Abstract
Fifteen percent of lung cancer cases occur in never-smokers and show characteristics that are molecularly and clinically distinct from those in smokers. Epidermal growth factor receptor (EGFR) gene mutations, which are correlated with sensitivity to EGFR-tyrosine kinase inhibitors (EGFR-TKIs), are more frequent in never-smoker lung cancers. In this study, microRNA (miRNA) expression profiling of 28 cases of never-smoker lung cancer identified aberrantly expressed miRNAs, which were much fewer than in lung cancers of smokers and included miRNAs previously identified (e.g., up-regulated miR-21) and unidentified (e.g., down-regulated miR-138) in those smoker cases. The changes in expression of some of these miRNAs, including miR-21, were more remarkable in cases with EGFR mutations than in those without these mutations. A significant correlation between phosphorylated-EGFR (p-EGFR) and miR-21 levels in lung carcinoma cell lines and the suppression of miR-21 by an EGFR-TKI, AG1478, suggest that the EGFR signaling is a pathway positively regulating miR-21 expression. In the never-smoker–derived lung adenocarcinoma cell line H3255 with mutant EGFR and high levels of p-EGFR and miR-21, antisense inhibition of miR-21 enhanced AG1478-induced apoptosis. In a never-smoker–derived adenocarcinoma cell line H441 with wild-type EGFR, the antisense miR-21 not only showed the additive effect with AG1478 but also induced apoptosis by itself. These results suggest that aberrantly increased expression of miR-21, which is enhanced further by the activated EGFR signaling pathway, plays a significant role in lung carcinogenesis in never-smokers, as well as in smokers, and is a potential therapeutic target in both EGFR-mutant and wild-type cases.
Keywords: apoptosis, microRNA, microarray, EGFR-TKI, therapeutic target
Approximately 10% to 25% of all lung cancer cases are not attributable to smoking (1, 2). Recent studies that pay specific attention to lung cancers in never-smokers have suggested that these cancers have characteristics distinct from those in smokers (2): G-to-T transversions of the p53 and K-ras mutations occur less frequently in lung adenocarcinomas from never-smokers than in those from smokers (3–6), and mutations of epidermal growth factor receptor (EGFR) gene are observed more frequently in never-smoker cases (6). A profiling of global gene expression in never-smoker lung cancers may provide novel molecular and clinical aspects in lung carcinogenesis. Although EGFR tyrosine kinase inhibitors (EGFR-TKIs), including gefitinib and erlotinib, currently are in clinical use and are preferentially effective in EGFR-mutant cases (7, 8), as many as 30% of EGFR-mutant cases and 90% of EGFR wild-type cases showed no therapeutic response to EGFR-TKIs (9). Therefore, identification of a new therapeutic target and development of a method to improve the EGFR-TKI therapy will be of critical importance for the better treatment of lung cancer.
MicroRNAs (miRNAs) are small, non-coding RNA molecules of about 18 to 25 nucleotides that frequently are located at chromosomal regions deleted or amplified in cancers, suggesting that miRNAs are a class of genes involved in human tumorigenesis (10). Expression levels of miRNAs are altered in various types of human cancers, including lung cancers (11–18). Recently, miRNAs have been demonstrated to be diagnostic and prognostic markers in leukemia, lung cancer, and colon cancer (13, 18–19). It also is suggested that miRNAs can be a therapeutic target in human cancers (20). We previously analyzed miRNA expression profiles of 104 lung cancers, 99 of which were from smokers, and found that high expression of miR-155, miR-21, and miR-106a, as well as low expression of let-7a, correlated with poor survival (18). In the present study we investigate a global expression profile of miRNAs in lung cancers from never-smokers. Comparisons of miRNA expression in never-smoker versus smoker cases and in EGFR wild-type versus mutant cases find different profiles of miRNA expression associated with smoking status and reveal EGFR-mediated regulation of miRNA expression. Our in vitro functional analyses also suggest that the inhibition of miR-21, whose up-regulation is associated with EGFR mutations, can be a potential therapeutic strategy in combination with EGFR-TKI treatment or by itself.
Results
MicroRNA Expression Profiles in Lung Cancers from Never-Smokers.
