Abstract
Proteins that can bind specifically to targets that also have an intrinsic property allowing for easy detection could facilitate a multitude of applications. While the widely used green fluorescent protein (GFP) allows for easy detection, attempts to insert multiple binding loops into GFP to impart affinity for a specific target have been met with limited success because of the structural sensitivity of the GFP chromophore. In this study, directed evolution using a surrogate loop approach and yeast surface display yielded a family of GFP scaffolds capable of accommodating 2 proximal, randomized binding loops. The library of potential GFP-based binders or ″GFAbs″ was subsequently mined for GFAbs capable of binding to protein targets. Identified GFAbs bound with nanomolar affinity and required binding contributions from both loops indicating the advantage of a dual loop GFAb platform. Finally, GFAbs were solubly produced and used as fluorescence detection reagents to demonstrate their utility.
Keywords: alternative scaffold, directed evolution, yeast surface display, thermal stability, loop randomization
Antibodies have long been a mainstay of biological and medical research, and the current use of antibodies as therapeutics has further expanded their portfolio of applications. More recently, to address various challenges such as the reduced stability and production yields of the antibody fragments that are frequently used in in vitro evolution platforms, and in large part as a result of intellectual property concerns, the field of alternative binding scaffolds has emerged (1). By mutagenizing solvent-exposed loop regions or inserting diverse loop repertoires into nonantibody protein scaffolds, specific binding attributes can be conferred to proteins that naturally have desirable properties, such as high stability and production titers. In this way, alternative scaffolds such as the 10th human fibronectin type III domain (2), anticalins (3–5), designed ankyrin repeat proteins (6), and Affibodies (7, 8), among others, have been developed to bind to targets with antibody-like affinity.
Green fluorescent protein (GFP) has also been explored as a potential alternative scaffold. To date, GFP has been used for a wide variety of different applications (9) including Ca2+ detection (10), visualization of protein–protein interactions (11), and as a reporter for protein folding (12). Considerable effort has also been expended in attempts to develop GFP as a binding scaffold that would have 2 potential advantages over the aforementioned alternative scaffolds. First, by combining binding attributes with the intrinsic fluorescence of the GFP protein, the proteins could act as single step detection reagents in applications such as fluorescence-based ELISAs, flow cytometry, and intracellular targeting/trafficking in live cells. Second, GFP fluorescence requires that the protein is properly folded (13) offering an in situ metric for folding fidelity, absent from other alternative scaffolds. Such a folding probe could assist both assessment of library fitness upon binding loop introduction, and subsequent selection of properly folded, soluble clones.
Several attempts have been made to confer binding capability to GFP by inserting binding loops into various solvent-exposed turns that connect the β-strands of the GFP β-barrel structure. The regions of GFP that are most amenable to insertion of amino acids have been determined (turns Gln-157-Lys-158 and Glu-172-Asp-173) (14, 15), although fluorescence is diminished substantially, and when random loops were inserted, the resultant library fluorescence decreased to 2.5% of wild type (14). Selection of GFP-inserted peptide libraries for targeting various intracellular compartments has also been performed (16). In addition, antibody heavy chain CDR3 sequences have been inserted into several loop regions of superfolder GFP, a GFP variant evolved for high stability and improved folding kinetics (17), to create libraries of single CDR3-inserted GFP. Results from this study indicated that insertion at many sites substantially reduces GFP fluorescence as seen previously with standard GFP variants (18). Three loop regions of the superfolder GFP, however, tolerated single loop CDR insertions (including Asp-173-Gly-174) such that it was possible to isolate fluorescent binders against protein targets using T7 phage display, with the best being a 470 nM lysozyme binder (19). This level of affinity is in the realm of that found for peptide binders (20), likely as a consequence of its single-binding loop design. Affinity of GFP-based binding proteins could therefore in principle benefit from display of multiple binding loops that could act together to form a cooperative binding interface. However, the lone examples of multiple loop insertion into GFP include insertion of hemagglutinin peptide (21) or random loops (22) into 2 loops on opposite faces of GFP. While suitable for the authors' goals, these insertion locations would not be ideal for forming a cooperative binding interface. Moreover, GFP fluorescence of the resulting clones in the case of the random loop libraries was not demonstrated (22). Thus, to date, robust fluorescent multiple loop-inserted GFP repertoires have not been described, even using the superfolder GFP as a template, likely because studies have used preexisting GFP variants that while bright and stable have not been optimized for binding loop insertion. Thus, in this study, the GFP scaffold itself was evolved to maintain its fluorescence properties in the presence of 2 inserted binding loops, and we demonstrated that scaffolds designed in this way were capable of accepting a diverse loop repertoire from which fluorescent binding proteins could be isolated.
Results
Effects of Single and Dual Loop Insertions on GFP Expression and Fluorescence.
