Abstract
Frontotemporal dementia (FTD) is the most common form of dementia before 60 years of age. Rare pathogenic mutations in CHMP2B, which encodes a component of the endosomal sorting complex required for transport (ESCRT-III), are associated with FTD linked to chromosome 3 (FTD3). Animal models of FTD3 have not yet been reported, and what signaling pathways are misregulated by mutant CHMP2B in vivo is unknown. Here we report the establishment of a Drosophila model of FTD3 and show the genetic interactions between mutant CHMP2B and other components of ESCRT. Through an unbiased genome-wide screen, we identified 29 modifier loci and found that serpin5 (Spn5), a largely uncharacterized serine protease inhibitor, suppresses the melanization phenotype induced by mutant CHMP2B in the fly eye. We also found that Spn5 is a negative regulator of the Toll pathway and functions extracellularly, likely by blocking the proteolytic activation of Spaetzle, the Toll receptor ligand. Moreover, Spn5 inhibited activation of the Toll pathway by mutant CHMP2B. Our findings identify Spn5 as a regulator of the Toll pathway and CHMP2B toxicity and show that the Toll pathway is a major signaling pathway misregulated by mutant CHMP2B in vivo. This fly model will be useful to further dissect genetic pathways that are potentially relevant to the pathogenesis and treatment of FTD.
Keywords: Drosophila, endosomal sorting complex required for transport (ESCRT), neurodegeneration, modifier screen
Frontotemporal dementia (FTD), a major clinical syndrome of frontotemporal lobar degeneration (FTLD), is a progressive neurodegenerative condition associated with focal atrophy of the frontal and/or temporal lobes (1, 2). Although FTD is the most common form of senile dementia in people under 60 years of age, the molecular pathogenesis remains poorly understood (3). In some FTD brains, tau neurofibrillary tangles are present in diseased neurons, and some tau mutations are indeed pathogenic (4, 5). Several new genes have been implicated in FTD with tau-negative pathology, including those encoding valosin-containing protein (VCP) (6), CHMP2B (7), progranulin (8, 9), and TDP-43 (10, 11). The molecular pathways affected by these mutations and how they contribute to disease progression remain unclear.
Although dominantly inherited CHMP2B mutations associated with FTD linked to chromosome 3 (FTD3) are rare (7, 12, 13), studies of CHMP2B neurotoxicity in cell culture models have been informative. CHMP2B is the ortholog of the yeast protein Vps2, a component of the endosomal sorting complex required for transport (ESCRT-III), which is involved in the biogenesis of multivesicular bodies and other biological processes (14). In undifferentiated PC12 cells, ectopic overexpression of CHMP2BIntron5, a mutant form of CHMP2B missing 35 aa at the C terminus, led to the accumulation of vesicular structures (7). In cultured rodent cortical neurons and other cell types, CHMP2BIntron5 caused dendritic retraction, autophagosome accumulation, and neuronal cell loss (15, 16). At the molecular level, CHMP2BIntron5 seems to have a toxic effect by forming an abnormal complex with mSnf7–2, another ESCRT-III component that failed to dissociate properly (15). Thus, it is likely that pathogenesis of FTD3 is through a gain-of-function mechanism. However, animal models of FTD3 have not been reported, and the signaling pathways that are misregulated in vivo remain to be identified.
In recent years, Drosophila models have been instrumental in uncovering molecular pathways that contribute to the pathogenesis of neurodegenerative diseases (17, 18). In this study, we modeled the effect of CHMP2B in human FTD3 using a gain-of-function approach, which is similar to the approach that gives effects in neuronal cell culture, by expressing normal or mutant CHMP2B in Drosophila with the Gal4-UAS system. We performed a genetic screen to identify modifiers of mutant CHMP2B toxicity. One of the enhancers we cloned is serpin5 (Spn5)—a largely uncharacterized member of a family of evolutionarily conserved serine protease inhibitors. The precise functions of many serpins remain unknown, although some play essential roles in various biological processes and human diseases, including the Toll pathway and innate immunity (19). Here we show that Spn5 is another negative regulator of the Toll signaling pathway in Drosophila, and that the Toll pathway is a major target of mutant CHMP2B toxicity in vivo.
Results
A Fly Model to Dissect the Toxicity of Mutant CHMP2B Associated with FTD3.
CHMP2BIntron5 and CHMP2BΔ10, 2 mutant CHMP2B proteins resulting from a single nucleotide mutation at a splicing site, were associated with FTD3 in a large Danish family (7). Our earlier studies indicated that in contrast to CHMP2BIntron5, CHMP2BΔ10 was a highly unstable protein and had no effect on dendritic morphology and neuronal survival in transfected rodent cortical neurons (15). Thus, it is likely that not all reported mutant CHMP2B proteins are pathogenic. In this study, we focus our attention on CHMP2BIntron5. To take advantage of Drosophila genetics as a powerful tool to investigate the toxicity of FTD3-associated mutant protein CHMP2BIntron5, we generated UAS transgenic flies expressing CHMP2BWT and CHMP2BIntron5 (Fig. 1A). These fly lines allow us to achieve spatial and temporal control of transgene expression using the UAS-Gal4 system (20). Ubiquitous expression of UAS-CHMP2BIntron5 using tubulin-Gal4 resulted in lethality at late embryonic stage. Pan-neuronal expression of CHMP2BIntron5 also resulted in lethality. These findings indicate that CHMP2BIntron5 expression disrupts critical functions in fly neurons.