We examined miRNA expression profiles in 28 matched pairs of lung cancer and noncancerous lung tissues from never-smokers (Table 1 and supporting information (SI) Table S1) using the Ohio State miRNA microarray version 3.0 (21). In class comparison analysis using National Cancer Institute Division of Treatment and Diagnosis Biometric Research Branch (BRB) array tools, 18 miRNAs were found to be differentially expressed in cancers compared with noncancerous tissues [P < 0.01 with a false-discovery rate (FDR) of < 0.15)] (Table 2). The expression profiles of these 18 miRNAs distinguished cancer and paired noncancerous tissues with a prediction accuracy of 84% using the 3-nearest-neighbor algorithm and an accuracy of 82% using the support vector machine algorithm within BRB array tools (10-fold cross validation repeated 100 times). Expression levels of 5 miRNAs were higher in cancer tissues, with miR-21 enriched the most, at 2.35-fold. Expression levels of 13 miRNAs were lower in cancers, with miR-486 and miR-126* repressed the most, at 0.45-fold. The validity of the analysis was supported by the identification of a single miRNA by 2 different probes (miR-21, miR-521, and miR-516a), of 2 mature miRNAs generated from a single stem-loop pre-miRNA (miR-126 and miR-126*), and of more than 1 miRNA chromosomally clustered and possibly co-regulated (miR-30a and miR-30c on 6q13; miR-30b and miR-30d on 8q24.22; and miR-516a, miR-520, and miR-521 on 19q13.41)]. The mRNA microarray data of never-smoker lung adenocarcinoma cases (22) (http://www.ncbi.nlm.nih.gov/geo/, accession number = GSE10072) also showed that 2 host genes, TMEM49 and EGFL7 (Table 2), were differentially expressed in cancer and noncancerous tissues in the same directions as their resident miRNAs (miR-21 and miR-126/126*, respectively). The expression levels of 3 miRNAs (miR-21, miR-126, and miR-486) were examined by real-time quantitative RT-PCR (qRT-PCR) (Fig. S1). MiR-21 expression was significantly higher in cancer tissues than in noncancerous tissues (P < 0.05, paired t-test) (Fig. S1A), and miR-126 and miR-486 were expressed at significantly lower levels in cancers (each P < 0.05, paired t-test) (Fig. S1 B and C), further validating the results of the microarray analysis.
Table 1.
Characteristics of never-smoker patients with non-small cell lung cancer
| Characteristic | No. of patients |
|---|---|
| Histology | |
| Adenocarcinoma | 22 (78%) |
| Squamous cell carcinoma | 4 (14%) |
| Adenosquamous cell carcinoma | 1 (4%) |
| Unclassified | 1 (4%) |
| Stage | |
| I | 21 (75%) |
| II–IV | 7 (25%) |
| Age | |
| ≤ 65 | 18 (64%) |
| > 65 | 10 (36%) |
| Gender | |
| Female | 18 (64%) |
| Male | 10 (36%) |
| Race | |
| Caucasian | 19 (68%) |
| African American | 3 (11%) |
| Asian (Japanese) | 6 (21%) |
| EGFR gene status | |
| Wild-type | 22 (79%) |
| Mutant | 6 (21%) |
Table 2.
miRNAs differentially expressed in lung cancer tissues and normal lung tissues from 28 never-smokers
| Mature miR | Probe | Location | P-value† | FDR‡ | Type§ | Ratio¶ | Host gene‖ |
|---|---|---|---|---|---|---|---|
| miR-21 | hsa-mir-21-prec-17 | 17q23.1 | 3.0E-04 | 0.01 | Up | 2.35 | TMEM49 |
| miR-21 | hsa-mir-21–1 | 17q23.1 | 9.6E-04 | 0.03 | Up | 2.22 | TMEM49 |
| miR-141 | hsa-mir-141-prec-1 | 12p13.31 | 0.001 | 0.03 | Up | 1.50 | Intergenic |
| miR-210 | hsa-mir-210-prec | 11p15.5 | 0.002 | 0.06 | Up | 1.51 | Intergenic |
| miR-200b | hsa-mir-200b | 1p36.33 | 0.008 | 0.11 | Up | 1.39 | Intergenic |
| miR-346 | hsa-mir-346 | 10q23.2 | 0.009 | 0.12 | Up | 1.14 | GRID-1 |
| miR-126* | hsa-mir-126*-1 | 9q34.3 | 3.5E-05 | 0.01 | Down | 0.45 | EGFL7 |
| miR-126 | hsa-mir-126 | 9q34.3 | 0.004 | 0.07 | Down | 0.69 | EGFL7 |
| miR-30a | hsa-mir-30a-prec-1 | 6q13 | 1.4E-04 | 0.01 | Down | 0.61 | C6orf155 |
| miR-30d | hsa-mir-30d-prec-2 | 8q24.22 | 1.5E-04 | 0.01 | Down | 0.57 | Intergenic |
| miR-486 | hsa-mir-486 | 8p11.21 | 2.7E-04 | 0.01 | Down | 0.45 | Intergenic |
| miR-129 | hsa-mir-129–2 | 11p11.2 | 2.8E-04 | 0.01 | Down | 0.77 | Intergenic |
| miR-451 | hsa-mir-451–1 | 17q11.2 | 4.8E-04 | 0.02 | Down | 0.46 | Intergenic |
| miR-521 | hsa-mir-521–2 | 19q13.41 | 0.005 | 0.08 | Down | 0.84 | Intergenic |
| miR-521 | hsa-mir-521–1 | 19q13.41 | 0.005 | 0.08 | Down | 0.80 | Intergenic |
| miR-138 | hsa-mir-138–1-prec | 3p21.33 | 0.006 | 0.10 | Down | 0.72 | Intergenic |
| miR-30b | hsa-mir-30b-prec | 8q24.22 | 0.006 | 0.10 | Down | 0.58 | Intergenic |
| miR-30c | hsa-mir-30c-prec | 6q13 | 0.007 | 0.11 | Down | 0.61 | C6orf155 |
| miR-516a | hsa-mir-516a-1 | 19q13.41 | 0.008 | 0.11 | Down | 0.89 | Intergenic |
| miR-516a | hsa-mir-516a-2 | 19q13.41 | 0.010 | 0.12 | Down | 0.90 | Intergenic |
| miR-520 | hsa-mir-520 h | 19q13.41 | 0.009 | 0.12 | Down | 0.84 | Intergenic |
†miRNA-microarray analysis was performed using pairs of tumors and corresponding normal tissues from 28 never-smokers (P < 0.01).
