Abstract
The t(8:21)(q22;q22) translocation is 1 of the most common chromosomal abnormalities linked to acute myeloid leukemia (AML). AML1-ETO, the product of this translocation, fuses the N-terminal portion of the RUNX transcription factor AML1 (also known as RUNX1), including its DNA-binding domain, to the almost entire transcriptional corepressor ETO (also known as MTG8 or RUNX1T1). This fusion protein acts primarily by interfering with endogenous AML1 function during myeloid differentiation, although relatively few genes are known that participate with AML1-ETO during leukemia progression. Here, we assessed the consequences of expressing this chimera in Drosophila blood cells. Reminiscent of what is observed in AML, AML1-ETO specifically inhibited the differentiation of the blood cell lineage whose development depends on the RUNX factor Lozenge (LZ) and induced increased numbers of LZ+ progenitors. Using an in vivo RNAi-based screen for suppressors of AML1-ETO, we identified calpainB as required for AML1-ETO-induced blood cell disorders in Drosophila. Remarkably, calpain inhibition triggered AML1-ETO degradation and impaired the clonogenic potential of the human t(8;21) leukemic blood cell line Kasumi-1. Therefore Drosophila provides a promising genetically tractable model to investigate the conserved basis of leukemogenesis and to open avenues in AML therapy.
Keywords: acute myeloid leukemia, genetic model, runx
Acute myeloid leukemia (AML) is characterized by the clonal growth of immature blood cells and is often associated with non-random chromosomal translocations that impair the function of key hematopoietic regulators (1). For instance, the t(8:21)(q22;q22) translocation, which is present in 10 to 15% of all cases of AML, affects the transcription factor AML1 (2). AML1 is required at multiple steps of hematopoiesis from the emergence of definitive hematopoietic stem cells to the differentiation of myeloid and lymphoid lineages (3). AML1 is a member of the RUNX family of transcription factors that are characterized by a highly conserved DNA binding domain. AML1-ETO, the product of the t(8;21) translocation, contains AML1 N-terminal portion, including its DNA binding domain, fused to the almost entire transcriptional corepressor ETO (4, 5). While it was proposed initially that AML1-ETO promotes leukemia at least in part by repressing AML1 target gene expression (6), the molecular mechanism of action of AML1-ETO is likely to be more complex since it can both repress or promote transcription depending on the target genes and the cellular context (7).
To gain insights into the function and mode of action of AML1-ETO, several animal models for t(8;21) leukemia have been developed using bone marrow transplantation, knock-in or transgenic techniques (8). These models supported the hypothesis that AML1-ETO dominantly suppresses the function of the endogenous AML1 protein in vivo (9–11). In addition, these works indicate that AML1-ETO inhibits myeloid differentiation and promotes self-renewal of hematopoietic progenitors (12–16). However, AML1-ETO by itself is not sufficient to cause leukemia in mouse (15, 17, 18) and secondary mutations are required for AML1-ETO-expressing cells to become leukemogenic (18, 19). Identifying the genes interacting with or required for AML1-ETO function remains a pivotal but difficult task in mammalian systems.
Several aspects of hematopoietic cell development have been conserved from flies to mammals (20), suggesting that Drosophila may provide an alternative model to study the effect of AML1-ETO on blood cell development. Previous work in Drosophila showed that AML1-ETO constitutively represses RUNX-dependent target gene expression during eye development (21). However, the functional consequences of expressing AML1-ETO in Drosophila blood cells have not been investigated yet. The 2 major classes of Drosophila blood cells (or hemocytes), the plasmatocytes and the crystal cells, functionally and structurally resemble vertebrate myeloid cells (20). Their progenitors arise in 2 successive waves: first in the embryonic head mesoderm and second in the larval lymph gland. In both cases, crystal cell development depends on the RUNX factor Lozenge (LZ) (22), which is expressed in a small subset of prohemocytes and induces their differentiation into crystal cells (23–25). It is interesting to note that, although the Drosophila genome code for 4 RUNX genes, only lz is known to participate in hematopoiesis. The parallels with AML1 function during myeloid differentiation (7) prompted us to analyze the effect of AML1-ETO on this Drosophila RUNX+ blood cell lineage.
Our results show that, reminiscent of what is observed in AML, AML1-ETO specifically inhibited the differentiation of the crystal cell lineage, and induced an increased number of circulating LZ+ progenitors. In addition, by performing a large scale RNA-interference screen for suppressors of AML1-ETO in vivo, we found that calpainB is required for AML1-ETO-induced blood cell disorders in Drosophila. Remarkably, calpain inhibition in human t(8;21) blood cells caused AML1-ETO degradation and impaired their clonogenic potential, suggesting that calpains play a key role together with AML1-ETO to induce leukemic cell growth. Together, this data indicates that Drosophila provides a powerful genetic model to explore the function of AML1-ETO and to discover genes that participate in AML development.