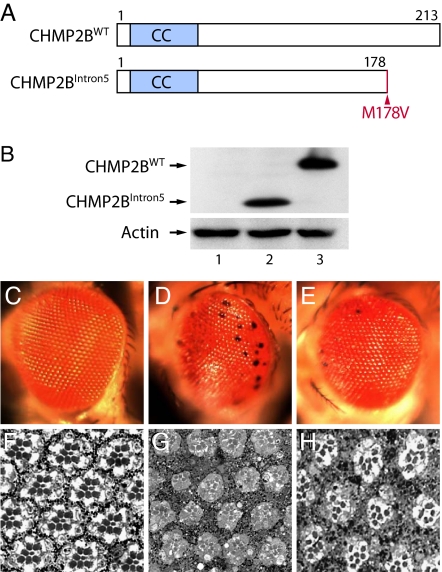
Fig. 1.
A fly model of FTD3. (A) Schematic representation of wild-type and the FTD3-associated mutant CHMP2B proteins. CC, coiled-coil domain. CHMP2BIntron5 lacks the C-terminal 35 aa, and methionine-178 is changed to valine. (B) Western blot analysis. Lane 1, GMR-Gal4; lane 2, GMR-Gal4:UAS-CHMP2BIntron5; lane 3, GMR-Gal4:UAS-CHMP2BWT. (C–H) CHMP2BIntron5 expression caused severe degeneration in the Drosophila eye. (C and F) GMR-Gal4 flies had normal external morphology (C) and internal retinal structure (F). (D and G) CHMP2BIntron5 expression in the eye caused black spots (D) and degeneration of internal structures in 1-day-old flies (G). (E and H) CHMP2BWT expression caused a mild black-spot phenotype (E) and with minor effect on the internal structure in 1-day-old flies (H).
To gain insight into the toxic effects of CHMP2BIntron5 expression, we used the Drosophila eye as the model system. Multiple independent insertion lines that expressed CHMP2BWT and CHMP2BIntron5 were generated and their expression levels compared. Two of them with comparable expression levels were selected for further genetic analysis (Fig. 1B). GMR-Gal4 (Fig. 1C), UAS-CHMP2BWT, or UAS-CHMP2BIntron5 flies (image not shown) did not have any obvious defects in eye morphology. The internal retinal structures of GMR-Gal4 flies also appeared to be normal with regular patterning of ommatidia containing precise numbers and arrangement of photoreceptors and pigment cells (Fig. 1F). In contrast, CHMP2BIntron5 expression caused a rough eye phenotype and the appearance of black spots (Fig. 1D). Expression of CHMP2BWT resulted in a much weaker eye phenotype (Fig. 1E). Moreover, CHMP2BIntron5 expression severely disrupted internal eye structures as well (Fig. 1G), whereas CHMP2BWT caused only mild retinal distortion (Fig. 1H). These phenotypes were confirmed in multiple independent transgenic lines for each construct. Expression of multiple CHMP2BΔ10 independent transgenic lines did not show any adverse effect in the fly eye, consistent with our earlier findings in cultured rodent cortical neurons (15). Thus, CHMP2BIntron5 causes severe retinal degeneration in an in vivo Drosophila eye model, thereby establishing a fly model of FTD3.
CHMP2BIntron5 Genetically Interacts with Drosophila Genes Encoding ESCRT-III Components.
To provide genetic evidence that the CHMP2BIntron5 phenotype is due to perturbation of endogenous ESCRT-III function, we performed genetic interaction experiments with genes encoding different components of Drosophila ESCRT-III, including shrub, vps2, and vps24. Previously we described Shrub as the fly homolog of the yeast ESCRT-III subunit Snf7, whose loss of function caused a defect in dendritic morphogenesis (21). A single copy of shrub4–1 did not cause an eye phenotype but significantly enhanced the CHMP2BIntron5 phenotype (Fig. 2 A–C). Previously we found in cultured rodent cortical neurons, CHMP2BIntron5 sequestered mSnf7–2, the mouse homolog of Shrub, resulting in cellular phenotypes identical to loss of mSnf7–2 (15). Indeed, reduction of Shrub activity by siRNA or ectopic expression of a dominant-negative fusion protein, Shrub-GFP (21), caused an eye phenotype similar to the CHMP2BIntron5 phenotype (Fig. 2).
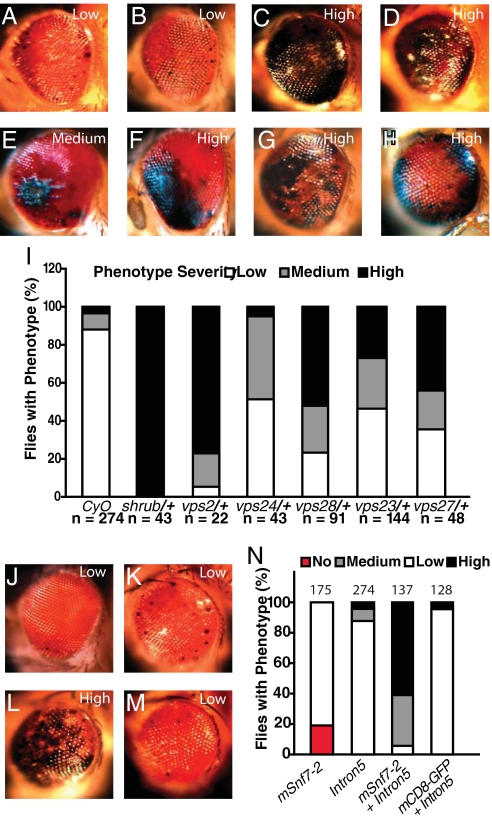
Fig. 2.