‡False-discovery rate (FDR) < 0.15.
§Up, up-regulated in tumors compared with normal tissue; down, down-regulated in tumors compared with normal tissue.
¶Ratio of tumor to normal tissue.
Differential miRNA Profiles in Lung Cancers from Never-Smokers Versus Smokers.
To identify cancer-associated changes in miRNA expression that are related to smoking status, we compared the miRNA expression profiles of the present never-smoker cases with those of 58 smoker lung adenocarcinoma cases in our previous study (18) and 23 additional cases of lung adenocarcinoma in smokers (Table S2). We identified 5 miRNAs that commonly were changed in expression in never-smoker and smoker cases, among which was the increased miR-21 (Table S3). Although only 2 miRNAs, miR-138 and let-7c, were changed significantly (both were down-regulated) in never-smoker cases, the altered expression of 36 miRNAs was preferentially associated with smoker cases (Table S3), probably reflecting the more extensive genetic and epigenetic changes in smoker-derived lung cancers (23). By qRT-PCR we validated the specific down-regulation of miR-138 in never-smoker adenocarcinomas and the up-regulation of miR-21 and the down-regulation of miR-126* irrespective of smoking status (Fig. S2). Interestingly, miR-138 is located at chromosome 3p21.33, a candidate locus that carries a lung cancer suppressor gene (24), and was reported to target the human telomerase reverse transcriptase gene (hTERT) (25) on which a variety of cellular and viral oncogenic mechanisms act (26). A role for this miRNA in the etiology of lung cancers from never-smokers deserves further investigation.
MiRNA Expression Profiles Associated with EGFR Gene Mutations.
The status of the EGFR gene was determined by DNA sequencing in the 28 lung cancer tissues from never-smokers, and 6 cases were found to have the activating mutations of EGFR in the tyrosine kinase domain (Table S1). The class comparison analysis of miRNA expression between 22 EGFR wild-type and 6 EGFR-mutant cases found 12 miRNAs that were significantly more or less abundant in EGFR-mutant cases (P < 0.01 with FDR < 0.15) (Table 3). Of the 12 miRNAs, 10 (miR-21, miR-210, miR-486, miR-126, miR-126*, miR-138, miR-521, miR-451, miR-30d, and miR-30a) were changed in the same direction as in cancer versus noncancerous tissues (Table 2), suggesting that EGFR mutations may reinforce the aberrant regulation of some miRNAs associated with lung carcinogenesis in never-smokers. MiR-21 and miR-486, which were most up-regulated and most down-regulated, respectively, in cancerous versus noncancerous tissues, again showed the greatest difference between EGFR-mutant and wild-type cancers (≈1.7-fold and 0.60-fold, respectively). Although the qRT-PCR data shown in Fig. S1 reflect a limited number of cases, limiting our ability to show a statistically significant difference between EGFR-mutant and wild-type cases in the expression of miR-21, miR-126, or miR-486, the 3 cases expressing the highest levels of miR-21 in cancer (cases 24, 25, and 28) had the activating mutation of EGFR (Fig. S1A and Table S1).
Table 3.
miRNAs differentially expressed in EGF-mutant and wild-type lung cancers from never-smokers
| Mature miR | Probe | Location | P-value† | FDR‡ | Type§ | Ratio§ |
|---|---|---|---|---|---|---|
| miR-21 | hsa-mir-21–1 | 17q23.1 | 0.001 | 0.05 | Up | 1.79 |
| hsa-mir-21-prec-17 | 17q23.1 | 0.001 | 0.04 | Up | 1.67 | |
| miR-210 | hsa-mir-210-prec | 11p15.5 | 0.007 | 0.13 | Up | 1.20 |
| miR-129 | hsa-mir-129–2 | 11p11.2 | 0.001 | 0.05 | Up | 1.06 |
| miR-486 | hsa-mir-486 | 8p11.21 | 0.001 | 0.04 | Down | 0.60 |
| miR-126 | hsa-mir-126–2 | 9q34.3 | 0.003 | 0.08 | Down | 0.69 |
| miR-126* | hsa-mir-126*-1 | 9q34.3 | 0.0005 | 0.04 | Down | 0.70 |
| miR-138 | hsa-mir-138–1-prec | 3p21.33 | 0.004 | 0.10 | Down | 0.69 |
| miR-521 | hsa-mir-521–1 | 19q13.41 | 0.005 | 0.11 | Down | 0.81 |
| hsa-mir-521–2 | 19q13.41 | 0.003 | 0.08 | Down | 0.82 | |
| miR-451 | hsa-mir-451–1 | 17q11.2 | 0.002 | 0.07 | Down | 0.81 |
| miR-141 | hsa-mir-141-prec-1 | 12p13.31 | 0.004 | 0.10 | Down | 0.85 |
| miR-30d | hsa-mir-30d-prec-2 | 8q24.22 | 0.001 | 0.04 | Down | 0.93 |
| miR-30a | hsa-mir-30a-prec-1 | 6q13 | 0.001 | 0.04 | Down | 0.95 |
Class comparison analysis was performed between 22 EGFR wild-type and 6 mutant tumors (P < 0.01).