The initial goal of this study was to evaluate the capability of monomeric yeast enhanced green fluorescent protein (GFPM) (23, 24) (see Materials and Methods for details) to accommodate dual loop insertions. The Glu-172-Asp-173 turn region was chosen as 1 insertion site since earlier studies have shown that GFP can retain its fluorescence upon insertions of various lengths at this location (14, 15) (Fig. 1A). The second location selected was turn Asp-102-Asp-103 because of its proximity (≈1.6 nm) to Glu-172-Asp-173 on the same face of the β-barrel allowing the eventual possibility of improved affinity for targets through cooperative antigen binding with both inserted loop regions (Fig. 1A). Surrogate binding loops in the form of CDRH3 and CDRL3 from the D1.3 anti-lysozyme antibody (25) were inserted into positions Glu-172/Asp-173 and Asp-102/Asp-103, respectively (Table S1 and Table S2), and the resultant constructs were displayed on the surface of yeast. While a single loop inserted at the Glu-172/Asp-173 site of GFPM (GFPM-XMH3) could retain GFP fluorescence and expression on the surface of yeast as expected (Fig. S1), single-loop insertion at Asp-102/Asp-103 (GFPM-ASL3) ablated all GFP fluorescence and much of the surface expression (Fig. 1B and Fig. S1). When both loops were inserted (GFPM-H3L3), very little surface display was detected and the protein possessed no fluorescence (Fig. 1B and Fig. S1).
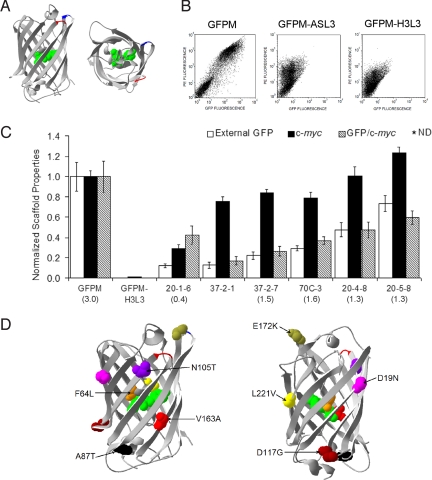
Fig. 1.
Directed evolution of a set of dual loop-compatible scaffolds. (A) Side view and top view of GFPM with loop insertion site Asp-102/Asp-103 highlighted in red and Glu-172/Asp-173 highlighted in blue. The chromophore residues are shown in space-fill green. The structure used was that of GFP with the S65T mutation pdb ID = 1EMA (38). (B) Flow cytometric dot plots for surface displayed GFPM, single loop-inserted GFPM-ASL3, and dual loop-inserted GFPM-H3L3. PE fluorescence (y axis) is indicative of full-length expression using the c-terminal c-myc epitope for detection and GFP fluorescence (x axis) is indicative of properly folded GFPM or its variants. The negative population lacking both PE and GFP fluorescence located in the lower left quadrant of dot plots in B is a nondisplaying population characteristic of yeast display. (C) Fluorescence (external GFP), full-length expression (c-myc), and fluorescence per molecule (GFP/c-myc) for the evolved scaffolds were normalized to non loop-inserted GFPM for comparison. All data were derived using flow cytometry of triplicate yeast transformants induced for display at 20 °C. It is important to note the GFP fluorescence measured via flow cytometry comprises an internal contribution from protein retained inside the cell and an external contribution from the displayed protein. Only the external contribution to the fluorescence is reported here (see SI Methods for details). Clones are named based on the selection they were recovered from, except for 20–5-8 which was recovered in both the 20 °C and 37 °C selections. ND denotes the absence of a detectable signal. Secretion yields for the various scaffolds using a baseline expression system are denoted beneath each scaffold in (mg/L). (D) GFPM structure with residues mutated in the 20–5-8 scaffold highlighted. Loop insertion site Asp-102/Asp-103 is highlighted in red and Glu-172/Asp-173 is highlighted in blue. Structure on right was generated by rotating the structure on the left 180° to allow a clearer view of several of the mutations.
Directed Evolution of a Family of Scaffolds that Can Accommodate Dual Loop Insertion.
We hypothesized that evolution of the GFPM-H3L3 dual surrogate loop-inserted scaffold for improved fluorescence and expression would yield a better folded and processed GFP scaffold capable of accommodating a diverse loop repertoire. Since GFP requires correct folding to become fluorescent (13) and very little nonfluorescent GFPM-H3L3 protein makes it through the yeast secretory pathway to the cell surface (Fig. 1B), the secretory quality control machinery provided a reasonable filter that limited the export of proteins lacking an intact chromophore environment (Fig. S1). Thus, the folding and processing fitness of GFPM-H3L3 was used as a convenient selection criterion. Scaffolds were mutagenized while keeping the inserted surrogate loops free of mutation (Fig. S3). Then, scaffolds that were both fluorescent and displayed were selected using a combination of 2 induction conditions over 4 rounds of directed evolution. For directed evolution rounds 1–4, scaffold libraries were selected after display induction at 20 °C since it has been shown previously that it is the optimum temperature for soluble expression of unmodified GFP using yeast (26). In parallel to 20 °C selections, for directed evolution rounds 2–4, the library was also selected after induction at 37 °C to apply a more restrictive selection pressure that requires the scaffold to be properly folded and processed even at a temperature that has been shown to have deleterious effects on yeast expression for unmodified GFP (26).
