
- 38.Schiske P. Image reconstruction by means of focus series. J Microsc. 2002;207:154. [PubMed] [Google Scholar]
- 39.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 40.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009 doi: 10.1016/j.molcel.2009.04.030. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.