‡False-discovery rate (FDR) < 0.15.
§Up, up-regulated in EGFR-mutant compared with wild-type tumors; down, down-regulated in EGFR-mutant compared with wild-type tumors.
¶Ratio of tumors with mutant EGFR to tumors with wild-type EGFR.
Expression of miR-21 and the Status of EGFR Signaling in Lung Cancer Cell Lines.
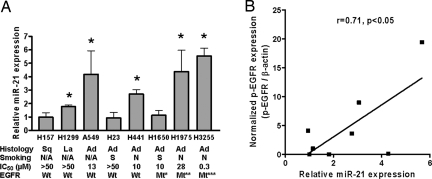
Because of its most remarkable increase in cancer as compared with noncancerous tissues and its association with EGFR mutations, an indicator of sensitivity to EGFR-TKIs (9), miR-21 was chosen for further analyses. To investigate a correlation between miR-21 expression levels and the status of EGFR signaling pathway, 8 non-small cell lung cancer (NSCLC) cell lines were examined in Western blot (Fig. S3 A, B, and C) and qRT-PCR analyses (Fig. 1A). Among them, 3 adenocarcinoma cell lines (H3255, H1975, and H1650) were mutant for EGFR, as reported (8, 27–29). These 3 EGFR-mutant cell lines had high levels of phosphorylated EGFR (p-EGFR), as well as increased amounts of total EGFR protein and induction of phosphorylated Akt (p-Akt) (Fig. S3C), consistent with the constitutive activation of the EGFR signaling pathway in these cells (28, 29). Of the 3 cell lines, H3255 and H1975, but not H1650, expressed elevated levels of miR-21 (Fig. 1A). Of 5 EGFR wild-type cell lines, 3, either with (H441) or without (A549 and H1299) detectable levels of p-EGFR (Fig. S3 A and B), also expressed significantly higher levels of miR-21 than seen in control untransformed cells (Fig. 1A). The quantitative comparison of miR-21 and p-EGFR levels showed a significant positive correlation between these 2 factors (Pearson's correlation, r = 0.71, P < 0.05) (Fig. 1B). These results suggest that the activated EGFR signaling pathway can be functionally associated with miR-21 up-regulation. It also was noteworthy that miR-21 expression and/or EGFR status correlated with sensitivity (indicated as IC50) to an EGFR-TKI, AG1478 (Fig. 1A); the 5 cell lines showing AG1478-inhibited cell proliferation either had mutant EGFR (H1650) or expressed > 2-fold increased levels of miR-21 (H441 and A549), or both (H3255 and H1975). We selected 2 lung adenocarcinoma cell lines derived from never-smoker cancers for the functional assays of miR-21: H3255 with high sensitivity to AG1478 (IC50, 0.3 μM), mimicking never-smoker lung cancer cases with mutant EGFR and the highest levels of miR-21 (e.g., case numbers 24, 25, and 28 in Fig. S1A and Table S1); and H441 with intermediate sensitivity to AG1478 (IC50, 10 μM), mimicking never-smoker lung cancer cases with wild-type EGFR but with significantly increased levels of miR-21 (e.g., case numbers 5 and 23 in Fig. S1A and Table S1).
Fig. 1.
MiR-21 expression in human lung cancer cell lines. (A) MiR-21 expression levels were analyzed by qRT-PCR and expressed relative to hTERT-immortalized normal human bronchial epithelial cells (HBET2) (defined as 1.0, not shown). Data were mean ± SD from 3 independent experiments. The suppressive effects of AG1478 on cell growth were determined by MTS assay and indicated as IC50. *, P < 0.05 when compared with HBET2, Student's t-test. Ad, adenocarcinoma; La, large cell carcinoma; Mt*, EGFR mutant ΔE746-A750 (in-frame deletion of codons 746 to 750); Mt**, L858R (substitution from leucine to arginine at codon 858) and T790M (substitution from threonine to methionine at codon 790); Mt***, L858R; N/A, information not available; N, derived from never-smoker cases; S, derived from smoker cases; Sq, squamous cell carcinoma; Wt, EGFR wild-type. (B) Correlation between miR-21 expression and p-EGFR levels (Pearson's correlation, r = 0.71, P < 0.05). The miR-21 data were from panel A, and the p-EGFR data were obtained by quantitatively analyzing the results shown in Fig. S3.
Activated EGFR Signaling Enhances miR-21 Expression.