From the first round, 3 unique clones (20–1-3, 20–1-6, and 20–1-8) were obtained that exhibited improved fluorescence or expression or both (Table 1 and Fig. 1C, compare with GFPM-H3L3). Among these 3, clone 20–1-6 had 3 mutations: D19N, F64L, and A87T, all of which were conserved in 20–1-3 and 20–1-8 (Table 1). To further improve the fluorescence and expression of the dual loop-inserted scaffold, the DNA sequences corresponding to 20–1-3, 20–1-6, and 20–1-8 were shuffled and mutated to obtain a second generation library of dual loop-inserted scaffolds. The best performing clones resulting from the second round, 37–2-1 and 37–2-7, were derived from the 37 °C selection scheme. Of the second-round mutants, clone 37–2-7 had the highest external GFP fluorescence, yielding fluorescence per molecule (GFP/c-myc) 28% that of unaltered GFPM, with substantially improved cell surface expression. Comparison of the mutations found in 37–2-7 to 20–1-6 indicated the conservation and importance of all 20–1-6 mutations plus the presence of 2 additional mutations, Y39H and V163A (Table 1). The GFP fluorescence per molecule was a bit lower for 37–2-7 than 20–1-6 indicating the additional mutations were primarily “expression mutations” (Fig. 1C). After additional mutagenesis of 37–2-1 and 37–2-7, the best clone from the third round of directed evolution, 20–4-8, arose from the 20 °C selection and had an approximate 2-fold increased fluorescence/molecule to reach 50% of the nonloop inserted GFPM parent (Fig. 1C). The mutant consisted of the combination of 37–2-1 and 37–2-7 mutations along with an additional mutation L221V. The best final round clone derived using 20–4-8 as a mutagenesis template, 20–5-8 (N105T and D117G), was identified in both the 20 °C and 37 °C sorts and possessed 60% of the fluorescence per molecule of GFPM (Fig. 1 C and D). To confirm that our yeast surface measurements of improved fluorescence/molecule correlated with improved function of soluble protein, we measured the corresponding brightness of GFPM, 37–2-7, and 20–5-8 as soluble, purified proteins. Indeed the fluorescence per molecule values of the soluble scaffold proteins correlated well with those measured on the yeast cell surface with 37–2-7 at 37% of GFPM (28% on surface) and 20–5-8 at 66% of GFPM (60% on surface) (Table S3).
Table 1.
Scaffold mutations
| 20–1-3 | 20–1-6 | 20–1-8 | 37–2-1 | 37–2-7 | 70C-3 | 20–4-8 | 20–5-8 | |
|---|---|---|---|---|---|---|---|---|
| K3E | x | |||||||
| D19N | x | x | x | x | x | x | x | x |
| K26E | x | |||||||
| Y39H | x | x | x | x | ||||
| F64L | x | x | x | x | x | x | x | x |
| A87T | x | x | x | x | x | x | x | x |
| N105T | x | |||||||
| D117G | x | |||||||
| Y151C | x | |||||||
| V163A | x | x | x | x | x | |||
| E172K | x | x | x | x | ||||
| L221V | x | x |
In addition to evolving folding and expression competence by 20 °C and 37 °C selections, it was hypothesized that direct selection of a more thermally stable surrogate loop-inserted scaffold could aid the maintenance of structural integrity by stabilizing the β-barrel fold and chromophore environment, leading to improved protein folding and processing. Thus, for the third round of directed evolution, using 37–2-1 and 37–2-7 as the templates for additional mutagenesis, a selection for loop-inserted scaffolds having increased resistance to thermal denaturation at 70 °C was also performed. The 70 °C selection, while yielding a significantly more thermally resistant scaffold, 70C-3 (half-life = 21 min), failed to significantly improve GFP fluorescence and expression properties above the 37–2-7 parent (half-life = 9 min) (Fig. 1C and Fig. S2). In contrast, the less stable 20–4-8 (half-life = 18 min) scaffold that arose from the same 37–2-1 and 37–2-7 parents via the 20 °C selection had substantially improved properties (Fig. 2A and Fig. S2). Interestingly, even from the 20 °C and 37 °C selections, there was a distinct trend toward scaffolds that along with improved fluorescence and expression, also exhibited gains in thermal stability matching or exceeding non loop-inserted GFPM (Fig. S2). Taken together, it appears that improved scaffold fluorescence and expression required an improvement in thermal stability, but improved thermal stability alone does not guarantee improved scaffold fluorescence or expression. Thus, the secretory pathway of yeast is providing an additional selection criterion that can help sample segments of the fitness landscape not wholly substituted by a single biophysical property such as resistance to thermal denaturation. All round 2–4 scaffolds were produced as soluble fluorescent proteins at levels between 45–55% of GFPM, corresponding to 1.3–1.6 mg/L (Fig. 1C), and thus represent potential scaffolds for dual random loop insertion in that they are expressed on the yeast surface and can be solubly produced as fluorescent proteins.
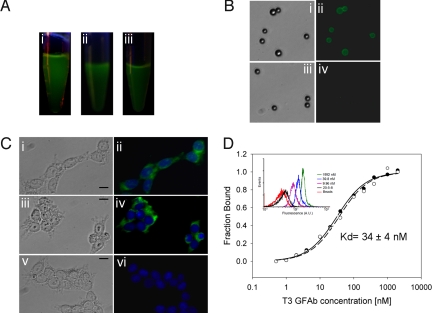
Fig. 2.
