Abstract
Carotenoid pigments in plants fulfill indispensable functions in photosynthesis. Carotenoids that accumulate as secondary metabolites in chromoplasts provide distinct coloration to flowers and fruits. In this work we investigated the genetic mechanisms that regulate accumulation of carotenoids as secondary metabolites during ripening of tomato fruits. We analyzed two mutations that affect fruit pigmentation in tomato (Lycopersicon esculentum): Beta (B), a single dominant gene that increases β-carotene in the fruit, and old-gold (og), a recessive mutation that abolishes β-carotene and increases lycopene. Using a map-based cloning approach we cloned the genes B and og. Molecular analysis revealed that B encodes a novel type of lycopene β-cyclase, an enzyme that converts lycopene to β-carotene. The amino acid sequence of B is similar to capsanthin-capsorubin synthase, an enzyme that produces red xanthophylls in fruits of pepper (Capsicum annum). Our results prove that β-carotene is synthesized de novo during tomato fruit development by the B lycopene cyclase. In wild-type tomatoes B is expressed at low levels during the breaker stage of ripening, whereas in the Beta mutant its transcription is dramatically increased. Null mutations in the gene B are responsible for the phenotype in og, indicating that og is an allele of B. These results confirm that developmentally regulated transcription is the major mechanism that governs lycopene accumulation in ripening fruits. The cloned B genes can be used in various genetic manipulations toward altering pigmentation and enhancing nutritional value of plant foods.
Carotenoids comprise a large group of pigments that are ubiquitous in nature. In all photosynthetic organisms they carry out indispensable functions in light harvesting systems and in photosynthetic reaction centers. In higher plants, carotenoids play additional roles in providing distinct yellow, orange, and red colors to certain organs, such as flowers and fruits, to attract animals for pollination and for dispersal of seeds. In these tissues unique carotenoids that are synthesized as secondary metabolites accumulate to high concentrations and are stored within the chromoplasts.
Carotenoids play an important role in human nutrition because of their pro-vitamin A activity. However, there are other significant health benefits that are attributed to carotenoids (1–3), which are thought to be associated with their activity as antioxidants. Carotenoids have many industrial applications as food and feed additives, in cosmetics and pharmaceuticals.
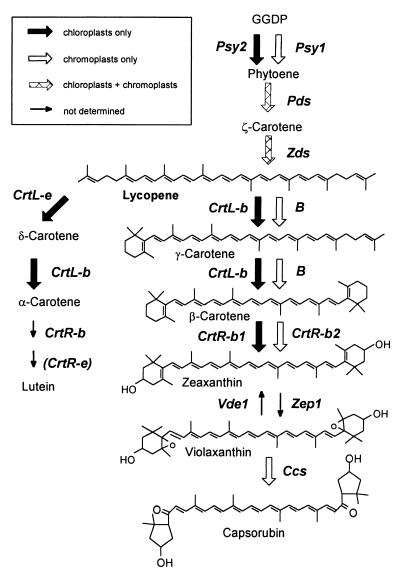
Carotenoids are produced via the general isoprenoid biosynthetic pathway that takes place in chloroplasts of photosynthetic tissues and in chromoplasts of fruits and flowers. Molecular characterizations unequivocally defined the enzymatic steps in the pathway (Fig. 1). All enzymes are nuclear-encoded and most of them have been cloned in recent years (4, 5). Tomato is a favorite model system for studying the regulation of carotenoid biosynthesis because of the dramatic color changes that occur during fruit development. In the early stages the fruits are green and consist of essentially the same carotenoids as green leaves, i.e., mainly β-carotene, lutein, and violaxanthin. At the “breaker” stage of ripening lycopene begins to accumulate and its concentration increases 500-fold in ripe fruits, reaching ca. 70 μg/g fresh weight (6). During this process the transcription of Psy and Pds, which encode phytoene synthase and phytoene desaturase, respectively, are up-regulated (7–9), whereas the mRNA of CrtL-b and CrtL-e, which encode lycopene β-cyclase and ɛ-cyclase, respectively, disappears (10, 11). Transcriptional regulation of carotenoid biosynthesis genes also appears to be the key regulatory mechanism in flowers (12–14).
Figure 1.
The carotenoid biosynthesis pathway in tomato. CrtL-b, lycopene β-cyclase; CrtL-e, lycopene ɛ-cyclase; CrtR-b, β-ring hydroxylase, CrtR-e, ɛ-ring hydroxylase; GGDP, geranylgeranyl diphosphate; Ggps, geranylgeranyl diphosphate synthase; Pds, phytoene desaturase; Psy, phytoene synthase; Vde, violaxanthin deepoxidase; Zds, ζ-carotene desaturase; Zep, zeaxanthin epoxidase.
Many single-gene mutations that affect fruit pigmentation have been isolated in tomato since the late 1940s (see: http://tgrc.ucdavis.edu/). The molecular basis of three of them has been determined: yellow-flesh (r), a loss-of-function mutation in the Psy-1 gene (15); Delta, a dominant mutation in the gene CrtL-e (11); and high-pigment-2 (hp-2), a mutation in the tomato homolog of the Arabidopsis signal transduction gene Det1 (16).