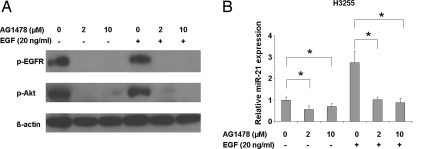
EGFR-mutant H3255 cells were treated with AG1478 in the presence or absence of EGF (Fig. 2). AG1478 at either 2 μM or 10 μM effectively inhibited the EGFR signaling under conditions with or without EGF ligand stimulation, as shown by diminished p-EGFR and p-Akt (Fig. 2A), consistent with the IC50 value of 0.3 μM in this cell line. The levels of miR-21 expression in the absence of EGF were significantly repressed by treatment with either concentration of AG1478 (P < 0.01, paired t-test) (Fig. 2B, Left). The addition of EGF resulted in ≈2.5-fold up-regulation of miR-21 expression, which still was inhibited back to the basal levels by treatment with either concentration of AG1478 (P < 0.05, paired t-test) (Fig. 2B, Right). These results indicate that miR-21 expression is positively regulated by the activated EGFR signaling in cancer cells with an activating EGFR mutation and that EGFR-TKIs can effectively repress the aberrantly increased miR-21. In H441 cells with wild-type EGFR, AG1478 at 10 μM (equivalent to the IC50 value in this cell line), but not at 2 μM, significantly repressed miR-21 expression (P < 0.05, paired t-test) (Fig. S4). Thus, the activated signaling from wild-type EGFR in H441 cells (Fig. S3B), probably through a self-produced TGF-alpha stimulation (28), also can be inhibited by AG1478, resulting in the repression of miR-21.
Fig. 2.
AG1478 represses miR-21 expression. H3255 lung adenocarcinoma cells, characterized by a high expression of miR-21 and EGFR mutation, were serum starved for 24 h and then were grown in either the presence or absence of AG1478 (2 μM or 10 μM) for 2 h with or without following exposure to 20 ng/mL EGF for 15 min. (A) The effect of AG1478 on p-EGFR and p-Akt expression. β-actin was a loading control. (B) MiR-21 expression levels analyzed by qRT-PCR after the AG1478 treatments (2 μM or 10 μM) with or without EGF ligand stimulation. MiR-21 expression levels were expressed as values relative to untreated cells in the absence of EGF. Data were mean ± SD from 4 independent experiments. *, P < 0.05, paired t-test.
Antisense Inhibition of miR-21 Induces Apoptosis in Cooperation with EGFR-TKI.
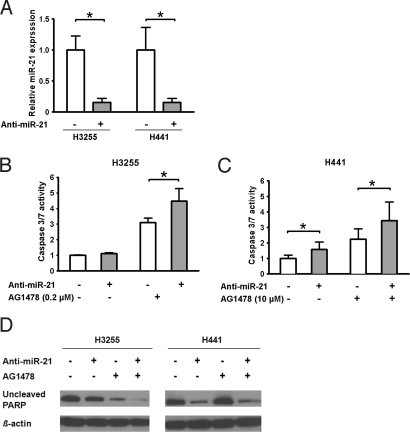
H3255 and H441 cells were transfected with an antisense oligonucleotide targeting miR-21 (anti-miR-21). The antisense-mediated repression of miR-21 in these cells was confirmed by qRT-PCR (Fig. 3A). Because miR-21 reportedly has an anti-apoptotic activity (30), we used an assay measuring caspase-3 and caspase-7 enzymatic activities to determine whether inhibition of miR-21 induces apoptosis in these cells (Fig. 3 B and C). In H3255 cells, anti-miR-21 alone did not induce apoptosis (Fig. 3B, Left). Notably, however, when used in combination with AG1478 at 0.2 μM (a concentration slightly lower than the IC50 value), anti-miR-21 significantly enhanced AG1478-induced apoptotic response (Fig. 3B, Right). In H441 cells, anti-miR-21 by itself resulted in a significant increase in apoptotic response (Fig. 3C, Left), although it was less effective than AG1478 treatment at 10 μM (a concentration equivalent to the IC50 value). Similar to the combinational effect observed in H3255 cells, anti-miR-21 further enhanced apoptotic response induced by 10 μM of AG1478 in H441 cells (Fig. 3C, Right). The effect of anti-miR-21 on apoptosis was substantiated further by Western blot analysis of poly (ADP-ribose) polymerase (PARP), a main cleavage target of caspase-3 in the apoptotic response (Fig. 3D). The amounts of uncleaved PARP were markedly decreased in H3255 cells treated with both anti-miR-21 and AG1478 and in H441 cells treated with anti-miR-21 in the presence or absence of AG1478, where anti-miR-21 caused significant increases in caspase 3/7 activities.
Fig. 3.
Inhibition of miR-21 enhances AG1478-induced apoptosis. (A) Cells were transfected with 40 nM of anti-miR-21 (+) or control oligonucleotide (anti-EGFP) (−) for 72 h and examined by qRT-PCR. The expression levels of miR-21 after transfection of anti-miR-21 were expressed as the relative values to control. Data were mean ± SD from 3 independent experiments. *, P < 0.05, paired t-test. (B, C) Cells (H3255 or H441) were transfected with 40 nM of anti-miR-21 (+) or anti-EGFP (−) for 72 h and then were grown in the presence or absence of 0.2 μM of AG1478 for 24 h (H3255) or 10 μM for 72 h (H441). The activities of caspase 3/7 were expressed as the values relative to the activities of cells without anti-miR-21 and AG1478. Data were mean ± SD from at least 4 independent experiments. *, P < 0.05, Student's t-test. (D) Uncleaved PARP was evaluated by Western blot analysis. Cells were transfected with anti-miR-21 or anti-EGFP as described previously and then were grown in the presence or absence of 2 μM of AG1478 for 72 h. β-actin was a loading control.