Properties of soluble, purified GFAbs. (A) Samples of purified protein illuminated with a hand-held UV lamp indicating the presence of fluorescent protein: i) GFPM, ii) 20–5-8, and iii) G6 GFAb. (B) Streptavidin-coated polystyrene beads were labeled with streptavidin-PE binding 1.3 GFAb (i and ii) or its parent scaffold 20–5-8 (iii and iv) and imaged by fluorescence microscopy. (C) Permeablized HEK-293 cells were incubated with: (i and ii) GAPDH binding G6 GFAb, (iii and iv) an anti-GAPDH antibody followed by fluorescent Alexa488 secondary antibody, and (v and vi) the parent scaffold 20–5-8, and cells were monitored for green fluorescence (GFAb-derived: i, ii, v, and vi or Alexa488-derived: iii and iv). DAPI was used to stain the nucleus (blue). (Scale bar, 20 μm.) (D) Affinity titration for secreted T3 GFAb on TrkB-loaded beads. Binding-dependent GFAb fluorescence was monitored by flow cytometry as a function of T3 concentration to generate a binding curve and associated KD. Representative flow cytometry histograms for various T3 dilutions are presented in the Inset. Also shown are no GFAb, bead-only controls, and the negligible binding of parent scaffold 20–5-8 to TrkB-loaded beads. Beads loaded with irrelevant antigen and labeled with T3 also exhibited negligible binding mirroring that shown for 20–5-8. Data from duplicate samples are depicted as open and filled circles along with their associated equilibrium binding model fits.
Development of Dual Random Loop GFPM Libraries.
The next step of the study was to determine whether the surrogate loop-evolved scaffolds could accommodate amino acid diversity at both positions simultaneously. We also wished to determine how the various scaffolds having differing stability, fluorescence, and expression properties affected the quality of the resultant libraries in terms of expressed diversity and overall fitness. For each scaffold, the inserted loop regions were replaced by loops of the same length (8 aa at 172 and 173 and 9 aa at 101 and 102) that had been randomized using the NNK oligonucleotide method (See Materials and Methods for details, Fig. S3). The expressed diversity of the dual random loop libraries built into 37–2-7, 70C-3, and 20–4-8 scaffolds was indistinguishable. When corrected for stop-codon probability and the negative displaying yeast population lacking plasmid (27), approximately 70% of the dual random loop-inserted GFPs not containing stop codons are displayed on the surface and possess fluorescence above background (Fig. S4A). Interestingly, although an improvement in thermal stability occurred when evolving 37–2-7 to 70C-3 and 20–4-8 (Fig. S2), there was no effect on the expressed diversity. However, the stability, folding, and processing attributes of the scaffold do appear to play an important role in library fitness for the expressed clones because the aggregate fluorescence, expression and stability properties of the libraries generally improve as the fitness of the scaffold improves (Fig. S4A and compare with scaffold trends in Fig. 1C). As an exception, the best scaffold in terms of fluorescence and expression properties, 20–5-8, only allowed 55% of the loop-inserted GFPs to be expressed. In addition, there was a drop in fluorescence and expression properties compared with the 20–4-8 library, indicating that gains provided by the evolution strategy may have been beginning to become specific to the amino acid content of the surrogate loops. Finally, to test the scaffold capability of accommodating different forms of loop diversity, a dual loop library in the 37–2-7 scaffold was also created using randomized loops possessing only tyrosine and serine residues as these have been shown to allow minimalist design of binding sites (28). Indeed, 37–2-7 also accommodated this form of diversity although the fluorescence and expression properties were a bit diminished compared with the fully randomized loops discussed above (Fig. S4A).
Selection and Characterization of Dual Loop-inserted GFP Binders (GFAbs).
For the selection of binders to various antigens, we combined the various scaffold libraries discussed above to form a selectable library of high fitness. Using flow cytometry, we recovered the top 20% of clones in terms of both GFP fluorescence and expression. The resulting GFAb library had an expressed diversity of 6 × 106 clones having high fitness averaging 40% of the fluorescence and 60% of the expression of nonloop inserted GFPM (Fig. S4B). This library was used for all binding selections discussed below. As proof-of-concept selections, binders against streptavidin-phycoerythrin and biotin-phycoerythrin conjugates were selected by flow cytometry, and several unique GFAbs were recovered against each target and affinity titrations on the surface of yeast yielded binding dissociation constants from 70 nM to micromolar (Fig. S4C and Table 2). Next, GFAbs were raised against the monomeric extracellular domain of a neurotrophin receptor (TrkB) and against glyceraldehyde 3-phosphate dehydrogenase (GAPDH). While GAPDH-binding GFAbs bound in the 18–500-nM range, GFAbs specific to TrkB gave monomeric binding dissociation constants as low as 3.2 nM (Fig. S4C and Table 2). Moreover, although no counterselections were performed to fine-tune specificity, binding of the high affinity T3 GFAb to 2 other tyrosine kinase neurotrophin receptors (TrkA and TrkC) having high homology to TrkB was not detected, indicating the capability for identifying isoform-specific GFAbs. As a whole, GFAb clones had properties ranging from 25–160% of the displayed fluorescence/molecule of the parent scaffold, and surface expression levels ranged from 50–150% of the scaffold (Table 2).
Table 2.