Results
AML1-ETO Inhibited Drosophila RUNX+ Blood Cell Lineage Differentiation.
When AML1-ETO was expressed in all embryonic hemocytes using the srp-gal4 driver, it did not appear to impair prohemocyte differentiation into plasmatocytes. Indeed plasmatocytes expressed normally differentiation markers like crq, migrated throughout the embryo and acquired the typical morphology of mature plasmatocytes (i.e., enlarged cells with phagocytic vacuoles) (Fig. 1B and Fig. S1). On the other hand, AML1-ETO almost completely abolished the expression of crystal cell differentiation markers such as the 3 prophenoloxidase (PO) genes, which are direct targets of LZ (Fig. 1F and Fig. S1) (25). Occasionally 1 or 2 PO-expressing cells were observed but they lacked the cytoplasmic “crystal“ inclusions characteristic of mature crystal cells (Fig. S1). Thus AML1-ETO expression in all hemocytes specifically blocks crystal cell differentiation. AML1-ETO effect on crystal cell differentiation was not caused by the absence of lz since its expression was normal (Fig. 1D). In addition, co-expressing lz and AML1-ETO with the srp-gal4 driver partially restored PO45/CG8193 expression in the prospective crystal cells (Fig. 1H) and the ectopic activation of PO45/CG8193 induced by LZ alone (Fig. 1G) was strongly reduced by AML1-ETO (Fig. 1H). While AML1-ETO competitively inhibited LZ-dependent transactivation of PO45/CG8193, it did not inhibit lz expression, which is normally maintained via an autoregulatory loop in the crystal cell lineage (25, 26). Hence, as observed in mammals (7), AML1-ETO does not behave exclusively as a transcriptional repressor of RUNX target genes in Drosophila blood cells in vivo.
Fig. 1.
AML1-ETO specifically inhibits LZ-dependent blood cell differentiation. (A–D) Pan-hematopoietic expression of AML1-ETO under the control of srp-gal4 does not affect plasmatocyte development (A and B: crq) but inhibits crystal cell differentiation (E and F: PO45/CG8193). This repression is not due to a reduction in lz expression (C and D: lz) but to the competition between AML1-ETO and LZ to regulate LZ target genes (G and H: PO45/CG8193). (A–H): Lateral views of stage 11 embryos. Genotypes are indicated in the lower part of each panel. Arrows in (G and H) indicate ectopic activation of PO45 induced by LZ in the plasmatocytes and posterior endoderm.
In humans, AML1-ETO is active in cells expressing AML1. Therefore we subsequently expressed it selectively in the Drosophila LZ+/RUNX+ cell lineage using the lz-gal4 driver, which recapitulates lz expression (22). In addition, a UAS-gfp reporter transgene was used to track LZ+ blood cells at the different embryonic and larval life stages. Consistent with the results above, AML1-ETO prevented crystal cell differentiation in the embryo and in the larval lymph gland, without suppressing LZ-GFP+ blood cell formation (Fig. 2B, F, J, and N). Finally, AML1-ETO strongly impaired the differentiation of circulating larval cells into mature crystal cells, which can be visualized through the cuticle as black cells either after heat activation or in a Black cell mutant context (Fig. 2R and Fig. S2). Importantly, neither the expression of the AML1 (AML1∂ETO) or ETO moiety of AML1-ETO impaired crystal cell differentiation (Fig. 2), demonstrating the essential contributions from both domains on LZ+ cells development. Therefore, as in humans, AML1-ETO prevented Drosophila RUNX+ blood cell lineage differentiation.
Fig. 2.
Both moieties of the AML1-ETO fusion protein are concomitantly required to block crystal cell differentiation. lz-gal4-driven expression of AML1-ETO, but not that of its AML1 (AML1∂ETO) or ETO moiety, inhibits crystal cell differentiation (A–D and I–L: PO45; Q–T: heat-revealed crystal cells). Formation and maintenance of the LZ+ cells (marked by lz-gal4, UAS-gfp) is not impaired (E–H, M–P, and U–X: GFP). (A–H) Dorsal views of stage 13 embryos. (I–P) Third instar larval lymph gland. (Q–X) Dorsal views of the posterior segments of third instar larvae. Genotypes are indicated in the upper part of the figure.
AML1-ETO Increased the Number of Committed RUNX+ Blood Cell Progenitors.