Discussion
This study shows molecular characteristics of lung cancers in never-smokers and smokers: (i) changes in expression of a relatively small number of miRNAs are involved in lung carcinogenesis in never-smokers; (ii) EGFR mutations may reinforce some of these changes in miRNA expression, e.g., an increase in miR-21; (iii) miR-138 on 3p21.33, a chromosomal region carrying a long-sought lung cancer suppressor gene, is down-regulated preferentially in never-smoker cases; and (iv) miR-21 is one of the most aberrantly increased miRNAs in both never-smoker and smoker cases. There was no significant difference in the expression levels of miR-21 when stage I cases (n = 21) were compared with stage II, III, and IV cases (n = 7) (data not shown), suggesting that increased miR-21 expression is an early event in lung carcinogenesis. These findings identified miR-21 as a major miRNA that may play an oncogenic role in lung carcinogenesis and prompted us to choose it as a candidate for molecular targets in treatment of lung cancers in never-smokers as well as in smokers. Given the relationship between EGFR mutations and miR-21 up-regulation, we also hypothesized that this miRNA might have implications in improving EGFR-TKI therapy, whose effectiveness is correlated with EGFR gene status and smoking history of the patients (2, 4–6, 31).
Although high levels of miR-21 expression have been reported in various types of human tumors (19, 30, 32), including lung cancer from both smokers (18) and never-smokers (this study), the mechanism that up-regulates miR-21 during carcinogenesis is not well understood. In addition to the miRNA microarray data showing higher levels of miR-21 in EGFR-mutant cases (Table 3), the in vitro analyses using NSCLC cell lines showed that the activated EGFR signaling up-regulates miR-21 expression. A statistically significant positive correlation was observed between miR-21 expression levels and p-EGFR levels in NSCLC cell lines (Fig. 1B). Furthermore, the treatment with the EGFR-TKI (AG1478) inhibited miR-21 expression in 2 NSCLC cell lines with elevated p-EGFR, EGFR-mutant H3255 (Fig. 2) and EGFR wild-type H441 (Fig. S4), providing a mechanistic link between the activated EGFR signaling pathway and the aberrant up-regulation of miR-21 and a therapeutic basis for inhibition of miR-21 in lung cancers with EGFR activation. STAT3 reportedly signals IL-6–induced up-regulation of miR-21 in multiple myeloma cells (33). However, siRNA-mediated knockdown of endogenous STAT3 expression did not affect miR-21 levels in H441, H1650, and H1975 cells (data not shown), suggesting that STAT3 does not play a primary role in the EGFR signaling-induced up-regulation of miR-21 in NSCLC cells. It remains to be examined whether activator protein-1 (AP-1), which is activated by the EGFR signaling (34) and activates the miR-21 transcription through the binding to the promoter (35), is responsible for the increased expression of miR-21 in NSCLC cells. Nevertheless, there also should be EGFR-independent mechanisms to control miR-21 expression, because miR-21 was expressed abundantly in A549 cells without EGFR mutation or p-EGFR (Figs. 1A and S3B), and no increased miR-21 expression was observed in H1650 cells with EGFR mutation and increased p-EGFR (Figs. 1A and S3C). An increased copy number or amplification of the chromosomal region carrying miR-21 (17q23.1) (36) may be an EGFR-independent mechanism.
Antisense oligonucleotide-mediated knockdown of miR-21 induced or enhanced apoptotic responses in 2 NSCLC cell lines, H3255 and H441 (Fig. 3) probably recapitulating some lung cancer cases from never-smokers. H3255 and H441 both expressed elevated levels of miR-21 (Fig. 1A) but had biologically and genetically different features. H3255 was highly responsive to EGFR-TKI (Fig. 1A), expressed high levels of p-EGFR (Figs. 2A and S3C), and had mutated and amplified EGFR (Fig. 1A) (37). H441 was less responsive to EGFR-TKI (Fig. 1A), expressed low levels of p-EGFR (Fig. S3B), and had wild-type EGFR (Fig. 1A). As previously reported in other types of human cancer cells (32, 33), the antisense inhibition of miR-21 by itself led to increased apoptotic response in H441 cells (Fig. 3 C and D), suggesting that miR-21 also can be a therapeutic target in lung cancers. Importantly, in both cell lines, anti-miR-21 significantly enhanced the apoptotic response induced by AG1478 (Fig. 3 B and C). The lack of effect of anti-miR-21 alone in H3255 cells (Fig. 3B) may indicate that the combinational use of anti-miR-21 and EGFR-TKI is required to attenuate effectively the constitutively activated EGFR signaling pathway to cell survival, which is evidenced by the highest levels of p-EGFR (Fig. S3C) and miR-21 (Fig. 1A). Although EGFR-TKIs are in wide clinical use for lung cancer (38), and inhibition of oncogenic miRNAs is a new promising approach in cancer therapy (39), this study reveals that the combination of these 2 therapeutic strategies can be significantly more effective than either alone. The finding is of particular importance in developing better treatment for lung cancer patients of non-Asian ethnicity, who tend to be less responsive to the current EGFR-TKI therapy (40). This study also has potential clinical implications in preventing and rescuing acquired EGFR-TKI resistance in NSCLC, an issue with important clinical relevance. Besides a secondary T790M mutation (41) and acquired MET amplification (42), selection of an EGFR wild-type subpopulation on a background of wild-type/mutant mixture leads to acquired EGFR-TKI resistance in NSCLC (43). The combinatorial use of EGFR-TKI and anti-miR-21 could prevent and rescue such acquired resistance caused by the selection for wild-type EGFR, because anti-miR-21 is effective on both EGFR wild-type and mutant tumor cells. Successful i.v. administration of a locked nucleic acid-modified antisense miRNA in primates (44) supports the feasibility of in vivo targeting miRNAs in therapy of human diseases.