This work focuses on the elucidation of the molecular basis of two color mutations in tomato, Beta and old-gold (og). Beta is a partially dominant, single-locus mutation that causes an orange color in the fully ripened fruit because of the accumulation of β-carotene at the expense of lycopene (Fig. 2). In the wild type, β-carotene constitutes 5–10% of total fruit carotenoids, whereas in Beta it is 45–50% and can exceed 90% in combination with the Beta-modifier (MoB) gene. The B allele originated in wild Lycopersicon species and was introduced into the cultivated tomato by crossings (17). Two recessive allelic mutations, old-gold (og) and old-gold-crimson (ogc), have the same phenotype of deep red fruits that lack β-carotene and tawny orange flowers (Fig. 2). These mutations have been identified independently in Lycopersicon esculentum (18). The loci B and ogc previously were found to be tightly linked on chromosome 6 of the tomato linkage map (19–21).
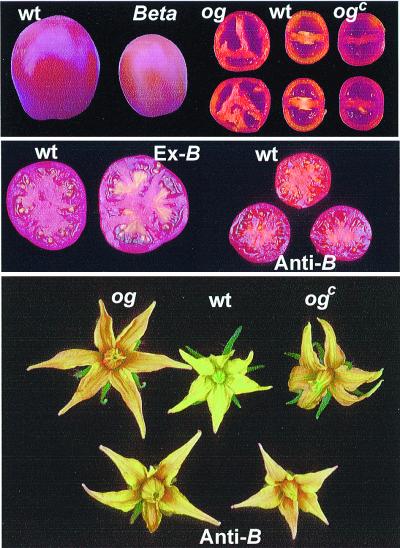
Figure 2.
Fruits and flowers of wild type (wt) and mutants Beta, old-gold (og), and old-gold crimson (ogc). Ex-B, transgenic expression of B in wt plants; anti-B, transgenic wt expressing of antisense of B.
We report here on the cloning and molecular characterization of the genes B, og, and ogc. We show that all three genes are allelic in a locus that encodes a novel fruit- and flower-specific lycopene β-cyclase, an enzyme that converts lycopene to β-carotene. The phenotype of Beta results from increased expression of the B gene during fruit ripening, whereas og and ogc are null mutants in the locus B.
Methods
Bacteria and Plants.
Escherichia coli strain XL1-Blue was used in all experiments that are described in this work. Agrobacterium tumefacience strain EHA105 was used for transformation of tomato plants.
Tomato (L. esculentum) CV M82 served as the wild-type strain in constructing the mapping population, as well as for the fruit-ripening measurements. The introgression lines IL 6–2 and IL 6–3 (22) were used as the source of Beta and for the fine mapping of B. Tomato cultivar VF-36 was used as the host for gene transformation. The og and ogc mutants lines LA3179 and LA348, respectively, were kindly provided by Roger Chetelat, the Tomato Genetics Resource Center, University of California, Davis.
Isolation of DNA from Tomato, Restriction Fragment Length Polymorphism (RFLP) Mapping, and Positional Cloning.
Genomic DNA was prepared from 5 g of leaf tissue as described (22). RFLP in the tomato genomic DNA was done by using markers CT-193, TG275, TG279, TG578 (23), and TM-16 (24).
A tomato genomic yeast artificial chromosome (YAC) library (25) was screened with markers TM16 and TG275, and three overlapping YAC clones, #310, #271 and #153, were identified. DNA sequences from the ends of the inserts in these YACs were amplified by PCR and were used as RFLP probes in screening DNA from recombinant plants. DNA fragments from the insert of YAC 310 were subcloned in the lambda vector λ-gt11 (24). Two phage clones, designated B1 and B3, cosegregated with B and mapped to the end of YAC 310. An ORF with similarity to the Ccs gene from pepper was identified in the insert of B1, after hybridization with Ccs and sequence analysis of the insert DNA. This ORF was used to screen a genomic library of wild-type tomato (cv VF36) in the lambda vector EMBL3 and a cosmid genomic library of Lycopersicon pennellii. A single positive phage clone and a single positive cosmid clone were isolated. Tomato (cv VF36) cDNA libraries from ripe fruits and flower tissues were screened by using B1 clone as a probe. Altogether 3 million phage plaques were screened. Three identical positive phages were cloned from a fruit library; each contained a 1,300-bp stretch of the cDNA, which lacked the 5′ end. More cDNAs were obtained from flowers of wild type (VF-36) and Beta (IL 6–3) by using reverse transcription (RT) followed by PCR. Sequence analysis was done as described (11).
Expression of B in Escherichia coli.
Plasmids pACCRT-EIB and pDCAR for expressing bacterial carotenoid biosynthesis genes in E. coli have been described (11, 26). To express B in E. coli a 1,502-bp DNA fragment, beginning from the initiation codon, was amplified by PCR from the cDNA clone of B and inserted in the EcoRV site of the vector pBluescript KS− in the sense orientation with respect to the lac promoter. The recombinant plasmid was designated pBPEN. In a similar way a 1,502-bp fragment from the wild-type allele, b, was inserted into pBluescript KS−, and the recombinant plasmid was designated pBESC.
Pigment Extraction and Analysis.