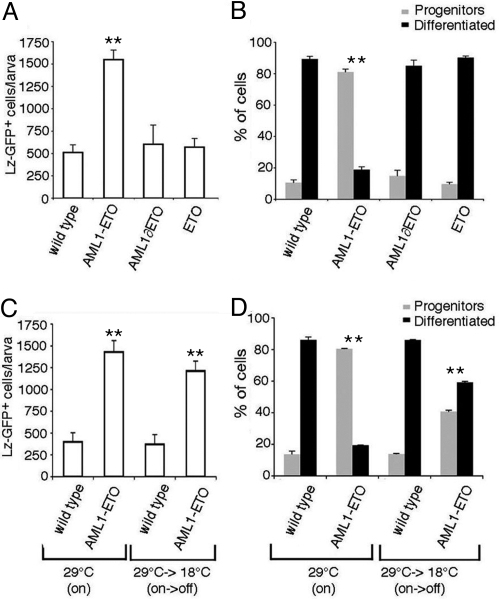
Circulating larval hemocytes are derived from embryonic blood cells. Consequently, circulating larval LZ+ cells are exposed to AML1-ETO for a longer term than in the embryo or in the lymph gland prompting us to further analyze their phenotype. Interestingly, when individual larvae were bled and the number of GFP+ blood cells was counted, we found that AML1-ETO induced greater than 3-fold increase in circulating LZ-GFP+ cells (Fig. 3A) (Student t test: P < 0.001). This increase was not linked to a global increase in hemocyte load as the number of circulating plasmatocytes remained similar in lz-gal4,UAS-gfp; UAS-aml1eto (4,120 ± 640; n = 10) and control larvae (4,096 ± 288; n = 10) (P > 0.2). In parallel the differentiation status of LZ-GFP+ circulating larval blood cells was examined by double fluorescent in situ hybridization and immunostaining against PO45 and GFP respectively (Fig. 3B and Fig. S2). Control larvae contained 87% of differentiated crystal cells (GFP+, PO45+) and 13% of crystal cell progenitors (GFP+, PO45−). This scheme was completely skewed in AML1-ETO-expressing larvae (P < 0.001) where we observed 19% of PO45+ cells and 81% of PO45− cells. Again, neither the expression of the AML1 or ETO moiety of AML1-ETO modified the number of circulating LZ-GFP+ cells or biased their differentiation ratio (P > 0.2). Hence, reminiscent of the preleukemic state induced by AML1-ETO in mammalian blood cells (12, 13, 15, 16), our results suggest that AML1-ETO promotes the maintenance of the Drosophila RUNX+ blood cells as progenitors and increases their proliferation capacity and/or survival rate.
Fig. 3.
AML1-ETO expression induces a preleukemic state. (A–D) lz-gal4, UAS-gfp third instar wandering larvae of the indicated genotypes were bled and the absolute number of GFP+ cells (A and C) as well as the proportion of LZ-GFP+ cells expressing the crystal cell differentiation marker PO45 (B and D) were determined. (A) AML1-ETO but not AML1∂ETO or ETO induces a net increase in LZ-GFP+ circulating blood cells as compared to control larvae. (B) In the presence of AML1-ETO, the ratio of progenitors (GFP+, PO45−) to differentiated (GFP+, PO45+) crystal cells is almost inverted. Neither AML1∂ETO nor ETO affects this ratio. (C and D) Switching off the expression of AML1-ETO at the early L3 stage does not suppress the increase in circulating LZ-GFP+ cells (C) at the wandering larvae stage but partially restore crystal cell differentiation (P < 0.001) (D). **, significant difference (Student's t test, P < 0.001) compared to the wild-type strain.
To test whether the LZ+ cells were maintained as progenitors by AML1-ETO, its expression was turned off in early third instar larvae by using a thermosensitive Gal80 transgene (27) and the absolute number of LZ-GFP+ cells as well as their differentiation status were assessed. The different temperature regimes used in these experiments did not affect crystal cell lineage development in control larvae (Fig. 3 C and D). Switching off AML1-ETO expression in early third instar modestly reduced the number of LZ-GFP+ cells, which remained 3-fold more abundant than in wild-type larvae (Fig. 3C), indicating that the effect of AML1-ETO on LZ-GFP+ cells accumulation takes place before that stage. On the contrary, there was a clear increase in the proportion of differentiated LZ-GFP+ cells, which raised from 19 to 59% (Fig. 3D). Thus, continuous AML1-ETO expression is required to prevent crystal cell differentiation and AML1-ETO maintains the majority of the LZ+ cells as committed crystal cell progenitors.
A Genetic Screen in Drosophila Identifies Suppressors of AML1-ETO.