Last, our lists of dysregulated miRNAs (Tables 2 and 3) include a number of other miRNAs that may have an oncogenic or tumor-suppressive role in lung carcinogenesis, e.g., up-regulated miR-141 (45), up-regulated miR-210 (46), down-regulated miR-126 (47), and down-regulated miR-486 (48). Further studies will address the roles of other individual miRNAs in lung carcinogenesis and a possible therapeutic relevance of overexpression and/or knockdown of multiple miRNAs.
In conclusion, lung cancers in never-smokers have a characteristic profile of miRNA expression. MiR-21 is a downstream effector of the activated EGFR signaling pathway and can be a therapeutic target in lung cancers with and without EGFR mutations. Antisense inhibition of miR-21 may improve clinical response to EGFR-TKI therapy.
Materials and Methods
Clinical Samples and Cell Lines.
Lung cancer tissues and corresponding noncancerous tissues were obtained from never-smokers who had undergone surgical resection between 2000 and 2004 at the University of Maryland Medical Center (n = 15) or the Mayo Clinic (n = 7) in United States or at the Hamamatsu University School of Medicine (n = 6) in Japan (Tables 1 and S1). Institutional review board approval and written informed consent from all patients were obtained at each collection site. H3255 was provided by Dr. Bahar of the National Cancer Institute. A549, H23, H441, H1650, H1975, H157, and H1299 were purchased from American Type Culture Collection (ATCC). hTERT-immortalized normal human bronchial epithelial cells (HBET2) were established in our laboratory.
Microarray Analysis.
Microarray analysis was performed as previously described (21, 49). Briefly, 5 μg of total RNA was used for hybridization on miRNA microarray chips containing 389 probes in triplicate (Ohio State microRNA microarray version 3.0). Processed slides were scanned using a PerkinElmer ScanArray XL5K Scanner. With statistical software R (http://www.r-project.org/), only spot values that were not flagged by the image quantification software GenePix Pro 6.0.1.00 (www.moleculardevices.com/pages/software/gn_genepix_pro.html) and whose foreground intensities were larger than background intensities were used. The remaining spots then were normalized by locally weighted scatterplot smoothing (LOESS), and duplicate spots were averaged. The preprocessed and normalized data then were imported into BRB-ArrayTools version 3.5.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html). Finally, 291 miRNAs with non-missing log values present in more than 75% of the samples were selected. More information is available in SI Materials and Methods.
Real-Time RT-PCR Analysis.
Expression of mature miRNAs was examined by qRT-PCR analysis using a TaqMan Human MicroRNA Assay kit and a PRISM 7700 Sequence Detector System (Applied Biosystems). RNU6B was an endogenous control (#4373381, Applied Biosystems). Gene expression data (mean ± SD from triplicate samples) were shown as 2−ΔΔCT (50).
Cell Treatment and Growth Inhibition Assay.
To evaluate the effect of AG1478 on the EGFR signaling pathway and miR-21 expression levels, lung cancer cell lines were serum starved for 24 h, incubated in the presence or absence of AG1478 (2 μM or 10 μM; Calbiochem) for 2 h, and then for an additional 15 min in the presence or absence of EGF (20 ng/mL; Promega). Growth inhibition was assessed by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl) 2H-tetrazolium, inner salt (MTS) assay (Dojindo). Cells (5,000/well) were seeded into 96-well plates, and increasing concentrations of AG1478 (0, 0.4, 2.0, 10, and 50 μM) were added. After incubation for 72 h at 37 °C, MTS was added and incubated for 2 h at 37 °C; then absorbance at 450 nm was measured. The IC50 value was defined as the concentration needed for 50% reduction of the growth.
Western Blot Analysis.
Cells were lysed in buffer containing 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 0.1% SDS, 1% Nonidet P-40, and 0.5% sodium-deoxycholate. Ten μg of proteins were separated by gel electrophoresis on 10% gels, transferred to nitrocellulose membranes, and detected by immunoblotting using a chemiluminescence system (GE Healthcare Bio-Sciences Corp.). The images were quantified using National Institutes of Health ImageJ1.40g (http://rsb.info.nih.gov/ij/). Antibodies are in SI Materials and Methods.
Oligonucleotide Transfection and Apoptosis Assay.