Extraction of carotenoids from bacteria and plants followed previously described protocols (11). Saponification of pigments was done in ethanol/KOH (60% wt/vol), 9:1 for 16 h at 4°C. The carotenoids were extracted with ether after addition of NaCl to a final concentration of 1.2%. Analysis by HPLC has been described (11). Carotenoids were identified by their characteristic absorption spectra and typical retention time, which corresponded to standard compounds of lycopene, β-carotene, and zeaxanthin. Quantification was done by integrating the peak areas by using the millenium chromatography software (Waters).
Measurement of mRNA by RT-PCR.
Protocols for RNA extraction and quantification by RT-PCR have been described (11). Total RNA was isolated from fruit, flowers (corolla) or leaves, by using the TRI-reagent protocol (Molecular Research Center, Cincinnati). The amplification procedure by PCR consisted of 20 cycles of 1 min at 95°C, 1 min at 59°C, and 1 min at 72°C. For the purpose of quantification 5 μCi of 32P-dCTP (specific activity 3,000 Ci mmol−1) was included in the PCR mixture. Various initial concentrations of mRNA, ranging over 9-fold difference, were used to demonstrate a linear ratio between concentration of the template cDNA (corresponding to the mRNA) and the final PCR products. To eliminate the possibility that genomic DNA contaminated the RNA samples, a control RT reaction was carried out without adding reverse transcriptase. The control samples were amplified by 22 cycles of PCR amplification.
The following primers were used for the PCR amplification: for Pds, 5′-TTGTGTTTGCCGCTCCAGTGGATAT-3′ (forward) and 5′-GCGCCTTCCATTGAAGCCAAGTAT-3′ (reverse); for CrtL-b, 5′-GGCTTCTCTAGATCTCTTGTTCAG-3′ (forward) and 5′-CCGATTCCAACGACTCTCTGA-3′ (reverse); for B, 5′-GGGTAATGAGCCATATTTAAGGG-3′ (forward) and 5′-TAGGATCCAGATCAAAGAAAGCG-3′ (reverse).
Products of the PCR amplification were separated by electrophoresis in 7% polyacrylamide gels. The gels were dried and autoradiographed. The amount of DNA was determined by measuring the radioactivity by using PhosphorImager (macbas 2.5, FUJIX, Tokyo) and exposure to x-ray film.
Construction of Transgenic Plants.
For transgenic expression of the B gene in L. esculentum we constructed a plasmid, pBPGEN, by inserting a 4,784-bp XhoII–PvuII fragment of genomic DNA from L. pennellii into the Ti vector pBI 101 (CLONTECH) that was cut with Ecl136II and BamHI. This insert contained the entire coding sequence of B plus 1,599 bp upstream of the initiating ATG and 1,687 bp downstream of the termination codon.
For antisense silencing of B in transgenic plants we constructed a Ti recombinant plasmid, pANTIB. A 420-bp fragment was amplified by PCR from the gene B by using the primers 5′-CATTTTCTACGAGCTCCACCTCCCTCC-3′ (forward) and 5′-TTTGGCCACATCTAGAGTGGTGAAGGG-3′ (reverse), which contain restriction sites of SacI and XbaI, respectively. This DNA, which starts 63 bp upstream of the initiation ATG codon, was inserted into the XbaI and SacI-digested pBI121 plasmid (CLONTECH). Thus the B sequence was placed under the cauliflower mosaic virus 35S promoter in the antisense orientation.
The pBI-derived plasmids, pANTIB and pBPGEN, were introduced into Agrobacterium tumefascience cells by electroporation. Tomato cotyledons were inoculated with Agrobacterium, following a standard protocol (27).
Results
Carotenoid Composition in Fruits and Flowers of Beta and old-gold.
The color of tomato fruits begins to change from green to red at the breaker stage of ripening. This transition is caused by the degradation of chlorophyll and the disappearance of xanthophylls and β-carotene, which occur concomitantly with the accumulation of lycopene. To characterize pigment composition in tomatoes the carotenoids were analyzed in the ripe stage of fruit development (red stage) in wild type, old-gold, and Beta (Fig. 2 and Table 1). The major carotenoid in wild-type fruits is lycopene (ca. 90%) with small amounts of β-carotene (5–10%), lutein, and phytoene. Fruits of old-gold essentially lack β-carotene whereas in Beta they contain up to 45% β-carotene. These data indicate that β-carotene biosynthesis in tomato fruits is conversely altered by B and og.
Table 1.