Expression, fluorescence and binding properties of selected GFAb clones
| Target | GFAb | Scaffold | Loop 102–103 | Loop 172–173 | KD, nM* | Expression, % of scaffold‖ | GFP/c-myc, % of scaffold‖ | Secreted Yield, % of scaffold†† | GFP/mol, % of scaffold‡‡ |
|---|---|---|---|---|---|---|---|---|---|
| TrkB | T1 | 37–2-7 | SKSRSLESV | GYLRWLFG | 60 | 79 ± 5 | 140 ± 20 | ||
| T3 | 20–5-8 | VINPFTVRS | NAWVVHRR | 3.2 ± 0.7 | 66 ± 4 | 77 ± 10 | 20 [0.30] | 43 | |
| T5 | 37–2-7 | PSWWSLFFP | STATGLFA | 29 ± 4 | 78 ± 4 | 160 ± 30 | 24 [0.32] | 170 | |
| GAPDH | G6 | 20–5-8 | RVSRFLLTT | KSRIISSQ | 18 ± 6** | 99 ± 7 | 73 ± 11 | 80 [1.1] | 120 |
| G7† | 20–5-8 | NVIYPFLYA | RVTKTKHK | > 500 | |||||
| G4† | 20–5-8 | RGVSKSFLL | QGITKGYK | > 500 | 130 ± 20 | 91 ± 31 | |||
| G8† | 20–5-8 | SHCLFRKCY | ARSIRMKV | > 500 | 140 ± 20 | 51 ± 18 | |||
| Strep-PE | 1.3‡ | 20–5-8 | AISRSFFST | IRNLKYTN | 70 ± 11 | 120 ± 10 | 67 ± 15 | 140 [1.9] | 41 |
| 2.7§ | 20–5-8 | VFSSLRYHV | LRMDDTNP | 140 ± 20 | 96 ± 7 | 61 ± 11 | 120 [1.6] | 60 | |
| 2.17§ | 20–5-8 | KARMRLFLM | EYGDTLLS | 110 | 85 ± 6 | 27 ± 6 | |||
| Biotin-PE | 2¶ | 20–5-8 + E115K | RGLFWPVLI | PNPVFRQN | 1400 | 58 ± 4 | 37 ± 9 | ||
| 5¶ | 20–5-8 | SCSWCLFTL | KNQMGMGK | 260 | 52 ± 3 | 47 ± 8 | |||
| 12¶ | 37–2-7 | KSHFTIFRT | DGWRRTTV | 250 | 87 ± 7 | 56 ± 8 | |||
| 16¶ | 20–5-8 | NCRWCTYYL | STSNWMRM | 190 | 56 ± 7 | 25 ± 5 |
*KD ± SD reported for triplicate titrations on yeast surface, otherwise KD from single sample titration reported.
†Clone binds both GAPDH and streptavidin phycoerythrin.
‡Clone binds streptavidin alone.
§Clone requires presence of both streptavidin and PE.
¶Clone binds PE alone.
‖Expression (c-myc) and fluorescence (external GFP/c-myc) properties measured on yeast surface normalized to the parent scaffold.
**Represents affinity estimate based on low concentration binding data as binding at higher concentrations (>100 nm) of GAPDH antigen exhibit the hook effect with decreased binding signal
††Values in brackets represent non-optimized shakeflask yields in mg/L.
‡‡GFP per molecule (brightness) for the soluble protein defined as the extinction coefficient x quantum yield. Brightness values are normalized to parent scaffold to show the effects of changes in loop sequence. Detailed values for extinction coefficient and quantum yield can be found in Table S3.
We also examined whether both loops in the putative binding interface contributed to the binding affinity of the selected clones. To accomplish this task, we individually grafted the binding loops for T3 back into the 20–5-8 scaffold such that single-loop GFAbs were created with the second loop being the surrogate loop. Affinity titrations of the single-binding loop variants were performed on the yeast surface and indicated that both loops contribute to the observed binding affinity. While the 20–5-8 scaffold possesses no binding affinity toward TrkB, addition of the 172–173 loop yields a GFAb with 300-nM binding affinity, addition of the 102–103 binding loop yields a GFAb of 19-nM affinity, while both loops as in the original T3 GFAb yield 2 nM affinity (Fig. S4C). The analogous constructs were also created for the G6 GFAb that binds GAPDH with titrations again indicating dual loop contribution to binding.
To further characterize the GFAbs, 5 were secreted from yeast and purified yields ranged from 0.3–2 mg/L (Table 2). All soluble, purified proteins retained fluorescence with per molecule values ranging from 40–120% of the parent scaffold (Fig. 2A, Table 2, and Table S3). Moreover, the purified GFAbs possess binding activity as demonstrated by several fluorescent labeling assays. Streptavidin-PE binding 1.3 GFAb specifically labeled streptavidin-linked beads with a fluorescent signal discernable from the parent scaffold (Fig. 2B), and binding could be competed with soluble streptavidin-PE antigen. In addition, the G6 GAPDH-binding GFAb was used as a single-step reagent to detect GAPDH present in the cytoplasm of HEK-293 cells (Fig. 2C). Finally, purified and soluble T3 GFAb was used in combination with TrkB-loaded beads and FACS for detailed binding-affinity measurement. Using solely the intrinsic fluorescence that results from T3 GFAb binding to the TrkB-loaded beads, the binding affinity of the soluble T3 GFAb was determined to be 34 nM (Fig. 2D). The nanomolar binding affinity of soluble T3 GFAb was also further confirmed by a solution-phase competition assay where both the T3 and TrkB were in soluble form (see SI Methods for details). In summary, for each of the 4 targets there was at least 1 GFAb clone with a solid combination of affinity, fluorescence, and expression properties, indicating the ability to isolate useful lead molecules from the high fitness dual loop-inserted GFP library.