Genetic interactions between CHMP2BIntron5 and genes encoding ESCRT components. (A) The majority of 1-day-old GMR-Gal4:UAS-CHMP2BIntron5/CyO flies showed a weak eye phenotype. (B) One-day-old GMR-Gal4:UAS-CHMP2BIntron5/+ flies had the same eye phenotype as GMR-Gal4:UAS-CHMP2BIntron5/CyO. (C–H) A single copy of the mutant alleles shrub4–1 (C), dvps2GS11024 (D), vps24EY04708 (E), vps28l (2)K16503 (F), vps23f00976 (G), and vps27D28 (H) enhanced the CHMP2BIntron5 phenotype in 1-day-old flies. (I) Phenotype severity in each genotype. n is the number of flies per genotype. (J) Mild eye phenotype in flies expressing UAS-mSnf7–2. (K) Representative eye phenotype caused by CHMP2BIntron5 expression. (L) Coexpression of UAS-CHMP2BIntron5 and UAS-mSnf7–2 enhanced the severity of the eye phenotype caused by expression of individual transgenes. (M) The CHMP2BIntron5 phenotype was not affected by coexpression of UAS-mCD8-GFP. (N) Phenotype severity in each genotype. n is the number of flies per genotype.
We also identified CG14542 and CG4618, both previously uncharacterized genes, encoding putative fly homologs of vps2 and CHMP2B. The presence of one copy of vps2GS11024 or Df(3R)ED210 that uncovers CG4618 within its breakpoints did not alone cause an eye phenotype, but considerably enhanced the CHMP2BIntron5 phenotype (Fig. 2D). Reduced vps24 activity with a mutant allele, vps24EY04708 (22), also enhanced the CHMP2BIntron5 phenotype (Fig. 2E). These genetic interaction studies support the notion that CHMP2BIntron5 has a gain-of-function effect to sequester endogenous Shrub or its homologs, leading to reduced normal function of ESCRT-III. We also tested components of other ESCRT complexes, such as Vps28 (23), Vps23 (24), and Vps27 (25). Single copies of mutant alleles of vps28 (Fig. 2F) or vps23 (Fig. 2G) (vps28l (2)K16503 or vps23f00976), both encoding components of ESCRT-I, or vps27D28 (Fig. 2H), encoding a component of ESCRT-0, all significantly enhanced the eye phenotype caused by CHMP2BIntron5 (Fig. 2I).
Our earlier findings showed that CHMP2BIntron5 and mSnf7–2 fail to dissociate and that dysfunctional ESCRT-III causes neuronal cell loss in cultured cortical neurons (15). Indeed, although expression of the mouse Shrub homolog mSnf7–2 alone caused a very mild eye phenotype (Fig. 2J), coexpression of mSnf7–2 in the Drosophila eye dramatically enhanced the CHMP2BIntron5 phenotype (Fig. 2L). The CHMP2BIntron5 phenotype (Fig. 2K) was not enhanced by coexpression of mCD8-GFP (Fig. 2M). These findings strongly support the notion that the abnormal complex containing CHMP2BIntron5 and mSnf7–2 is toxic and recapitulates the toxicity seen in the mammalian system (15), further validating our fly model. A toxic protein complex with a pathogenic gain of function has also been reported for glutamine repeat-containing ataxin 1 associated with spinocerebellar ataxia type 1 (SCA1) (26).
Genetic Screen Identifies Spn5 as a Major Dominant Enhancer of CHMP2BIntron5 Toxicity.
Our genetic interaction studies showed that Shrub-GFP and CHMP2BIntron5 phenotypes are sensitive to partial loss of other genetic factors. Therefore we conducted an F1 genetic screen to identify enhancers of the Shrub-GFP phenotype first (Fig. 3A). Briefly, we recombined GMR-Gal4 and UAS-Shrub-GFP onto the second chromosome, and the resulting flies exhibited an eye phenotype identical to that caused by CHMP2BIntron5 (Fig. S1). The GMR-Gal4, UAS-shrub-GFP/CyO stock was crossed to all 257 individual deletion stocks in the DrosDel Deletion Collection (University of Cambridge, Cambridge, U.K.). This collection represents a deletion coverage of ≈75% of the Release 5.1 Drosophila genome (27). Scoring of the phenotype was based on a comparison with a GMR-Gal4, UAS-shrub-GFP/CyO outcrossed to the w1118 strain. Twenty-nine enhancers were identified and classified as strong (+++), medium (++), or weak (+). After this initial primary screen, we crossed some of these enhancers with flies expressing CHMP2BIntron5 under the control of GMR-Gal4. For instance, the CHMP2BIntron5 phenotype was significantly enhanced by Df(2L)ED1243/+ and Df(3R)ED5664/+ (Fig. 3B).
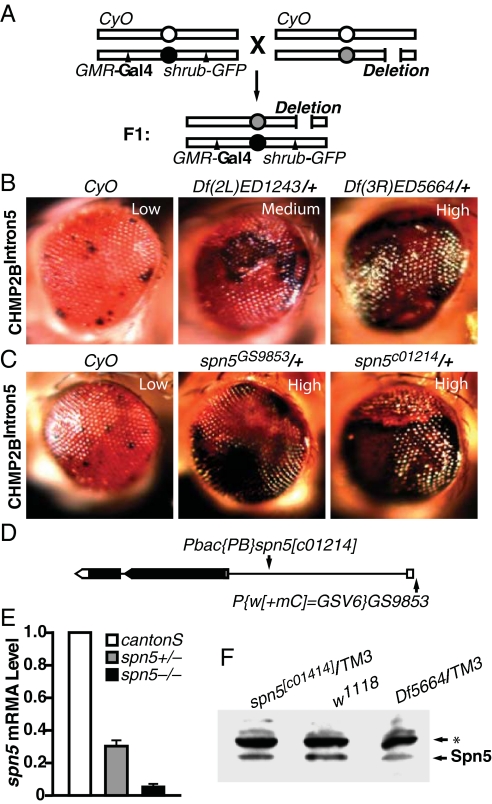
Fig. 3.