Cells were transfected with the oligonucleotides (at a final concentration of 40 nM) using LipofectAMINE 2000 reagent (Invitrogen). After 72 h, the cells were incubated in the presence or absence of 0.2 μM of AG1478 for 24 h (H3255) or 10 μM of AG1478 for 72 h (H441). Activities of caspase-3 and caspase-7 were analyzed using ApoONE Homogeneous Caspase 3/7 Assay (Promega). Each experiment was done in triplicate and at least 4 times independently. The data were shown as mean ± SD. Oligonucleotide sequences are in SI Materials and Methods.
Statistical Analysis.
The paired t-test identified miRNAs that were differentially expressed in lung cancer tissues and normal lung tissues (P < 0.01, FDR < 0.15). We also identified miRNAs that were differently expressed in EGFR-mutant and wild-type lung cancers using the F-test (P < 0.01, FDR < 0.15). The paired t-test was used to analyze differences in miRNA expression in tumors and corresponding normal tissues for qRT-PCR data. Graphpad Prism v5.0 (Graphpad Software Inc.) analysis was used for the Pearson's correlation. All statistical tests were 2-sided, and statistical significance was defined as P < 0.05.
Supplementary Material
Acknowledgments.
We thank Dr. Kensuke Kumamoto for helpful discussions, Drs. Raymond T. Jones, Andrew Borkowski, and Mark J. Krasna for sample collection and pathology reports, and Audrey Salabes for interviewing the lung cancer patients. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microRNA microarray data have been deposited in the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/, GSE14936).
This article contains supporting information online at www.pnas.org/cgi/content/full/0905234106/DCSupplemental.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Mountzios G, Fouret P, Soria JC. Mechanisms of disease: Signal transduction in lung carcinogenesis—a comparison of smokers and never-smokers. Nature Clinical Practice Oncology. 2008;5:610–618. doi: 10.1038/ncponc1181. [DOI] [PubMed] [Google Scholar]
- 3.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 4.Vähäkangas KH, et al. p53 and K-ras mutations in lung cancers from former and never-smoking women. Cancer Res. 2001;61:4350–4356. [PubMed] [Google Scholar]
- 5.Mounawar M, et al. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non-small cell lung cancers in relation to smoking history. Cancer Res. 2007;67:5667–5672. doi: 10.1158/0008-5472.CAN-06-4229. [DOI] [PubMed] [Google Scholar]
- 6.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nature Reviews Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 7.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 8.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 9.Uramoto H, Mitsudomi T. Which biomarker predicts benefit from EGFR-TKI treatment for patients with lung cancer? Br J Cancer. 2007;96:857–863. doi: 10.1038/sj.bjc.6603665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inamura K, et al. let-7 microRNA expression is reduced in bronchioloalveolar carcinoma, a non-invasive carcinoma, and is not correlated with prognosis. Lung Cancer. 2007;58:392–396. doi: 10.1016/j.lungcan.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 14.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 17.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 18.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. J Am Med Assoc. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weidhaas JB, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CG, Calin GA, Volinia S, Croce CM. MicroRNA expression profiling using microarrays. Nature Protocols. 2008;3:563–578. doi: 10.1038/nprot.2008.14. [DOI] [PubMed] [Google Scholar]
- 22.Landi MT, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE. 2008;20:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 25.Mitomo S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horikawa I, Barrett JC. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis. 2003;24:1167–1176. doi: 10.1093/carcin/bgg085. [DOI] [PubMed] [Google Scholar]
- 27.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 28.Tracy S, et al. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 29.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 30.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toyooka S, et al. The impact of sex and smoking status on the mutational spectrum of epidermal growth factor receptor gene in non small cell lung cancer. Clin Cancer Res. 2007;13:5763–5768. doi: 10.1158/1078-0432.CCR-07-0216. [DOI] [PubMed] [Google Scholar]
- 32.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 33.Löffler D, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 34.Li J, et al. Differential requirement of EGF receptor and its tyrosine kinase for AP-1 transactivation induced by EGF and TPA. Oncogene. 2003;22:211–219. doi: 10.1038/sj.onc.1206102. [DOI] [PubMed] [Google Scholar]
- 35.Fujita S, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Łastowska M, et al. Breakpoint position on 17q identifies the most aggressive neuroblastoma tumors. Genes Chromosomes Cancer. 2002;34:428–436. doi: 10.1002/gcc.10089. [DOI] [PubMed] [Google Scholar]
- 37.Engelman JA, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepherd FA, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 39.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: A potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thatcher N, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: Results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;24:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 42.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 43.Jiang SX, et al. EGFR genetic heterogeneity of nonsmall cell lung cancers contributing to acquired gefitinib resistance. Int J Cancer. 2008;123:2480–2486. doi: 10.1002/ijc.23868. [DOI] [PubMed] [Google Scholar]
- 44.Elmén J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 45.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 46.Foekens JA, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B, et al. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009 doi: 10.1016/j.lungcan.2009.01.010. in press. [DOI] [PubMed] [Google Scholar]
- 48.Gal H, et al. MIR-451 and imatinib mesylate inhibit tumor growth of glioblastoma stem cells. Biochem Biophys Res Commun. 2008;376:86–90. doi: 10.1016/j.bbrc.2008.08.107. [DOI] [PubMed] [Google Scholar]
- 49.Liu CG, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmittgen TD, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.