Discussion
In this investigation, we demonstrate that it is possible to create fluorescent dual-loop inserted GFAb scaffolds capable of binding to various antigens with nanomolar affinity. Examination of the mutations in the evolved scaffolds reveals some cross-over with mutants previously identified to impart fluorescence and stability to GFP (Table 1). The mutation F64L has been uncovered while evolving GFP for higher fluorescence when produced in E. coli, and the mutation improves the fluorescence per molecule in addition to shifting the excitation maximum to 488 nm (29). The same study yielded the S65G and S72A mutations used in our enhanced GFPM starting template. Moreover, our selection for improved fluorescence per molecule was performed using an excitation of 488 nm, suggesting how the F64L mutation may function. DNA shuffling was previously used to improve the fluorescence of GFP largely by increasing solubility in bacteria, and the cycle 3 mutant identified in that study included the mutation V163A (30). The V163A mutant is present in all of our dual loop library scaffolds and was likely isolated because the selection pressure used here included improved folding and processing through the yeast secretory pathway which could be a function of solubility. Another relevant comparison is the superfolder GFP that has been used quite extensively in binding loop insertion studies as described in the introduction. This protein was raised by attaching GFP to an insoluble protein (H-subunit ferritin) and evolving the GFP for enhanced fluorescence and solubility of the complex when expressed in E. coli (17). In addition to F99S, M53T, V163A, F64L, and S65T present in the superfolder starting template, 6 additional superfolder mutations: S30R, Y39N, N105T, Y145F, I171V, and A206V were introduced that improved the forward folding kinetics of GFP and its chemical stability. Of the 6 superfolder mutations, Y39N (Y39H, this study) and N105T are found in our scaffolds. Interestingly, these 2 mutations appeared to function differently in superfolder GFP, Y39N improved refolding kinetics, while N105T improved refolding stability, both of which could be argued to assist processing through the yeast secretory pathway. In addition, 5 scaffold mutations (A87T, D19N, E172K, L221V, and D117G) exist, 2 of which were found in the very first round of directed evolution, which to our knowledge have not been reported as assisting fluorescence or expression properties of GFP or its variants. It is interesting to note that 2 of these mutations, A87T and D117G, along with N105T, are located in or near turns involved in the 2 beta strands that are part of the Asp-102-Asp-103 insertion position that was most deleterious upon surrogate loop insertion (Fig. 2B). Another of these mutations E172K is part of the Glu-172-Asp-172 loop-insertion site (Fig. 2B). Thus, these additional mutations are likely partially a consequence of the filter provided by the advanced secretory pathway of yeast that was an important part of the selection procedure, and partially a result of our approach to engineer a dual loop-compatible scaffold using the surrogate loop approach.
Yeast display is well suited for the engineering of fluorescent proteins because it is possible to select libraries using antigen and GFAb fluorescence as dual simultaneous criteria. This approach therefore results in selection of only fluorescent binders, something that could not be guaranteed using phage display without the aid of labor-intensive secondary confirmations (18). Moreover, while there is not a perfect quantitative agreement between surface-displayed and secreted GFAb stability, fluorescence and binding properties (Figs. 1C and 2D, Table 2, Fig. S2, and Table S3), surface display fitness of GFAbs certainly correlates with GFAbs that can be produced solubly while maintaining solid fluorescence and binding properties. The GFAb affinities for the selected clones ranged from low nanomolar to micromolar and were similar to those seen for binding clones isolated from other nonimmune antibody and alternative scaffold libraries (2, 31, 32). Interestingly, compared with the highest affinity single loop-inserted GFP binder previously reported (470 nM) (19), the dual-loop scaffold appeared capable of providing an extra level of binding affinity as loop-swapping experiments with 2 individual clones selected against different antigens indicated that each loop contributed to the measured KD value. Moreover, it is expected that standard evolutionary techniques could be used to fine-tune the specificity, affinity and fluorescence properties of our lead GFAb molecules as desired. Although we constrained the randomized loop length to that of the surrogate loops for our lead library, it may prove useful for the fine-tuning of GFAb properties to include a component of loop length diversity as this approach has proven fruitful for antibody affinity maturation and for artificial scaffold maturation (28, 33). Moreover, although the extracted GFAb binders all originated from the NNK-based loop libraries rather than the binary code YS library, further refinement of the amino acid content of the binding loops could also be a target for library optimization (34). Finally, successful development of dual-loop GFAb scaffolds could enable a wide range of other applications given the range of GFP spectral variants that have been developed (9), and the surrogate loop approaches used here could in principle be applied to other structurally homologous fluorescent proteins like the monomeric red fluorescent protein family (35).
Materials and Methods
Strains and Media.
Surface display was performed using the standard surface display yeast strain EBY100 (MATa AGA1::GAL1-AGA1::URA3 ura3–52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL). Saccharomyces cerevisiae strain BJ5464 (MATα ura3–52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) was used for protein secretion. Yeast cells were grown in minimal SD-CAA medium (20 g/L dextrose, 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 10.19 g/L Na2HPO4.7H2O, and 8.56 g/L NaH2PO4.H2O) and protein display or secretion was induced using SG-CAA (20 g/L galactose replacing dextrose). BSA (BSA) was added at 1 g/L as a nonspecific carrier for protein secretion studies. The E. coli strains XL1-Blue (Stratagene) and DH5α (Invitrogen) were used for molecular cloning. Luria–Bertani medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, pH 7.5, and 50 μg/mL ampicillin) was used for bacteria growth and plasmid amplification.
Plasmids.