A genetic screen identified spn5 as a strong enhancer of the CHMP2BIntron5 phenotype. (A) Schematic representation of the genetic screen to identify enhancers of the CHMP2BIntron5 eye phenotype. (B) Representative genetic enhancers of the CHMP2BIntron5 phenotype. (C) Enhancement of the CHMP2BIntron5 phenotype by one copy of spn5 mutant alleles was comparable to that of Df(3R)ED5664/+. (D) Schematic representation of the spn5 gene structure. Introns, exons, and ORF are shown as lines, boxes, and shaded area, respectively. (E) qRT-PCR analysis of the levels of spn5 transcripts in first instar larvae. (F) Western blot analysis of head homogenates with anti-Spn5 antibody. Three independent experiments showed a decrease of approximately 40–50% in heterozygotes. The asterisk denotes a nonspecific band recognized by the Spn5 antibody.
Because Df(3R)ED5664 exhibited the most dramatic enhancement of the CHMP2BIntron5 phenotype, we set out to identify the gene(s) responsible for the genetic interaction. This deficiency has breakpoints at 88D1–88E3 on the left arm of the third chromosome and covers about 57 genes. We first obtained smaller deficiency lines that partially overlap with Df(3R)ED5664 and narrowed the enhancer region down to 88E1–88E3, an overlapping region between Df(3R)ED10564 and Df(3R)ED10566 that contains 21 genes (Fig. S2). We then tested all available mutant lines that disrupt individual genes within 88E1–88E3. A PiggyBac P-element insertion at the gene serpin5 (spn5, CG18525), spn5c01214 (Fig. 3D) significantly enhanced the CHMP2BIntron5 phenotype (Fig. 3C). This effect was confirmed by an independent P-element insertion affecting spn5 (spn5GS9853) (Fig. 3C). To confirm that the spn5c01214 allele did affect Spn5 expression, we examined the level of spn5 mRNA by real-time PCR (RT-PCR) at the first instar larval stage. Indeed, the level of spn5 mRNA was markedly reduced (Fig. 3E). Moreover, a polyclonal antibody we generated against recombinant Spn5 N terminus (amino acids 1–176) detected the reduced Spn5 protein level of ≈50% in the heads of spn5c01214/+ or Df(3R)ED5664/+ heterozygous adult flies (Fig. 3F). These findings indicate that spn5 is the gene located in Df(3R)ED5664 whose partial loss of activity enhanced the CHMP2BIntron5 phenotype.
Overexpression of Spn5 Suppresses the CHMP2BIntron5 Phenotype Extracellularly.
To further examine the role of Spn5 in CHMP2BIntron5 toxicity, we performed rescue experiments. We generated transgenic flies containing UAS-spn5 and ectopically expressed the transgene by GMR-Gal4. Overexpression of spn5 did not cause an eye phenotype in a wild-type background, but completely suppressed the enhancement effect of Df(3R)ED5664/+ on the CHMP2BIntron5 phenotype (Fig. 4 A and B), further confirming that loss of a copy of spn5 located in Df(3R)ED5664 is indeed responsible for the observed enhancement. Expression of spn5 also rescued the CHMP2BIntron5 phenotype enhanced by spn5c01214/+ (Fig. 4 C and D). Ectopic expression of Spn5 also suppressed the external eye phenotype caused by CHMP2BIntron5 in the absence of enhancers (Fig. 4 E and F). Overexpression of Spn5 did not dramatically rescue the photoreceptor degeneration phenotype, suggesting that not all aspects of CHMP2B toxicity can be rescued by Spn5.
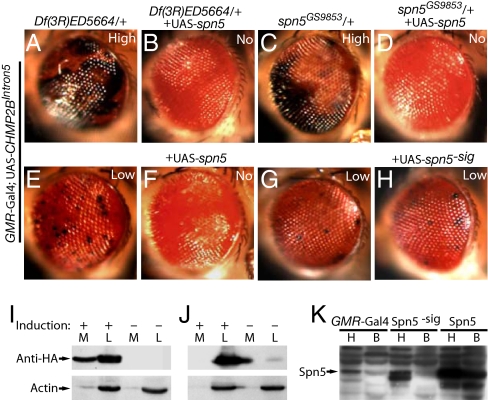
Fig. 4.
Spn5 suppresses the CHMP2BIntron5 eye phenotype extracellularly. (A and B) Enhancement of the CHMP2BIntron5 phenotype by Df(3R)ED5664/+ (A) was suppressed by coexpression of UAS-spn5 (B). (C and D) Enhancement of the CHMP2BIntron5 phenotype by spn5GS9853/+ (C) was suppressed by coexpression of UAS-spn5 (D). (E and F) The CHMP2BIntron5 phenotype itself (E) was completely suppressed by coexpression of UAS-spn5 (F). (G and H) Expression of Spn5 without the signal peptide (H) did not suppress the CHMP2BIntron5 phenotype in sibling flies (G). (I) Spn5-HA was secreted into the medium from transfected S2 cells. M, medium; L, cell lysates. (J) The secretion of Spn5-HA was dependent on the signal peptide. (K) Spn5 expressed in the eye was detected in the body, which was also dependent on the presence of the putative signal peptide. H, head; B, body; Spn5−sig, Spn5 without the putative signal peptide.