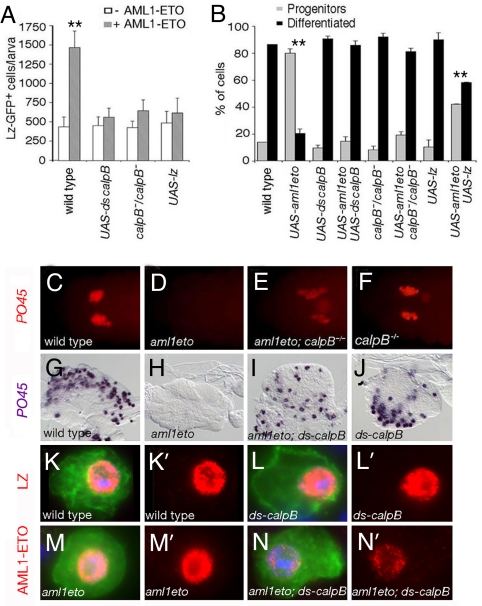
Correlating with previous observations (21), we observed that lz-gal4-driven expression of AML1-ETO induced 100% of lethality at the pupal stage (Fig. S3). In addition, similar to what was observed with AML1-ETO-associated blood cell phenotypes (Fig. 1 and 4 A and B), this lethality was partially rescued by increasing LZ dosage (Fig. S3). We therefore used this phenotype to screen for suppressors of AML1-ETO. To identify these suppressors, we chose to target gene knockdown in AML1-ETO-expressing cells by a UAS-based RNAi strategy (see Materials and Methods). We screened UAS-dsRNA transgenic lines targeting around 1,500 genes and recovered 8 candidates, among which a UAS-dsRNA line against calpainB (calpB) (28) that we studied in detail. Calpains are a large family of Ca2+-dependent proteases conserved throughout evolution (29). In humans, they consist of 14 members with ubiquitous or tissue specific isoforms that influence many aspects of cell physiology such as cell migration, proliferation and apoptosis. However, it is not known whether calpains contribute to the development of leukemia, our findings therefore prompted us to investigate this hypothesis.
Fig. 4.
calpB is required for AML1-ETO function. (A) Absolute number of circulating LZ-GFP+ cells in third instar larvae. (B) Ratio of circulating progenitors (GFP+, PO45−) to differentiated (GFP+, PO45+) crystal cells in third instar larvae. (C–F) PO45 expression in stage 13 embryos. (G–J) PO45 expression in third instar larval lymph gland. The phenotypes induced upon expression of AML1-ETO in the LZ+ blood cell lineage are suppressed when AML1-ETO is coexpressed with a UAS-dsRNA against calpB or when it is expressed in a calpB−/− mutant background (P < 0.001). Similarly, coexpressing LZ with AML1-ETO significantly suppressed AML1-ETO-induced LZ+ cell increase and differentiation bias (P < 0.001). **, significant difference (Student's t test, P < 0.001) compared to the wild-type strain. (K-N) GFP (green) and LZ or AML1-ETO (red) expression in circulating larval blood cells from lz-gal4, UAS-gfp (K), lz-gal4, UAS-gfp; UAS-dsCalpB (L), lz-gal4, UAS-gfp; UAS-aml1eto (M), and lz-gal4, UAS-gfp; UAS-aml1eto;UAS-dsCalpB (N) larvae. LZ expression is not affected by calpB loss of function whereas AML1-ETO levels are strongly decreased. Nuclei were stained with DAPI (blue). (K'-N′) show the red channel from panels (K–N).
Firstly, we asked whether calpB knock-down, which suppressed AML1-ETO-induced lethality, also suppressed AML1-ETO-induced blood cell disorders. As shown in Fig. 4 A and B, down-regulating calpB by dsRNA in circulating larval LZ-GFP+ cells did not impinge on their development. However, the co-expression of calpB dsRNA with AML1-ETO almost completely restored both the absolute number of LZ-GFP+ cells (Fig. 4A) and the ratio of differentiated crystal cells to progenitors (Fig. 4B) (P < 0.001). These results suggest that calpB down-regulation is sufficient to inhibit AML1-ETO function in circulating larval blood cells. Next, we generated a null allele of calpB (see Materials and Methods and Fig. S4). calpB mutation specifically suppressed AML1-ETO-induced phenotypes in circulating LZ-GFP+ larval cells (P < 0.001) and did not interfere with normal crystal cell lineage development (Fig. 4 A and B). In addition, calpB down-regulation also relieved the AML1-ETO-induced differentiation block in the embryo and larval lymph gland (Fig. 4 C–J). All together, these results demonstrate that calpB is required for AML1-ETO activity in Drosophila RUNX+ blood cells. To get insights into the possible mechanism of action of CalpB, we assessed its expression. As shown in Fig. S4, CalpB is specifically expressed in the LZ+ blood cells where it localizes mainly into the nucleus. We then asked whether CalpB regulates the levels or subcellular localization of AML1-ETO or LZ. Down-regulation of calpB in circulating larval LZ-GFP+ cells did not affect LZ (Fig. 4L and Fig. S5), but strongly decreased AML1-ETO levels (Fig. 4N and Fig. S5). Thus, suppression of AML1-ETO-induced blood cell phenotypes by loss of function of calpB is not due to an increase in LZ activity and more likely reflects that CalpB is required to stabilize AML1-ETO.