Carotenoid composition in fruits and flowers of various tomato lines
| Lines | Phytoene/ phytofluene | Lycopene | β-car. | Lutein | β,β-xanthophylls | |
|---|---|---|---|---|---|---|
| Fruits | WT | 1 | 90 | 7 | 2 | 0 |
| B | 3 | 50 | 46 | 1 | 0 | |
| og | 3 | 96 | <1 | 1 | 0 | |
| Ex-B#1 | 12 | 72 | 16 | 0 | 0 | |
| Ex-B#2 | 5 | 79 | 16 | 0 | 0 | |
| Ex-B#3 | 4 | 68 | 28 | 0 | 0 | |
| Ex-B#4 | 11 | 55 | 34 | 0 | 0 | |
| Ex-B#5 | 4 | 59 | 37 | 0 | 0 | |
| Ex-B#6 | 7 | 50 | 43 | 0 | 0 | |
| Anti-B#1 | 4 | 95 | <1 | 1 | 0 | |
| Anti-B#2 | 2 | 95 | 2 | 1 | 0 | |
| Anti-B#3 | 2 | 94 | 3 | 1 | 0 | |
| Anti-B#4 | 1 | 97 | 1 | 1 | 0 | |
| Flowers | WT | 0 | 0 | 1 | 5 | 94 |
| og | 6 | 13 | 0 | 4 | 77 | |
| Anti-B#1 | 8 | 10 | 0 | 6 | 76 | |
| Anti-B#2 | 6 | 5 | 0 | 5 | 84 | |
| Anti-B#3 | 4 | 3 | 0 | 5 | 88 | |
| Anti-B#4 | 6 | 12 | 0 | 4 | 78 |
WT, L. esculentum CV VF36; ex-B, L. esculentum (VF36) expressing a transgenic B (numbers refer to individual transformants); anti-B, L. esculentum CV VF36 expressing antisense B. Fruit carotenoids were extracted from the placenta.
The yellow xanthophylls constitute 99% of the total carotenoids that accumulate in tomato flowers. No difference was detected between flowers of wild type and Beta. However, the petals and anthers of og accumulate ca. 20% of lycopene and its precursor intermediates.
Map-Based Cloning of the B Gene.
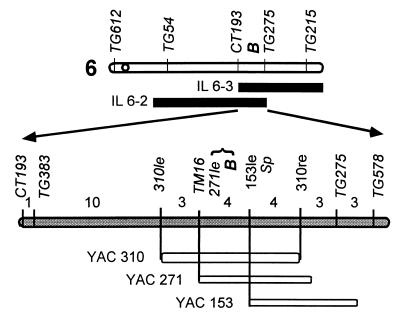
B was previously mapped to chromosome 6 of tomato in proximity to the self-pruning (sp) locus (19, 24). IL 6–2 and IL 6–3 both carried the B locus, indicating that the target gene maps to a genomic region that overlaps between the lines. IL6–2 and IL 6–3 (BB; SpSp) (22) were crossed with L. esculentum cv M-82 (bb;spsp), and the F1 hybrids were selfed to create F2 populations segregating for the Beta phenotype. A total of 1,335 F2 plants were scored for the B and sp phenotypes and for the RFLP markers CT193 and TG578 that flank the target trait (23, 24). Twenty-eight recombinant plants were identified between the flanking markers. The recombinants were further screened with RFLP probes surrounding B (Fig. 3). One RFLP marker, TM16, that cosegregated with B was used to screen the YAC library and identify YAC clones #310 and #271; YAC 153 was isolated by using TG275 as a probe. Further mapping with the YAC ends established the location of B to a relatively small region in the 200-kb YAC 310 that spanned 11 recombinants (Fig. 3).
Figure 3.
Positional cloning of the gene B. Fine mapping of chromosome 6 was done with RFLP markers indicated above the map (centromere position appears as a circle). Chromosomal segments in IL 6–3 and IL 6–2 that overlap B are depicted as black bars. The genetic map, based on the number of recombinants between CT193 and TG578, was derived from scoring a population of 1,335 F2 plants obtained from a cross between Beta and the wild-type M-82. The relative positions of YAC 310, YAC 271, and YAC 153 are given below the map as empty bars.
DNA fragments from the insert of YAC 310 were subcloned in the lambda vector λ-gt11. An ORF with similarity to the Ccs gene from pepper (28) was identified in the insert of one clone, B1, after hybridization with Ccs and sequence analysis of the insert DNA. This DNA was used to clone the homologous genomic and cDNA sequences from wild-type L. esculentum (cv VF36) and L. pennellii. The 1,502-bp cDNAs from VF36 (b allele) and L. pennellii (B) were subcloned in the plasmid vector pBluescript KS−, and the recombinant plasmids were designated pBESC and pBPEN, respectively. DNA sequence comparison between cDNA and genomic sequences revealed no intron interference in the genomic sequence of the gene B. DNA blot hybridization with total genomic DNA indicated that B exits in a single copy in the tomato genome (data not shown).
B Encodes Lycopene β-Cyclase.
Lycopene-accumulating E. coli cells, carrying the plasmid pACCRT-EIB (11) were cotransfected with plasmids pBESC or pBPEN, which express the cDNA of b (wild type) or B, respectively. Carotenoids were extracted and analyzed by HPLC. Expression of either b or B brought about synthesis of β-carotene that reached up to 10% of the total carotenoids in E. coli (data not shown). There was no significant difference in the β-cyclase activity between the two alleles.
Expression of the B-type lycopene cyclase was also tested in E. coli that accumulated δ-carotene. E. coli cells carrying either pBESC or pBPEN together with plasmid pDCAR (11) produced up to 28% α-carotene (β,ɛ-carotene). These results confirm that B encodes a bona fide lycopene β-cyclase, which catalyzes the symmetric formation of two β-ionone rings on the linear lycopene molecule to produce β-carotene and also can participate in the formation of α-carotene.
Sequence Analysis of B Alleles from Wild Type and Beta.