Serpins are protease inhibitors that function either intracellularly or extracellularly (19). Like most other Drosophila serpins, Spn5 contains a putative N-terminal secretion signal and is widely expressed in many tissues (28). However, the exact function of Spn5 and its secretory properties have not been characterized. To determine if Spn5 suppresses the CHMP2BIntron5 phenotype through an intracellular or extracellular pathway, we first examined whether Spn5 can be secreted. In S2 cells transiently transfected with a metal-inducible Spn5-HA construct, endogenous Spn5 was mainly present in the medium fraction. Upon induction, Spn5-HA was also detected in the medium (Fig. 4I). Thus, both endogenous and ectopically expressed Spn5 can be secreted from S2 cells. As expected, Spn5 without the signal peptide was not secreted (Fig. 4J). The same notion was confirmed in vivo. We expressed Spn5 with the signal peptide in the eye using the GMR-Gal4 and detected Spn5 accumulation in the body by Western blot, indicating that Spn5 was secreted and transported through the Drosophila hemolymph (Fig. 4K). To further confirm this result, we expressed Spn5-HA in the eye and did detect Spn5 in the body using the HA antibody, which is more specific than the Spn5 antibody used in Fig. 4K (Fig. S3). However, when Spn5 without the signal peptide was expressed in the eye, the levels of Spn5 in the body were not increased (Fig. 4K). Correspondingly, ectopic expression of Spn5 without the secretion signal failed to rescue the melanization phenotype caused by CHMP2BIntron5 (Fig. 4H). These findings demonstrate that Spn5 exerts its activity extracellularly to suppress the effect of CHMP2BIntron5.
Spn5 Is a Negative Regulator of the Toll Pathway.
The ability of Spn5 to suppress the CHMP2BIntron5 phenotype prompted us to examine whether other fly serpins could do so as well. Spn4, the fly homolog of the mammalian neuroserpin, seems to be an intracellular regulator of the subtilisin-like proprotein convertase furin (29). Ectopic expression of Spn4 failed to suppress the CHMP2BIntron5 phenotype. Another well-studied fly serpin is Necrotic (Nec, also known as Spn43Ac), which regulates proteases that cleave and hence activate Spaetzle, the ligand for the Toll receptor (30). Similar to Spn5, partial reduction of Nec activity through 2 mutant alleles, nec10 or nec2, markedly enhanced the CHMP2BIntron5 phenotype (Fig. S4). This enhancement was rescued by ectopic expression of UAS-nec, indicating that Nec modulates the CHMP2BIntron5 phenotype (Fig. S4). These findings suggest that Nec and Spn5 may regulate the same proteolytic cascade.
To determine whether Spn5 also regulates the Toll pathway, we analyzed lysates from spn5c01214 homozygous mutant larvae by Western blot. The level of Toll increased (Fig. S5A), as did Spaetzle precursor, probably by a positive feedback regulation of the Toll pathway, as in nec mutants (30). To further examine the effect of Spn5 on the Toll pathway, we performed quantitative RT-PCR to measure the induction of the antifungal peptide drosomycin, a direct target and transcriptional readout of Toll pathway activation. In Spn5 homozygous mutant larvae, drosomycin mRNA expression was increased 9-fold (Fig. S5B), further supporting the notion that Spn5 is a unique regulator of the Toll pathway.
Activation of Toll pathway could lead to the initiation of the melanization cascade (31). De novo synthesis of melanin in response to tissue damage occurs through the melanization cascade, a series of enzymatic reactions involving serine proteases that activate phenol oxidase (PO), which catalyzes the conversion of phenolic substrates to quinones, which then polymerize to form melanin (32). The melanization reaction in the hemolymph can be blocked by mutations in the Drosophila gene Black cells (Bc), which encode PO, and Bc/Bc larvae do not have PO activity (33). To determine if the black spots in the Drosophila eyes expressing CHMP2BIntron5 are melanin deposits, we crossed GMR-Gal4, UAS-CHMP2BIntron5 flies with Bc mutant flies. Indeed, in the Bc1/+ background, the CHMP2BIntron5-mediated black spot phenotype was markedly suppressed (Fig. S6). In an in vitro assay (34), reduced Spn5 activity increased PO enzymatic activity (Fig. S5C) and spontaneous melanization in 100% of spn5c01214 homozygous mutant larvae (Fig. S5 D and E). spn5c01214/Df(3R)ED5664 larvae exhibited the same phenotype as spn5c01214 homozygous larvae (Fig. S5F), confirming that the melanotic spots are due to loss of Spn5 activity. These spn5 mutants failed to survive beyond the larval stage.
CHMP2BIntron5 Activates the Toll Pathway.
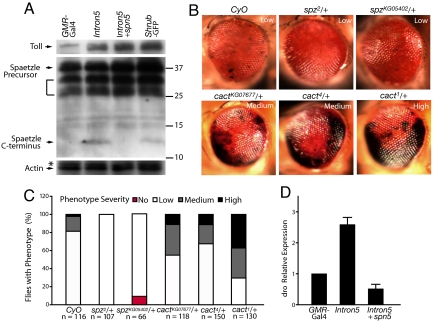
Our findings that spn5 showed a strong genetic interaction with CHMP2BIntron5 and that spn5 is a unique regulator of the Toll pathway led us to examine whether the Toll pathway is a major target of CHMP2BIntron5 toxicity. We found that the level of Toll in head homogenates was on average 3.4-fold higher with CHMP2BIntron5 expression (Fig. 5A). Expression of Shrub-GFP resulted in a similar eye phenotype (Fig. S2) and also increased Toll accumulation by 4.7-fold (Fig. 5A).