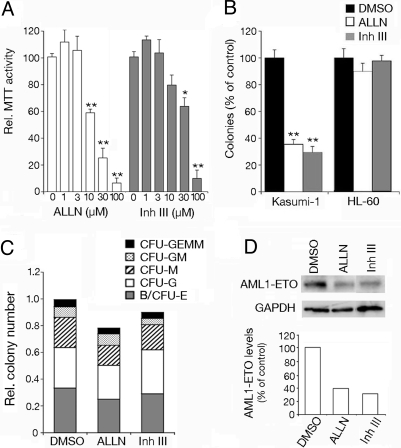
Finally we asked whether calpains might interfere with AML1-ETO function in human cells. Kasumi-1 cells are derived from an AML patient carrying the t(8;21) translocation and constitutively express AML1-ETO (30). Inhibiting AML1-ETO activity in these cells both reduces their growth rate and their capacity to form clones (13, 31). Interestingly, Kasumi-1 cell viability was decreased in a dose dependent manner upon treatment with 2 different calpain inhibitors, ALLN and calpain inhibitor III (Fig. 5A). To test the impact of calpains on clonogenicity, Kasumi-1 cells were incubated with mild doses of calpain inhibitors and cultured in semisolid medium. Both inhibitors severely reduced the number of colonies formed by Kasumi-1 cells whereas they did not affect colony formation by HL-60 cells, which are derived from a patient with acute promyelocytic leukemia (Fig. 5B). Furthermore, primary blood cells treated with calpain inhibitors showed similar capacity to form colonies and to differentiate compared to untreated cells (Fig. 5C). Finally, consistent with our observations in Drosophila, calpain inhibition in Kasumi-1 cells was paralleled by diminished levels of AML1-ETO protein (Fig. 5D). All together, our results indicate that calpains may play a key role in combination with AML1-ETO to induce leukemia.
Fig. 5.
Calpain inhibition reduces the clonogenicity of Kasumi-1 cells. (A) Relative viability of Kasumi-1 cells treated with increasing doses of ALLN or calpain inhibitor III (Inh III). (B) Relative colony numbers obtained upon treatment of Kasumi-1 or HL-60 cells with 10 μM ALLN or 30 μM Calpain Inhibitor III. (A and B) Significant differences between control and treated cells are indicated: *, P< 0.01; **, P < 0.001. (C) Colony forming activity and differentiation potential of primary blood cells treated with 10 μM ALLN or 30 μM calpain inhibitor III. B/CFU-E, blast/colony-forming unit erythroid; M, macrophage; G, granulocyte; GM, granulocyte-macrophage; and GEMM, granulocyte-erythrocyte-macrophage-megakaryocyte. (D) Western blots showing AML1-ETO or GAPDH expression in Kasumi-1 cells treated with 10 μM ALLN or 30 μM calpain inhibitor III. The relative levels of AML1-ETO (normalized to GAPDH) are indicated in the lower part of the panel.
Discussion
The development of cellular and in vivo models to study genes involved in human diseases is critical for understanding their mechanism of action and identifying potential therapeutic targets. Thus far, AML1-ETO has been mostly studied in vertebrate blood cells either in vitro or in vivo (8). Our report constitutes a demonstration that the Drosophila hematopoietic system provides a paradigm to dissect the function of AML1-ETO in vivo and stands as an alternate genetic model to investigate the conserved basis of leukemogenesis.
Notwithstanding the evolutionary distance between human and fly, key features of AML1-ETO activity can be recapitulated in Drosophila blood cells. AML1-ETO expression in the Drosophila RUNX+ lineage gives rise to phenotypes that are reminiscent of a preleukemic state, namely a differentiation blockage and the presence of an abnormally high number of progenitors. These results parallel those obtained in mammalian models either in vivo or ex vivo indicating that AML1-ETO inhibits myeloid differentiation and promotes self-renewal of hematopoietic progenitors (12, 13, 15, 16, 32). Consistent with results showing that AML1-ETO functions at least in part by binding to AML1 target genes (7), all of the phenotypes induced by AML1-ETO in Drosophila could be partially rescued by increasing the dose of LZ. Notably, AML1-ETO did not inhibit lz transcription indicating that it does not affect crystal cell differentiation by preventing the expression of this lineage programming transcription factor. Since lz transcription is autoactivated in blood cells (25), this also demonstrates that AML1-ETO does not exclusively behave as a constitutive transcriptional repressor of RUNX target genes, contrary to what has been proposed previously (21). Although LZ-responsive cis-regulatory module in PO45 and lz are relatively similar (25), it appears that AML1-ETO distinguishes between these 2 genes to differentially regulate their expression in the same cells. This constitutes an interesting model to study the distinct transcriptional responses to AML1-ETO. Indeed, the mechanism by which AML1-ETO can either activate or repress transcription in a cell- and gene-dependent manner is still largely unexplained (7).