Starting with pRS 316-yEGFP (26) which encodes a yeast codon-optimized variant of GFP (yEGFP) that also possesses the fluorescence enhancing mutations S65G and S72A (29), site directed mutagenesis (Stratagene) was used to change alanine at position 206 to lysine to convert yEGFP into its monomeric form (24) (GFPM), and this plasmid was denoted pRS 316-GFPM (Table S1). Next, insertion mutagenesis (Stratagene) was used to insert 2 unique restriction sites AflII and SpeI between amino acids Asp-102 and Asp-103 to give pRS 316-GFPM-AS (Table S2). Two more unique restriction sites, XbaI and MluI, were inserted between amino acids Glu-172/Asp-173 to yield the plasmid pRS 316-GFPM-XM. Insertion of restriction sites at both locations resulted in the plasmid pRS 316-GFPM-ASXM. These ORFs were then transferred to the pCT yeast surface display plasmid using NheI and BamH1 restriction sites (26). Synthesized oligonucleotides (IDT DNA) encoding the 9 aa of CDRL3 and 8 aa of CDRH3 from the single-chain D1.3 antibody (25) were inserted at positions Asp-102/Asp-103 and/or Glu-172/Asp-173, respectively, to yield 3 more plasmids: pCT-GFPM-ASL3, pCT-GFPM-XMH3, and pCT-GFPM-H3L3 (Table S2). Finally, pCT-GFPM-H3L3 was transferred to the pCT-ESO plasmid using NheI and BamH1 restriction sites as this plasmid is better suited for library mutagenesis (36).
Scaffold and Dual Random Loop Library Creation.
To create the scaffold libraries, the inserted loop regions were kept constant while the remaining GFPM scaffold was mutated (see Fig. S3 for schematic, Table S1 for primers used, and SI Methods for experimental details). To create the dual random loop libraries, the scaffold including the added restriction sites was left unaltered while the 9 aa inserted at Asp-102/Asp-103 and the 8 aa inserted at Glu-172/Asp-173 were simultaneously randomized using the NNK method (see Fig. S3 for schematic, Table S1 for primers used, and SI Methods for experimental details).
Library Screening and Selection.
For scaffold evolution, all libraries were grown in selective SD-CAA medium at 30 °C to an OD600 of 1.0 and were induced at 20 °C or 37 °C in SG-CAA for 12–16 h. For 70 °C selections, induced yeast were first subject to thermal denaturation for 30 min before sorting. The yeast display libraries were labeled with anti-c-myc antibody (1:100 dilution) (Covance) followed by anti-mouse phycoerythrin (PE) (1:40 dilution) (Sigma Aldrich). The library was first enriched for GFP positive clones for 2 to 3 rounds of sorting and the last round involved a stringent gate selecting cells with both GFP fluorescence and the presence of the c-myc epitope tag. All sort experiments were performed on a Becton Dickinson FACSVantage SE flow cytometric sorter at the University of Wisconsin Comprehensive Cancer Center. The recovered clones were sequenced using the PNL6 primer (31) (University of Wisconsin Biotechnology Center).
For selection of GFAbs, the dual random loop libraries using scaffolds 37–2-7, 70C-3, 20–4-8, and 20–5-8 were pooled together, induced, and labeled with anti c-myc antibody (Covance) followed by anti mouse PE (Sigma Aldrich). The double GFP and PE positive pool was collected and this pooled library was the source library for all binding selections. The intrinsic fluorescence of streptavidin PE and biotinylated PE was used for recovery of GFAbs against these ligands, whereas tyrosine kinase receptor B (TrkB) (R&D Systems) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Sigma Aldrich) were first biotinylated for detection of ligand binding (see SI Methods for selection details). All of the binders isolated were tested for affinity to secondary reagents and no cross-reactivity to an irrelevant biotinylated ligand (500 nM hen egg lysozyme) was detected. Any detected binding to secondary reagents is noted in Table 2.
Protein Secretion and Purification.
Scaffold and GFAb ORFs were subcloned from the pCT-ESO display construct to the pRS316-GFP expression vector by NheI-BamHI digest (26). BJ5464 transformed cells were grown for 72 h in SD-CAA at 30 °C. The media was switched to SG-CAA supplemented with BSA for 72 h at 20 °C. Protein purification was performed using a Ni-NTA column (Qiagen) as described earlier (26). Relative secretion levels were determined by western blot analysis using the anti-c-myc antibody as described earlier (26). Fluorescence properties of the purified protein as reported in Table S3, were measured as described in the SI Methods.
Characterization of GFAbs.
Binding affinity was determined using yeast surface display (37). Yeast displaying GFAbs were incubated with different concentrations of ligand for 1 hour on ice. Binding was subsequently detected using the secondary reagent combinations detailed above and quantified by flow cytometry. The dissociation constant was obtained by fitting the binding curve to a 2-parameter equilibrium binding model as previously described (37). To determine whether both loops contributed to binding, unique restriction sites upstream and downstream of the ORF were used to singly reinsert the surrogate loop into position 102/103 or 172/173. The effect of loops on affinity of TrkB binder T3 and GAPDH binder G6 were measured using yeast surface display. For details regarding the measurement of soluble GFAb properties in Fig. 3, please see SI Methods.
Supplementary Material
Acknowledgments.
The authors thank Dr. Nitin Agarwal and Viraj Kamat for their assistance with experimental design. This work was funded by the National Institutes of Health Grant NS052649 and the University of Wisconsin Graduate School. Y.K.C. is a recipient of a Genomic Sciences Training Program Fellowship funded through the National Institutes of Health, 5T32HG002760.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902828106/DCSupplemental.