Fig. 5.
The Toll pathway is a major target of CHMP2BIntron5 toxicity. (A) The levels of the Toll receptor and Spaetzle precursor were increased in head homogenates of flies expressing CHMP2BIntron5, CHMP2BIntron5 and Spn5, or Shrub-GFP. The bracket indicates other possible Spaetzle precursor forms recognized by this antibody as reported by Chang and Morisato (36). Values on right are molecular mass in KD. The Western blot to detect Spaetzle was overexposed to show its C-terminal fragment more clearly. Quantification for the relative levels of Spaetzle precursor was done on lighter exposures from 3 independent experiments. (B) One copy of the spz2 and spzKG05402 mutant alleles partially suppresses the CHMP2BIntron5 phenotype, and one copy of cact1, cact4, or cactKG07677 mutant alleles enhances the CHMP2BIntron5 phenotype. (C) Extent of suppression or enhancement of the phenotype. n is the number of flies examined per genotype. (D) qRT-PCR analysis of the levels of drosomycin transcripts in fly head homogenates with different genotypes.
Binding of proteolytically cleaved C-terminal fragment of the ligand, Spaetzle, activates the Toll transmembrane receptor, which in turn activates the transcription of spaetzle mRNA (30, 35). Thus, the accumulation of Toll might be accompanied by increased accumulation of Spaetzle C-terminal fragment. Indeed, the level of the C-terminal fragment of Spaetzle was significantly higher in the eye when CHMP2BIntron5 was expressed (Fig. 5A). The level of Spaetzle precursor was also increased by an average of 2.3-fold (Fig. 5A), probably due to the positive feedback regulation of the Toll pathway (30). As reported previously (36), Spaetzle precursors expressed in adults appeared as multiple bands on Western blots (Fig. 5A). Disruption of ESCRT-III function by Shrub-GFP expression had a similar effect, with an increase of 2.4-fold (Fig. 5A). Ectopic expression of Spn5 suppressed the production of the C-terminal fragment of Spaetzle without reducing the accumulation of the Toll receptor in the presence of CHMP2BIntron5 (Fig. 5A). This finding strongly suggests that the Spn5 regulates CHMP2B toxicity through the Toll pathway.
To further test this notion, we performed genetic interaction experiments. The presence of one copy of 2 mutant spaetzle alleles, spz2 and spzKG05402, markedly reduced the number of flies with melanin deposits caused by CHMP2BIntron5 (Fig. 5 B and C). Cactus, the Drosophila IκB protein, negatively regulates the Toll pathway by binding to and preventing nuclear translocation of the dorsal-dif complex, the Drosophila homologs of the Rel/NF-κB transcription factors (35). The presence of a copy of different cactus mutant alleles, cact1, cact4, or cactKG07677, significantly enhanced the CHMP2BIntron5 phenotype (Fig. 5 B and C). We also performed biochemical analysis. The level of drosomycin mRNA, a direct transcriptional target of the Toll pathway, was elevated in fly eyes expressing CHMP2BIntron5, which was suppressed by Spn5 expression (Fig. 5D). Moreover, head homogenates of flies expressing CHMP2BIntron5 in the eyes only had higher PO activity than controls (Fig. S6). This increased PO activity caused by CHMP2BIntron5 was also suppressed by Spn5 (Fig. S6), further demonstrating the inhibitory effects of Spn5 on activation of the Toll pathway by CHMP2BIntron5.
Discussion
Using a newly established Drosophila model of FTD3 and an unbiased genetic screen, we show that the Toll pathway is a major in vivo target activated by CHMP2BIntron5, a mutant protein associated with FTD3. We also show that Spn5 is a negative regulator of the Toll pathway and suppresses the CHMP2BIntron5 phenotype. These findings, although made in a fly model, raise the possibility that its mammalian counterpart, the Toll-like receptor/NF-κB pathway, is also a potential major target of CHMP2BIntron5 toxicity. Moreover, the establishment of the fly model will allow further genetic dissection of CHMP2BIntron5 toxicity in vivo.
Spn5 belongs to the superfamily of serine protease inhibitors that exert tight regulation of proteolytic cascades important for many biological processes, such as the complement cascade, inflammation, and innate immunity in different organisms. In Drosophila, there are 29 serpins, and the precise functions of most of them are not well understood (19). Several fly serpins regulate the Toll pathway as loss of activities in Spn43Ac, Spn-27A, and Spn77Ba lead to the activation of this important signaling pathway (30, 31, 37, 38). Our findings show that secreted Spn5 also regulates the Toll pathway, apparently by controlling the proteolytic processing of Spaetzle. Ectopic expression of Spn5 also abolished the increased production of the Spaetzle C-terminal fragment caused by CHMP2BIntron5 (Fig. 5). It remains to be determined which serine protease in the proteolytic cascade is the direct target of Spn5.