Nucleotide sequence analysis of the insert in pBESC (b) revealed an ORF of 499 codons, which encodes a polypeptide of a calculated molecular mass of 56.4 kDa (GenBank accession no. AF254793). The amino acid sequence of the polypeptide encoded by b is 53.2% identical to the tomato lycopene β-cyclase, CRTL-B (10) (see Fig. 7, which is published as supplementary material on the PNAS web site, www.pnas.org). Surprisingly, the amino acid sequence of this polypeptide is 86.1% identical to the enzyme capsanthin-capsorubin synthase (CCS) from pepper (Capsicum annum) (28). Comparison of the amino acid sequence of B from L. pennellii and b from L. esculentum showed that they are 98% identical with only 12 conservative amino acid substitutions. Sequence analysis of the genomic clone, revealed a 1,497-bp ORF, not interrupted by introns.
Expression of B During Fruit Ripening in Wild-Type and Beta Plants.
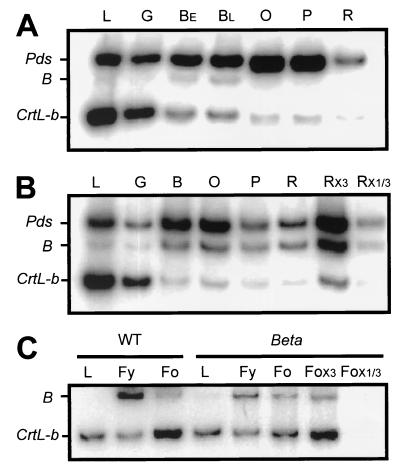
Previously, we have shown that while the expression of the genes Psy and Pds is up-regulated during tomato fruit ripening, the mRNA of CrtL-b (lycopene β-cyclase) and CrtL-e (lycopene ɛ-cyclase) decrease dramatically (10, 11). In flowers the expression of Psy, Pds, and CrtL-b is up-regulated whereas CrtL-e is not expressed (11). To determine the regulation of expression of B in tomato we measured the mRNA level of B in leaves and flowers and at different stages of fruit ripening. The results indicate that in wild-type plants B is expressed exclusively in chromoplast-containing tissues of flowers and fruits (Fig. 4 A and C). Expression in flowers is high, whereas in fruits it is low and limited to a short period around the breaker stage. However, the level of mRNA of B in fruits of Beta increases dramatically during the breaker stage and remains high during later developmental stages (Fig. 4B). No significant change in mRNA levels was observed in the flowers of Beta compared with the wild type (Fig. 5C). It is interesting to note that in flowers the two lycopene β-cyclases, B and CrtL-b, are simultaneously expressed at a similar proportion.
Figure 4.
Expression of B, CrtL-b, and Pds in fruits and flowers of tomato. The relative amount of mRNA of B, CrtL-b, and Pds was measured concomitantly by RT-PCR from total RNA isolated from different stages of fruit ripening and flower development of wild-type (WT) L. esculentum (M82) and from the mutant Beta. PCR-amplified DNA fragments were separated by PAGE and autoradiographed. (A) WT fruits. (B) Beta fruits. (C) Flowers of wild type and B. Fruit ripening stages: G, mature green; B, breaker; BE, early breaker; BL, late breaker; O, orange; P, pink; R, red; Rx3 and Rx1/3 are samples that contained three times or one-third the total RNA from red fruit. L, leaf. Flowering stages: Fy, preanthesis flower; Fo, postanthesis flower.
Figure 5.
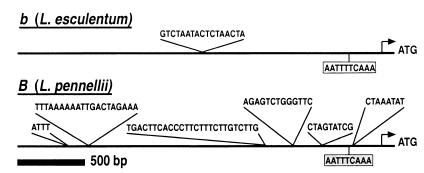
Organization of the genomic sequences upstream to the promoters of B and b.
Comparison of the Promoters in the Wild-Type and B Alleles.
The difference in the levels of mRNA of B, between fruits of the wild-type and Beta, could possibly be determined by transcriptional regulation of the B gene. To further investigate this possibility we analyzed the DNA sequence of the promoter regions of b (wild type) and B alleles. Comparison of the two alleles revealed that in contrast to the high conservation in the coding sequence, significant differences exist in their promoter regions. Although the overall nucleotide sequence is 97% conserved between B and b along 2,520 nts upstream to the coding sequence, there are six additional sequence elements that exist in B but are absent from b, and one sequence element that exists in b but not in B (Fig. 5).
Transgenic Expression of B in Wild-Type Tomato.
A 4,784-bp fragment of genomic DNA, which contains the coding sequence of B from L. pennellii together with 1,599 bp upstream to the initiation codon and 1,687 bp downstream of the stop codon, was introduced into wild-type L. esculentum (cv VF-36). Fruits of transgenic plants appeared with a red-colored pericarp, but contained bright orange placenta and collumela (Fig. 2). The composition of carotenoid in these tissues was typical for B fruits, up to 43% β-carotene and 50% lycopene (Table 1). Measurement of mRNA in different tissues of the transgenic fruits showed the B is up-regulated specifically in the placenta and collumela (data not shown). This result confirmed that B is indeed responsible for the B phenotype.