Whereas the differentiation block induced by AML1-ETO is clearly attributable to inhibition of LZ activity, whose function in promoting crystal cell differentiation is well established (22–25), its growth promoting activity is more surprising and may reflect an unexpected RUNX-dependent control of blood cell number in Drosophila. In mammals, AML1 haplo-insufficiency causes blood cell progenitor expansion (33, 34) and the growth-promoting activity of AML1-ETO was attributed to inhibition of AML1 activity as well as to additional gain of function mechanisms (7). We identified calpB as required for AML1-ETO activity in Drosophila blood cells and our results indicate that calpain activity participates in the growth of AML1-ETO-positive human cells. Thus the sustained growth of AML1-ETO-expressing cells depends on a similar pathway in fly and human. In humans, calpains have been linked to several pathologies including neurodegenerative diseases and, in contrast to our results, calpain inhibitors have been shown to promote cell survival in these models (29). However, calpain activation has also been associated with cancer progression and cell transformation (35). Unlike relatively promiscuous degradative proteases, calpains cleave a restricted set of protein substrates and use complex substrate-recognition mechanisms involving multiple determinants including PEST score (36). In addition, most substrates are cleaved in a limited fashion resulting in stable protein fragments. The C-terminal region of ETO exhibits a high PEST score, it is therefore tempting to speculate that calpains might cleave AML1-ETO to generate a proteolytic fragment similar to AML1-ETO9, a more potent inducer of leukemia than full length AML1-ETO (37). However, we did not observe any cleavage of AML1-ETO by CalpB in vitro. Yet, we found that calpain inhibition induced a decrease in AML1-ETO protein levels. This effect was post-transcriptional since AML1-ETO mRNA expression was maintained. Hence, loss of calpain activity leads to AML1-ETO degradation, suggesting that calpains impinge on AML1-ETO function by regulating its stability. Alternatively, calpains may promote leukemogenesis by cleaving other substrates linked to AML development such as ß-catenin or by regulating integrin signaling (35, 38). Understanding the mechanism of calpain action clearly requires further investigation; nonetheless our results hold promise that calpain inhibitors might be used as therapeutic agents in leukemia treatment.
In conclusion, our results establish that Drosophila can be used to identify conserved pathways impinging on AML1-ETO activity. Full length AML1-ETO is not sufficient to induce AML in mice (15, 17, 18) and secondary mutations are required to induce leukemic transformation (18, 19). To better understand the mode of action of AML1-ETO and to identify new therapeutic targets, it is important to discover the genes and pathways required for or collaborating with AML1-ETO during leukemogenesis. Here, we used a loss of function strategy to find suppressors of AML1-ETO. The use of UAS-dsRNA allowed us to target gene knockdown in the cells expressing AML1-ETO thereby bypassing possible deleterious or promiscuous effect of a systemic loss of function approach. Similarly, one could perform gain of function screens and/or look for enhancers of AML1-ETO. It is anticipated that the genetically amenable model exposed herein will prove valuable to identify the core regulatory network that contributes to the development of AML.
Materials and Methods
Fly Strains and Crosses.
We used the following D. melanogaster strains: lz-gal4, UAS-gfp, Bc1, tub-Gal80ts/TM2 (Bloomington Stock Center), srp-gal4 (23), UAS-lz (P. Gergen), UAS-aml1eto, UAS-aml1∂eto (R. Mann). Additional UAS-eto and UAS-aml1eto transgenic lines were generated by P-element-mediated germline transformation after subcloning the corresponding region of AML1-ETO into pUAST. Unless specified, crosses were performed at 25°C. To conditionally switch-off AML1-ETO expression, lz-gal4, UAS-gfp; UAS-aml1eto; tub-Gal80ts larvae were collected at 29°C until early L3 stage and transferred at 18°C for an additional 36 h before being processed for analysis. For the genetic screen, lz-gal4 females were crossed to each UAS-dsRNA line (National Institute of Genetics). The emerging lz-gal4/y; UAS-dsRNA/+ males were crossed to UAS-aml1eto females and the resulting progeny was screened for the presence of adult females. The 2 piggyback elements PBac{RB}CG6709e02786 (Bloomington Stock Center) and PBac{RB}calpBe04062 (Harvard Stock Center) were used as parental stocks to generate a null calpB mutant allele using a FLP/FRT excision strategy (39). The deletion of the calpB locus was verified by PCR.
Hemocytes Counts.
In lz-gal4, UAS-gfp/+ third instar larvae, circulating hemocytes were collected on 1-well glass slide in 20 μL PBS by opening 1 female at the level of the posterior segment. Fifteen microliters of the bleed were transferred to a haemocytometer and the number of LZ+ (GFP positive) or circulating blood cells was counted. A minimum of 8 females of each genotype was scored.
In Situ Hybridization and Immunostaining.