The identification of Spn5 as a strong modifier of the CHMP2BIntron5 phenotype led us to examine the Toll signaling pathway, which plays an essential role in innate immunity in Drosophila (39). Indeed, the Toll pathway is activated by CHMP2BIntron5, which is probably due to the abnormal sorting of the Toll receptor in the endocytic pathway. In mammals, Toll-like receptors (TLRs) are expressed predominantly in the immune system and are essential for generating innate immune responses; however, some are expressed in the nervous system and have been implicated in neurodegeneration (40, 41). For instance, TLR8 is expressed in neurons, and its activation promotes neuronal cell death in vitro (42). Conversely, reduced TLR4 activity protects cultured neurons from Aβ toxicity (43, 44). However, whether TLRs play a role in age-dependent neurodegeneration in vivo is poorly understood. Although the fly eye and human brain differ dramatically in their anatomy and physiology, our finding that the Toll receptor is a major target misregulated by CHMP2BIntron5 in vivo may have important implications for our understanding of FTD pathogenesis. It is conceivable that misregulation of TLRs may be at least in part responsible for the adverse effect of CHMP2BIntron5 on neuronal survival in vivo. If so, it will be interesting to determine which of the dozen or so TLRs mediate the neurotoxicity of CHMP2BIntron5 in mouse models. Moreover, it will be important to explore whether modulation of the TLR family could serve as a potential therapeutic target for FTD.
Experimental Procedures
Fly Stocks.
D. melanogaster strains were raised on a standard cornmeal and yeast diet at 25 °C unless otherwise stated. Canton S and w1118 were used as wild-type controls. Fly lines were obtained from the Bloomington Drosophila Stock Center, the Kyoto Institute of Technology, the Harvard Drosophila Stock Center, and the Drosdel Deletion Collection, University of Cambridge. For genetic interaction studies, lines containing GMR-Gal4 and UAS-CHMP2BWT or UAS-CHMP2BIntron5 elements were recombined onto the second chromosome. UAS-CHMP2BIntron5, shrub4–1, UAS-shrub-RNAi, UAS-shrub-GFP, UAS-mSnf7–2, UAS-mCD8::GFP stocks were previously described (15, 21). To quantify the CHMP2BIntron5 eye phenotype, we arbitrarily classified the eye phenotype with or without enhancers into 3 groups: strong (+++), medium (++), or weak (+). This classification was based on the relative size of the eye surface with black spots, ranging from approximately 50–70% or more of the eye surface (+++) to a dozen or so scattered spots (+).
Generation of Transgenic Fly Lines.
To generate UAS-spn5, UAS-spn5 without the secretion signal, and UAS-CHMP2BWT transgenic flies, the primers listed in Table S1 were used to clone into the pUAST vector, which in turn was sequenced and microinjected into wild-type (w1118) flies to generate transgenic lines.
Expression in S2 Cells.
S2 cells were cultured at 25 °C in Schneider's Drosophila Medium (GIBCO) supplemented with 10% heat-inactivated FBS. For transient transfection, S2 cells were transfected with a mixture of the pRmHa vector (0.5 μg) and Cellfectin reagent (10 μL; Invitrogen). To induce expression, 20 mL of 100 mM CuSO4/well (final concentration 1 mM) was added for 24 h. Cells were harvested and the medium was filtered through a 0.22-mm filter and analyzed by Western blot.
Antibody Generation and Western Blots.
Anti-Spn5 polyclonal antibody was generated by immunizing rabbits with peptide fragment spanning amino acids 1–176 (SKD Biotechnology). Rabbit CHMP2B antibody was generated by Covance using purified GST-CHMP2B protein. For protein expression analysis, fly heads were homogenized in the lysis buffer, and 15–25 mg of protein was separated on a 10% SDS gel and blotted onto a PVDF membrane. The membrane was probed with antibodies against CHMP2B (1:1,000; J.A.L.), HA (1:3,000; Sigma), Toll (1:250; Santa Cruz Biotechnology), Spaetzle (1:1,000; refs. 36 and 37), or actin (1:1,000; Abcam).
Phenol Oxidase (PO) Assay.
PO activity was assayed as described (34) with minor modifications. Briefly, fly heads and larvae were homogenized in PBS containing protease inhibitors. A total of 25 mg homogenate was added to 100 mL of l-DOPA saturated solution in 20 mM phosphate buffer (pH 6.6) and incubated at 37 °C for 1–2 h in the dark. Enzyme activity was measured by recording absorbance at 490 nm with Beckman DU640B spectrophotometer.
Quantitative Real-Time PCR.
Total RNA from fly heads and larvae was extracted with TRIzol reagent (Invitrogen). cDNAs were synthesized from total RNA (0.6 mg) with TaqMan Reverse Transcription Reagent (Applied Biosystems) and amplified with SYBR Green reagent (Biosciences) and the primers described below on an ABI7700 sequence detection system (Applied Biosystems). The primers used were listed in Table S1. A standard curve was generated for each reaction set. Expression was normalized to RP49 values and calculated with the DDCt method.
Supplementary Material
Acknowledgments.
We thank the stock centers of Bloomington, Szeged, Cambridge, and Harvard; the Drosophila Genetic Resource Center, Kyoto Institute of Technology; and J.-M. Reichhart, C. Hashimoto, H. Keshishian, and D. Morisato for fly lines and reagents. We also thank J. Fish for help with histology, S. Ordway for editorial assistance, Y. Lu for Western blot in Fig. S5, and lab members for discussion. S.T.S. acknowledges funding from the Medical Research Council (Grant G0400580) and the help of J. Roote. This work is supported by grants from the National Institutes of Health (F.-B.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903134106/DCSupplemental.