The Phenotypes of og and ogc Are Caused by Annulment of B.
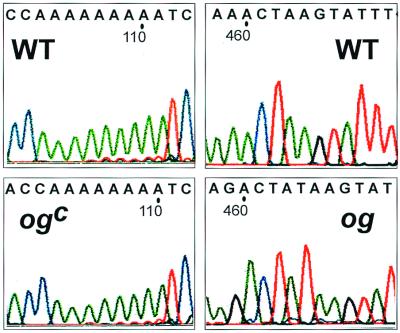
Previously og was mapped to chromosome 6 in close proximity to B (20, 21). We cloned the b gene from old-gold and old-gold-crimson plants and analyzed their sequence. Two distinct frame-shift mutations were detected in the coding sequence of these genes in codons (og)155 and 3 + (ogc) (Fig. 6). Consequently, no functional enzyme could be produced in these mutants.
Figure 6.
Frame-shift mutations in the B gene from old-gold (og) and old-gold-crimson (ogc). WT, wild type.
To demonstrate that loss of the B function is responsible for the og phenotype we have taken a gene-silencing approach to eliminate the B activity in wild-type tomato. To this end we expressed a 398-bp fragment from the b cDNA in reverse orientation by using the cauliflower mosaic virus 35S promoter in transgenic wild-type tomato plants (cv VF-36). Transgenic plants expressing antisense b had normal growth characteristics. The only phenotypic changes were detected in the pigmentation of fruits and flowers. In all plants the color of the flowers was altered from yellow to tawny-orange, typical of old-gold (Fig. 2). This change was caused by accumulation of up to 12% of lycopene (Table 1). The deep red appearance of the fruits in the transgenic plants was identical to the typical phenotype of og (Fig. 2), because of the elimination of β-carotene and increase in lycopene concentration (Table 1). The phenotype of the antisense B was inherited as a single dominant gene, which cosegregated with the transgenic insertion of the antisense B sequence (data not shown). Taken together these results establish that β-carotene biosynthesis in tomato fruits is carried out by the lycopene β-cyclase that is encoded by the gene B, and that og and ogc are null alleles of b.
Discussion
The Molecular Basis of Beta.
Several lines of evidence prove that the gene B encodes Beta: (i) The B sequence cosegregated with the Beta phenotype. (ii) The property of the B-encoded enzyme as lycopene β-cyclase fits the phenotype of β-carotene accumulation in fruits of Beta. (iii) The detection of elevated mRNA level of B in Beta fruits explains the higher β-carotene accumulation. (iv) Transgenic expression of B in wild type plants results in a phenotype that is similar to Beta in the inner tissues of the fruits.
The sequence of B is highly conserved with the wild-type allele b, and both function equally well when expressed in E. coli. The major difference between the alleles was found in the pattern of their expression during fruit ripening; whereas b mRNA appears at low level only at the breaker stage, B is highly expressed during ripening from breaker until the ripe stage. The results of transgenic expression of B in wild-type (VF36) plants confirm the existence of transcriptional regulation. Indeed, the major sequence discrepancy between the two alleles exists in the promoter region (Fig. 5). It is possible that some or all of the six additional sequence elements that exist upstream to the promoter in B, but not in b, are involved in regulating the increased transcription of B. Alternatively, an additional sequence that exists upstream to the b promoter could mediate suppression of expression in the wild-type fruits. Likewise, the B allele contains an 8-bp sequence (ATTTCAAA), which previously was found to be a cis element that is required for ethylene responsiveness (29), whereas in the b allele this sequence is slightly different (ATTTTCAAA). The involvement of these sequences in the regulation of B transcription needs to be investigated further.
The phenotype of fruits from transgenic plants that express B is slightly different from that of Beta. Although β-carotene is accumulated evenly in the Beta fruits, in the transgenic plants elevated β-carotene was found only in the columella and the placenta. Because high β-carotene correlates with increased in B mRNA (data not shown), it is possible that the promoter of B that was used in this experiment did not contain all of the upstream cis elements that exist in the authentic B gene. Thus the rate of expression in various parts of the fruit was altered.
The Lycopene β-Cyclase Gene Family in Plants.
There are two genuine lycopene β-cyclase enzymes in tomato: CRTL-B, which was previously characterized (10) and B, which is described in this report. The polypeptides of these enzymes are of the same size, but their amino acid sequence is only 53% identical. Interestingly, the amino acid sequence of B is more similar (86.1% identical) to CCS from pepper (Capsicum annum), an enzyme that converts antheraxanthin and violaxanthin to capsanthin and capsorubin, respectively. These xanthophylls are responsible for the typical red color of pepper fruits. When expressed in E. coli CCS exhibits low activity of lycopene β-cyclase (30). Similarly to B, Ccs is expressed only in chromoplast-containing tissues and neither gene contains introns. Moreover, the Ccs locus is located in a similar position on the genetic linkage map of pepper as B is in the tomato genetic map (31). Taken together, these data suggest that the genes Ccs and B originated from a common ancestor, most probably a lycopene β-cyclase gene, by a gene duplication that had taken place in the Solanecae before the divergence of Lycopersicon from Capsicum. Lack of introns in both genes supports the possibility that the gene duplication event was achieved by retrotransposition. Both genes are expressed exclusively in chromoplasts. However, whereas in tomato the duplicated gene has kept its original catalytic function of lycopene β-cyclization, the second cyclase in pepper acquired, during evolution, a new enzymatic activity of a similar nature. A deletion mutation in the Ccs gene is responsible for the yellow fruit recessive phenotype in pepper (31).