References
- 1.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Lipovsek D, et al. Evolution of an interloop disulfide bond in high-affinity antibody mimics based on fibronectin type III domain and selected by yeast surface display: Molecular convergence with single-domain camelid and shark antibodies. J Mol Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 3.Korndorfer IP, Schlehuber S, Skerra A. Structural mechanism of specific ligand recognition by a lipocalin tailored for the complexation of digoxigenin. J Mol Biol. 2003;330:385–396. doi: 10.1016/s0022-2836(03)00573-4. [DOI] [PubMed] [Google Scholar]
- 4.Schlehuber S, Beste G, Skerra A. A novel type of receptor protein, based on the lipocalin scaffold, with specificity for digoxigenin. J Mol Biol. 2000;297:1105–1120. doi: 10.1006/jmbi.2000.3646. [DOI] [PubMed] [Google Scholar]
- 5.Vogt M, Skerra A. Construction of an artificial receptor protein (“anticalin”) based on the human apolipoprotein D. Chembiochem. 2004;5:191–199. doi: 10.1002/cbic.200300703. [DOI] [PubMed] [Google Scholar]
- 6.Zahnd C, et al. A designed ankyrin repeat protein evolved to picomolar affinity to Her2. J Mol Biol. 2007;369:1015–1028. doi: 10.1016/j.jmb.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Nord K, Nord O, Uhlen M, Kelley B, Ljungqvist C, Nygren PA. Recombinant human factor VIII-specific affinity ligands selected from phage-displayed combinatorial libraries of protein A. Eur J Biochem. 2001;268:4269–4277. doi: 10.1046/j.1432-1327.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- 8.Nord K, Gunneriusson E, Ringdahl J, Stahl S, Uhlen M, Nygren PA. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 10.Miyawaki A, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 11.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 13.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 14.Abedi MR, Caponigro G, Kamb A. Green fluorescent protein as a scaffold for intracellular presentation of peptides. Nucleic Acids Res. 1998;26:623–630. doi: 10.1093/nar/26.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi N, Yanagawa H. Design of generic biosensors based on green fluorescent proteins with allosteric sites by directed evolution. Febs Letters. 1999;453:305–307. doi: 10.1016/s0014-5793(99)00732-2. [DOI] [PubMed] [Google Scholar]
- 16.Peelle B, Lorens J, Li W, Bogenberger J, Payan DG, Anderson DC. Intracellular protein scaffold-mediated display of random peptide libraries for phenotypic screens in mammalian cells. Chem Biol. 2001;8:521–534. doi: 10.1016/s1074-5521(01)00031-x. [DOI] [PubMed] [Google Scholar]
- 17.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 18.Kiss C, et al. Antibody binding loop insertions as diversity elements. Nucleic Acids Res. 2006;34:15. doi: 10.1093/nar/gkl681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai M, et al. Using T7 phage display to select GFP-based binders. Protein Eng Des Sel. 2008;21:413–424. doi: 10.1093/protein/gzn016. [DOI] [PubMed] [Google Scholar]
- 20.Craig L, Sanschagrin PC, Rozek A, Lackie S, Kuhn LA, Scott JK. The role of structure in antibody cross-reactivity between peptides and folded proteins. J Mol Biol. 1998;281:183–201. doi: 10.1006/jmbi.1998.1907. [DOI] [PubMed] [Google Scholar]
- 21.Zhong JQ, Freyzon Y, Ehrlich DJ, Matsudaira P. Enhanced detection sensitivity using a novel solid-phase incorporated affinity fluorescent protein biosensor. Biomol Eng. 2004;21:67–72. doi: 10.1016/j.bioeng.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Chen S-S, Yang Y, Barankiewicz T. Selection of IgE-binding aptameric green fluorescent protein (Ap-GFP) by the ribosome display (RD) platform. Biochem Biophys Res Commun. 2008;374:409–414. doi: 10.1016/j.bbrc.2008.06.123. [DOI] [PubMed] [Google Scholar]
- 23.Cormack BP, Bertram G, Egerton M, Gow NAR, Falkow S, Brown AJP. Yeast-enhanced green fluorescent protein (yEGFP): A reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 24.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 25.Bajorath J, Harris L, Novotny J. Conformational similarity and systematic displacement of complemenarity-determining region loops in high-resolution antibody X-ray structures. J Biol Chem. 1995;270:22081–22084. doi: 10.1074/jbc.270.38.22081. [DOI] [PubMed] [Google Scholar]
- 26.Huang D, Shusta EV. Secretion and surface display of green fluorescent protein using the yeast Saccharomyces cerevisiae. Biotechnol Prog. 2005;21:349–357. doi: 10.1021/bp0497482. [DOI] [PubMed] [Google Scholar]
- 27.Rakestraw A, Wittrup KD. Contrasting secretory processing of simultaneously expressed heterologous proteins in Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:896–905. doi: 10.1002/bit.20780. [DOI] [PubMed] [Google Scholar]
- 28.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. High-affinity single-domain binding proteins with a binary-code interface. Proc Natl Acad Sci USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 30.Crameri A, Whitehorn EA, Tate E, Stemmer WPC. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 31.Feldhaus MJ, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 32.Wang XX, Cho YK, Shusta EV. Mining a yeast library for brain endothelial cell-binding antibodies. Nat Methods. 2007;4:143–145. doi: 10.1038/nmeth993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbreth RN, Esaki K, Koide A, Sidhu SS, Koide S. A dominant conformational role for amino acid diversity in minimalist protein-protein interfaces. J Mol Biol. 2008;381:407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange, and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 36.Piatesi A, et al. Directed evolution for improved secretion of cancer-testis antigen NY-ESO-1 from yeast. Protein Expression Purif. 2006;48:232–242. doi: 10.1016/j.pep.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 37.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 38.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.