Immunostaining and in situ hybridization on embryos and third instar larval lymph glands were performed as described (25). For circulating blood cells, female third instar larvae were thoroughly washed in PBS and ethanol 75% and bled onto polylysine-coated 16-chamber slides (Nunc). Air dried hemocytes were then fixed for 20 min in 4% paraformaldehyde in PBS and rinsed twice with PBS-0.1% Tween-20 (PBST). For in situ hybridization coupled to immunostaining, the slides were preincubated for 1 h at 60°C in hybridization buffer (HB: 50% formamide, 2× SSC, 1 mg/mL Torula RNA, 0.05 mg/mL heparin, 2% Roche blocking reagent, 0.1% CHAPS, 5 mM EDTA, and 0.1% Tween-20). After incubation overnight at 60°C in a humid chamber with PO45 anti-sense probe diluted in HB, slides were washed with HB and PBST solutions, blocked with PBST-1% BSA and incubated overnight at 4°C with sheep anti-DIG (1:2000; Roche) and rabbit anti-GFP (1:200; Torrey) primary antibodies. The in situ hybridization signal was developed with Fast Red substrate (Roche) before incubation with a goat anti-rabbit Alexa Fluor 488 (1/400; Molecular Probes) secondary antibody. Slides were finally washed with PBS and mounted in 50% glycerol-PBS for examination. For fluorescent immunostaining, slides were blocked in PBS-0.3% Triton-1% BSA, incubated with mouse anti-LZ (1:100; DSHB) or rabbit anti-AML1 (1:100; Calbiochem) for 1 h before incubation with secondary antibodies coupled to Alexa Fluor 555 (1/400; Molecular Probes). Slides were finally washed and mounted in Vectashield-DAPI mounting medium for analysis.
Cell Culture and Calpain Inhibitors Treatment.
Kasumi-1, HL-60 cell lines and primary blood were grown in RPMI-1640 medium (Sigma) supplemented with 2 mM L-glutamine and 10% FBS. Primary blood cells were obtained from bone marrow biopsies of paediatric acute lymphoblastic leukemia patients at the end of treatment. The calpain inhibitors N-acetyl-leucyl-leucyl-norleucinal (ALLN) and calpain inhibitor III (Carbobenzoxy-valinyl-phenylalaninal) (Calbiochem) were prepared as a 20 mM stock solution in DMSO. AML1-ETO protein levels were analyzed by immunoblotting as described using an ETO antibody (C-20, SC9737, Santa Cruz Biotechnology) (31). Cell viability was measured by MTT assay: cells were seeded into flat-bottomed 96-well plates at a density of 50,000 cells in 100 μL/well. After treatment with calpain inhibitors for 16 h, 10 μL MTT (Methylthiazolyldiphenyl-tetrazolium bromide; Sigma) dye (5 mg/mL in PBS) was added to each well for 4 h at 37°C. Cells were then lysed with 100 μL solubilization solution (0.1 N HCL, 10% Triton in isopropanol). The absorbance values were measured at 570 nm with 650 nm as a reference wavelength. Cell numbers were calculated by control cell-dilution series. For colony formation assays, Kasumi-1, HL-60, or primary blood cells were incubated for 16 h with calpain inhibitors or DMSO, then 2,500 cells were plated in 250 μl semisolid medium (containing RPMI 1640, 20% FCS, and 0.56% methylcellulose) in 48-well plates. Primary cells (5 × 104) were seeded in 1.1 ml in methocult medium (Stemcell Technologies). Colonies consisting of more than 20 cells were counted 6 days after plating for Kasumi-1 and HL-60 cells and 14 days after plating for primary cells.
Acknowledgments.
We thank M. Crozatier, D. Morello, S. Plaza, L. Vandel, and N. Vanzo for comments on the manuscript; G. Beale for carefully reading it; Toulouse RIO imaging platform for assistance with confocal microscopy; B. Augé for excellent technical support; the National Institute of Genetics Fly Stock Center; and P. Gergen (Stony Brook University, Stony Brook, NY), R. Mann (Columbia University, New York, NY), and G. Mouchiroud (Centre de Génétique Moléculaire et Cellulaire, Lyon, France) for fly stocks and plasmids. This work was supported by grants from the Centre National de la Recherche Scientifique, Association pour la Recherche sur le Cancer (ARC), Association for International Cancer Research, Agence Nationale de la Recherche, and Fondation de France (L.W. and M.H.) and by Leukaemia Research and Kay Kendall Leukaemia Fund (O.H.). D.O. and V.G. are supported by fellowships from the ARC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902449106/DCSupplemental.