The Role of B Gene in Tomato.
The phenotype of transgenic plants that expressed antisense B resembled that of the mutants og and ogc, which are null mutations in the B locus. In both cases there was no phenotypic manifestation of the mutation, neither biochemical nor developmental, in leaves and stems, indicating that B does not play an indispensable role in vegetative tissues under normal growth conditions. However, distinct phenotypic changes occurred in these plants in chromoplast-containing tissues, i.e., fruits, petals, and anthers. In fruits, β-carotene, which in the wild type is found at a low level, was abolished and lycopene accumulated to a higher concentration. The origin of β-carotene that is present at low concentration (5–10%) in ripe tomato fruits has been enigmatic. Possible explanations are that either it is a leftover from the green stages or, alternatively, synthesized de novo during fruit ripening. Our results prove that β-carotene is synthesized de novo during tomato fruit development by the B lycopene cyclase only. Even if a low amount of CrtL-b mRNA does appear in the fruit it clearly does not provide a functional enzyme. A similar phenomenon has been found in the phytoene synthase gene Psy2, which does not contribute to carotenoid synthesis during fruit ripening although it is transcribed at a low level (32).
In flowers that lacked the B cyclase, a substantial amount of lycopene and other carotenoids that are produced earlier in the pathway accumulated in addition to the yellow β,β xanthophylls. This phenotype, together with the mRNA expression data, confirms that both lycopene cyclases, CRTL-B and B, function in flowers for the production of β-carotene.
The Evolution of Lycopene Accumulation in Tomato Fruits.
There are nine related species in the genus Lycopersicon (33). Six species have green fruits that contain chlorophyll and carotenoid pigments similar to leaves. The two wild species that synthesize carotenoids in the fruits as secondary metabolites are L. cheesmannii, which accumulates β-carotene, and L. pimpinellifolium, which accumulates lycopene. These species are phylogenetically the closest to L. esculentum (33).
Accumulation of carotenoids as secondary metabolites in fruits and flowers occurs by up-regulation of the pathway at the gene expression level. The green-fruited species L. pennellii was found to carry the recessive r gene, which confers yellow-flesh fruits in L. esculentum because of a null mutation in Psy1. Psy1 encodes a chromoplast-specific phytoene synthase, whereas Psy2 encodes a phytoene synthase that is constitutively expressed in all tissues. Occurrence of r agrees with lack of carotenoid accumulation in green-fruited species. Intercrossing with L. esculentum showed that all of the green-fruit species possess the dominant Del allele. Del is a dominant gene that in L. esculentum determines the accumulation of δ-carotene in the fruits. This gene encodes a lycopene ɛ-cyclase that is expressed only in photosynthetic tissues of wild-type tomatoes but is sharply up-regulated during ripening in the fruits of the Delta mutant (11). All of the green-fruited species and L. cheesmannii carry the B allele.
The existence of r, B, and Del in wild tomato species suggests a hypothetical scenario for the evolution of fruit color in tomato. Because flowers of the green-fruited species accumulate β,β-carotenoids it is obvious that they all maintain a chromoplast-regulated carotenogenesis. We propose that the gene expression mechanism that developmentally up-regulates carotenogenesis in the flowers was recruited by the fruit developmental program in the common ancestor of L. esculentum, L. cheesmannii, and L. pimpinellifolium. Later in evolution, a mutation that occurred in the ancestor of L. esculentum and L. pimpinellifolium reduced the expression of B in fruits but not in flowers and thus determined lycopene accumulation.
Biotechnological Implications of B.
Cloning genes for the carotenoid biosynthesis enzymes has paved the way for genetic manipulations of the pathway in crop plants to improve their nutritional value by achieving a higher content or a better composition of carotenoids. Two successful attempts to alter the pathway have been recently reported (34, 35). The B-type lycopene cyclase can be used in similar genetic manipulations to increase β-carotene content in fruits and flowers or to increase lycopene by eliminating its activity in tomato fruits.
Supplementary Material
Acknowledgments
We thank Dr. Ron Vunsh for technical assistance in plant transformation and Prof. Klaus Kloppstech for valuable comments on the manuscript. This work was supported by Grant 578/97 from the Israel Science Foundation and by the Ministry of Science and was carried out under the auspices of the Avron Even-Ari Minerva Center.