References
- 1.Boxer AL, Trojanowski JQ, Lee VY-M, Miller BL. Frontotemporal lobar degeneration. In: Beal MF, Lang AE, Ludolph AC, editors. Neurodegenerative Diseases: Neurobiology, Pathogenesis and Therapeutics. Cambridge, U.K.: University of Cambridge; 2005. pp. 481–493. [Google Scholar]
- 2.Neary D, Snowden J, Mann D. Frontotemporal dementia. Lancet Neurol. 2005;4:771–780. doi: 10.1016/S1474-4422(05)70223-4. [DOI] [PubMed] [Google Scholar]
- 3.Vossel KA, Miller BL. New approaches to the treatment of frontotemporal lobar degeneration. Curr Opin Neurol. 2008;21:708–716. doi: 10.1097/WCO.0b013e328318444d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong M, et al. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science. 1998;282:1914–1917. doi: 10.1126/science.282.5395.1914. [DOI] [PubMed] [Google Scholar]
- 5.Hutton M, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 6.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 7.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 8.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 9.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 10.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 11.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 12.Momeni P, et al. Sequence analysis of all identified open reading frames on the frontal temporal dementia haplotype on chromosome 3 fails to identify unique coding variants except in CHMP2B. Neurosci Lett. 2006;410:77–79. doi: 10.1016/j.neulet.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 13.Lindquist SG, et al. Frontotemporal dementia linked to chromosome 3 (FTD-3)–current concepts and the detection of a previously unknown branch of the Danish FTD-3 family. Eur J Neurol. 2008;15:667–670. doi: 10.1111/j.1468-1331.2008.02144.x. [DOI] [PubMed] [Google Scholar]
- 14.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JA, et al. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 16.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 18.Shulman JM, Shulman LM, Weiner WJ, Feany MB. From fruit fly to bedside: Translating lessons from Drosophila models of neurodegenerative disease. Curr Opin Neurol. 2003;16:443–449. doi: 10.1097/01.wco.0000084220.82329.60. [DOI] [PubMed] [Google Scholar]
- 19.Reichhart JM. Tip of another iceberg: Drosophila serpins. Trends Cell Biol. 2005;15:659–665. doi: 10.1016/j.tcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 21.Sweeney NT, Brenman JE, Jan YN, Gao FB. The coiled-coil protein Shrub controls neuronal morphogenesis in Drosophila. Curr Biol. 2006;16:1006–1011. doi: 10.1016/j.cub.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 22.Bellen HJ, et al. The BDGP gene disruption project: Single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sevrioukov EA, Moghrabi N, Kuhn M, Krämer H. A mutation in dVps28 reveals a link between a subunit of the endosomal sorting complex required for transport-I complex and the actin cytoskeleton in Drosophila. Mol Biol Cell. 2005;16:2301–2312. doi: 10.1091/mbc.E04-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd TE, et al. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 26.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryder E, et al. The DrosDel deletion collection: A Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charron Y, et al. The serpin Spn5 is essential for wing expansion in Drosophila melanogaster. Int J Dev Biol. 2008;52:933–942. doi: 10.1387/ijdb.072419yc. [DOI] [PubMed] [Google Scholar]
- 29.Osterwalder T, et al. Drosophila serpin 4 functions as a neuroserpin-like inhibitor of subtilisin-like proprotein convertases. J Neurosci. 2004;24:5482–5491. doi: 10.1523/JNEUROSCI.5577-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levashina EA, et al. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- 31.Ligoxygakis P, et al. A serpin mutant links Toll activation to melanization in the host defence of Drosophila. EMBO J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerenius L, Söderhäll K. The prophenoloxidase-activating system in invertebrates. Immunol Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 33.De Gregorio E, et al. An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell. 2002;3:581–592. doi: 10.1016/s1534-5807(02)00267-8. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Wang Y, Yu XQ, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta. A bacteria-inducible serine proteinase containing two clip domains. J Biol Chem. 2003;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- 35.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 36.Chang AJ, Morisato D. Regulation of Easter activity is required for shaping the Dorsal gradient in the Drosophila embryo. Development. 2002;129:5635–5645. doi: 10.1242/dev.00161. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto C, et al. Spatial regulation of developmental signaling by a serpin. Dev Cell. 2003;5:945–950. doi: 10.1016/s1534-5807(03)00338-1. [DOI] [PubMed] [Google Scholar]
- 38.Tang H, Kambris Z, Lemaitre B, Hashimoto C. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev Cell. 2008;15:617–626. doi: 10.1016/j.devcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan CA, Anderson KV. Drosophila: The genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen MD, Julien JP, Rivest S. Innate immunity: The missing link in neuroprotection and neurodegeneration? Nat Rev Neurosci. 2002;3:216–227. doi: 10.1038/nrn752. [DOI] [PubMed] [Google Scholar]
- 41.Okun E, et al. Toll-like receptors in neurodegeneration. Brain Res Rev. 2008;16:1006–1011. doi: 10.1016/j.brainresrev.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Y, et al. Toll-like receptor 8 functions as a negative regulator of neurite outgrowth and inducer of neuronal apoptosis. J Cell Biol. 2006;175:209–215. doi: 10.1083/jcb.200606016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kilic U, et al. TLR-4 deficiency protects against focal cerebral ischemia and axotomy-induced neurodegeneration. Neurobiol Dis. 2008;31:33–40. doi: 10.1016/j.nbd.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Tang SC, et al. Toll-like receptor-4 mediates neuronal apoptosis induced by amyloid beta-peptide and the membrane lipid peroxidation product 4-hydroxynonenal. Exp Neurol. 2008;213:114–121. doi: 10.1016/j.expneurol.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.