References
- 1.McCormack E, Bruserud O, Gjertsen BT. Review: Genetic models of acute myeloid leukaemia. Oncogene. 2008;27:3765–3779. doi: 10.1038/onc.2008.16. [DOI] [PubMed] [Google Scholar]
- 2.Peterson LF, Zhang DE. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- 3.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 4.Erickson P, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 5.Miyoshi H, et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutterbach B, Hiebert SW. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene. 2000;245:223–235. doi: 10.1016/s0378-1119(00)00014-7. [DOI] [PubMed] [Google Scholar]
- 7.Elagib KE, Goldfarb AN. Oncogenic pathways of AML1-ETO in acute myeloid leukemia: Multifaceted manipulation of marrow maturation. Cancer Lett. 2007;251:179–186. doi: 10.1016/j.canlet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller AM, Duque J, Shizuru JA, Lubbert M. Complementing mutations in core binding factor leukemias: From mouse models to clinical applications. Oncogene. 2008;27:5759–5773. doi: 10.1038/onc.2008.196. [DOI] [PubMed] [Google Scholar]
- 9.Yergeau DA, et al. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 10.Okuda T, et al. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 11.Kalev-Zylinska ML, et al. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- 12.Mulloy JC, et al. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood. 2002;99:15–23. doi: 10.1182/blood.v99.1.15. [DOI] [PubMed] [Google Scholar]
- 13.Martinez N, et al. The oncogenic fusion protein RUNX1-CBFA2T1 supports proliferation and inhibits senescence in t(8;21)-positive leukaemic cells. BMC Cancer. 2004;4:44. doi: 10.1186/1471-2407-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwieger M, et al. AML1-ETO inhibits maturation of multiple lymphohematopoietic lineages and induces myeloblast transformation in synergy with ICSBP deficiency. J Exp Med. 2002;196:1227–1240. doi: 10.1084/jem.20020824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Guzman CG, et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol. 2002;22:5506–5517. doi: 10.1128/MCB.22.15.5506-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenske TS, et al. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci USA. 2004;101:15184–15189. doi: 10.1073/pnas.0400751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhoades KL, et al. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood. 2000;96:2108–2115. [PubMed] [Google Scholar]
- 18.Higuchi M, et al. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- 19.Yuan Y, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crozatier M, Meister M. Drosophila haematopoiesis. Cell Microbiol. 2007;9:1117–1126. doi: 10.1111/j.1462-5822.2007.00930.x. [DOI] [PubMed] [Google Scholar]
- 21.Wildonger J, Mann RS. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development. 2005;132:2263–2272. doi: 10.1242/dev.01824. [DOI] [PubMed] [Google Scholar]
- 22.Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science. 2000;288:146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- 23.Waltzer L, Ferjoux G, Bataille L, Haenlin M. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 2003;22:6516–6525. doi: 10.1093/emboj/cdg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fossett N, Hyman K, Gajewski K, Orkin SH, Schulz RA. Combinatorial interactions of Serpent, Lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc Natl Acad Sci USA. 2003;100:11451–11456. doi: 10.1073/pnas.1635050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferjoux G, Auge B, Boyer K, Haenlin M, Waltzer L. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev Biol. 2007;305:726–734. doi: 10.1016/j.ydbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Muratoglu S, Hough B, Mon ST, Fossett N. The GATA factor Serpent cross-regulates lozenge and u-shaped expression during Drosophila blood cell development. Dev Biol. 2007;311:636–649. doi: 10.1016/j.ydbio.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 28.Jekely G, Friedrich P. Characterization of two recombinant Drosophila calpains. CALPA and a novel homolog, CALPB. J Biol Chem. 1999;274:23893–24900. doi: 10.1074/jbc.274.34.23893. [DOI] [PubMed] [Google Scholar]
- 29.Croall DE, Ersfeld K. The calpains: Modular designs and functional diversity. Genome Biol. 2007;8:218. doi: 10.1186/gb-2007-8-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asou H, et al. Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood. 1991;77:2031–2036. [PubMed] [Google Scholar]
- 31.Dunne J, et al. siRNA-mediated AML1/MTG8 depletion affects differentiation and proliferation-associated gene expression in t(8;21)-positive cell lines and primary AML blasts. Oncogene. 2006;25:6067–6078. doi: 10.1038/sj.onc.1209638. [DOI] [PubMed] [Google Scholar]
- 32.Heidenreich O, et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101:3157–3163. doi: 10.1182/blood-2002-05-1589. [DOI] [PubMed] [Google Scholar]
- 33.Song WJ, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 34.Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 35.Franco SJ, Huttenlocher A. Regulating cell migration: Calpains make the cut. J Cell Sci. 2005;118:3829–3838. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- 36.Tompa P, et al. On the sequential determinants of calpain cleavage. J Biol Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- 37.Yan M, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–959. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 38.Rios-Doria J, Kuefer R, Ethier SP, Day ML. Cleavage of beta-catenin by calpain in prostate and mammary tumor cells. Cancer Res. 2004;64:7237–7240. doi: 10.1158/0008-5472.CAN-04-1048. [DOI] [PubMed] [Google Scholar]
- 39.Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]