Abbreviations
- RFLP
restriction fragment length polymorphism
- YAC
yeast artificial chromosome
- RT
reverse transcription
- CCS
capsanthin-capsorubin synthase
- IL
introgression line
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF254793).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190177497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190177497
References
- 1.Mayne S T. FASEB J. 1996;10:690–701. [PubMed] [Google Scholar]
- 2.Gann P H, Ma J, Giovannucci E, Willett W, Sacks F M, Hennekens C H, Stampfer M J. Cancer Res. 1999;59:1225–1230. [PubMed] [Google Scholar]
- 3.Giovannucci E. J Natl Cancer Inst. 1999;91:317–331. doi: 10.1093/jnci/91.4.317. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham F X, Gantt E. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- 5.Harker M, Hirschberg J. Methods Enzymol. 1998;297:244–263. [Google Scholar]
- 6.Fraser P D, Truesdale M R, Bird C R, Schuch W, Bramley P M. Plant Physiol. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pecker I, Chamovitz D, Linden H, Sandmann G, Hirschberg J. Proc Natl Acad Sci USA. 1992;89:4962–4966. doi: 10.1073/pnas.89.11.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corona V, Aracri B, Kosturkova G, Bartley G E, Pitto L, Giorgetti L, Scolnik P A, Giuliano G. Plant J. 1996;9:505–512. doi: 10.1046/j.1365-313x.1996.09040505.x. [DOI] [PubMed] [Google Scholar]
- 9.Bramley P. Pure Appl Chem. 1997;69:2159–2162. [Google Scholar]
- 10.Pecker I, Gabbay R, Cunningham F X, Hirschberg J. Plant Mol Biol. 1996;30:807–819. doi: 10.1007/BF00019013. [DOI] [PubMed] [Google Scholar]
- 11.Ronen G, Cohen M, Zamir D, Hirschberg J. Plant J. 1999;17:341–351. doi: 10.1046/j.1365-313x.1999.00381.x. [DOI] [PubMed] [Google Scholar]
- 12.Giuliano G, Bartley G E, Scolnik P. Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann V, Harker M, Pecker I, Hirschberg J. Nat Biotechnol. 2000;18:888–892. doi: 10.1038/78515. [DOI] [PubMed] [Google Scholar]
- 14.Schledz M, Al-Babili S, von Liting J, Haubruck H, Rabbani S, Kleinig H, Beyer P. Plant J. 1996;10:781–792. doi: 10.1046/j.1365-313x.1996.10050781.x. [DOI] [PubMed] [Google Scholar]
- 15.Fray R G, Grierson D. Plant Mol Biol. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- 16.Mustilli A C, Fenzi F, Ciliento R, Alfano F, Bowler C. Plant Cell. 1999;11:145–157. doi: 10.1105/tpc.11.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomes M L, Quackenbush F W, Kargl T E. Bot Gaz. 1956;117:248–253. [Google Scholar]
- 18.Thompson A E, Tomes M L, Wann E V, McCollum J P, Stoner A K. Proc Am Soc Hortic Sci. 1965;86:610–616. [Google Scholar]
- 19.Lincoln R E, Porter J W. Genetics. 1950;35:206–211. doi: 10.1093/genetics/35.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson A E, Tomes M L, Erickson H T, Wann E V, Armstrong R J. Proc Am Soc Hortic Sci. 1967;91:495–504. [Google Scholar]
- 21.Tomes M L, Erickson H T, Barman R J. Tomato Genet Cooperative Rep. 1969;19:27–28. [Google Scholar]
- 22.Eshed Y, Zamir D. Genetics. 1995;141:1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanksley S D, Ganal M W, Prince J C, de Vicente M C, Bonierabale M W, Broun P, Fulton T M, Giovanonni J J, Grandillo S, Martin G B, et al. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. Development (Cambridge, UK) 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- 25.Martin G B, Ganal M W, Tanksley S D. Mol Gen Genet. 1992;233:25–32. doi: 10.1007/BF00587557. [DOI] [PubMed] [Google Scholar]
- 26.Cunningham F X, Chamovitz D, Misawa N, Gantt E, Hirschberg J. FEBS Lett. 1993;328:130–138. doi: 10.1016/0014-5793(93)80980-9. [DOI] [PubMed] [Google Scholar]
- 27.McKormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- 28.Bouvier F, Hugueney P, d'Harlingue A, Kuntz M, Camara B. Plant J. 1994;6:45–54. doi: 10.1046/j.1365-313x.1994.6010045.x. [DOI] [PubMed] [Google Scholar]
- 29.Xu R L, Goldman S, Coupe S, Deikman J. Plant Mol Biol. 1996;31:1117–1127. doi: 10.1007/BF00040829. [DOI] [PubMed] [Google Scholar]
- 30.Hugueney P, Badillo A, Chen H C, Klein A, Hirschberg J, Camara B, Kuntz M. Plant J. 1995;8:417–424. doi: 10.1046/j.1365-313x.1995.08030417.x. [DOI] [PubMed] [Google Scholar]
- 31.Lefebvre V, Kuntz M, Camara B, Palloix A. Plant Mol Biol. 1998;36:785–789. doi: 10.1023/a:1005966313415. [DOI] [PubMed] [Google Scholar]
- 32.Fraser P D, Kiano J W, Truesdale M R, Schuch W, Bramley P M. Plant Mol Biol. 1999;40:687–698. doi: 10.1023/a:1006256302570. [DOI] [PubMed] [Google Scholar]
- 33.Miller J C, Tanksley S. Theor Appl Genet. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- 34.Shewmaker C K, Sheehy J A, Daley M, Colburn S, Ke D Y. Plant J. 1999;20:401–412. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- 35.Ye X, Al Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.