Abstract
Double-strand breaks (DSBs) represent the most severe DNA lesion a cell can suffer, as they pose the risk of inducing loss of genomic integrity and promote oncogenesis in mammals. Two pathways repair DSBs, nonhomologous end joining (NHEJ) and homologous recombination (HR). With respect to mechanism and genetic requirements, characterization of these pathways has revealed a large degree of functional separation between the two. Nej1 is a cell-type specific regulator essential to NHEJ in Saccharomyces cerevisiae. Srs2 is a DNA helicase with multiple roles in HR. In this study, we show that Nej1 physically interacts with Srs2. Furthermore, mutational analysis of Nej1 suggests that the interaction was strengthened by Dun1-dependent phosphorylation of Nej1 serines 297/298. Srs2 was previously shown to be recruited to replication forks, where it promotes translesion DNA synthesis. We demonstrate that Srs2 was also efficiently recruited to DSBs generated by the HO endonuclease. Additionally, efficient Srs2 recruitment to this DSB was dependent on Nej1, but independent of mechanisms facilitating Srs2 recruitment to replication forks. Functionally, both Nej1 and Srs2 were required for efficient repair of DSBs with 15-bp overhangs, a repair event reminiscent of a specific type of HR called single-strand annealing (SSA). Moreover, absence of Rad51 suppressed the SSA-defect in srs2 and nej1 strains. We suggest a model in which Nej1 recruits Srs2 to DSBs to promote NHEJ/SSA-like repair by dismantling inappropriately formed Rad51 nucleoprotein filaments. This unexpected link between NHEJ and HR components may represent cross-talk between DSB repair pathways to ensure efficient repair.
Keywords: nonhomologous end joining, single strand annealing
Saccharomyces cerevisiae Srs2 (Hpr5) belongs to the widely represented SF1 group of helicases and functions in homologous recombination (HR), which along with nonhomologous end joining (NHEJ), constitute the 2 conserved pathways of DNA double-strand break (DSB) repair as reviewed in refs. 1–3. HR is characterized by the use of an undamaged homologous sequence as template to guide the repair process. Srs2 (suppressor of rad6) was originally identified through inactivating mutations that partly suppressed the UV light (UV) sensitivity of rad6 and rad18 mutant strains (4). RAD6 and RAD18 facilitate DNA replication across lesions in the postreplicative repair (PRR) pathway (5, 6). Inactivating mutations in the SRS2 gene also lead to mitotic hyperrecombination (7, 8). Subsequent characterization of the biochemical activity of Srs2 revealed that it has the ability to remove Rad51 from single-stranded DNA (ssDNA) (9, 10). Removal of Rad51 from ssDNA limits homologous recombination (HR) at a fundamental step in the pathway, namely, strand invasion of the template molecule. When RAD6/RAD18-dependent PRR is compromised, removal of Srs2 leads to increased UV resistance because more of the resulting DNA lesions can be channeled into Rad51-dependent HR pathways. Thus, both the mitotic hyper-recombination, and UV suppression phenotypes are explained by the ability of Srs2 to restrict HR by limiting Rad51 nucleation on ssDNA. Consistent with this model, many srs2 phenotypes are suppressed by deleting the RAD51 gene (11–13).
The roles of Srs2 in HR are more diverse than simply limiting the process. Different resolutions of recombination intermediates, termed Holliday junctions, lead to the formation of either cross-over or noncross-over products. In vegetative cells, cross-overs are usually prevented. Avoiding cross-overs may be important because loss-of-heterozygosity and chromosomal rearrangements, events linked to cancer formation in mammals, can occur as a result. Strains lacking Srs2 show increased cross-over levels during mitosis. It was suggested that the helicase activity of Srs2 melts the so-called D-loop, by unwinding the elongating invading strand from the template strand. The result is that the invading strand flips back and anneals with the other end of the DSB on the same chromosome, preventing the formation of Holliday junctions. This pathway is called synthesis-dependent strand annealing (SDSA). According to this model, Srs2 actively promotes SDSA, hence preventing cross-overs (14–16). Srs2 also has a role in promoting an HR pathway known as single-strand annealing (SSA) (17, 18). Recombination between flanking direct repeats by the SSA pathway produce a deletion of the intervening DNA. SSA is strongly dependent on Rad52, facilitating the annealing between repeats. In contrast, the absence of Rad51 in fact facilitates SSA, suggesting that SSA and Rad51-dependent gene conversion may compete (19). Using SSA substrates with long (25 kb) intervening DNA suggested that the major role of Srs2 in completing SSA was in fact to ensure recovery of the cell cycle after repair was completed (20). However, also in this case, deletion of RAD51 rescued the srs2 defect in recovery.
In the case of the PRR pathway, a model for recruitment of Srs2 to DNA lesions has been suggested. Post-translational sumoylation and ubiquitination of proliferating cell nuclear antigen (PCNA), a processivity factor for DNA polymerase, contributes to the PRR pathway in an essential manner. Sumoylated PCNA interacts better with Srs2 than unmodified PCNA, and this SUMO-modification has thus been suggested to be responsible for recruiting Srs2 to DNA already bound by PCNA (21, 22). Since this occurs in the absence of DNA damage, the PCNA-SUMO-Srs2 interaction was suggested to be a guarding mechanism that prohibits potentially detrimental recombination during DNA replication.
DSB repair by NHEJ differs from HR in that it does not rely on extensive homology for repair, but ligates the 2 severed ends in a manner that often generates small deletions or insertions. In S. cerevisiae, NHEJ is a back-up pathway, with HR repairing most DSBs, whereas in mammals NHEJ appears to be the predominant DSB repair pathway. The central component of the NHEJ pathway is a specialized DNA ligase, Dnl4 in yeast and LIG4 in mammals. Dnl4/LIG4 interacts with Lif1/XRCC4 that in turn interacts with Nej1/XLF. This entire complex is required for NHEJ in both yeast and mammals. An additional requirement for NHEJ in budding yeast is the Mre11 complex, composed of Mre11, Rad50, and Xrs2 (23–25). A unique property of the Mre11 complex is its involvement in both the NHEJ and HR pathways. The Mre11 complex is also an important DNA damage sensor (26, 27). During NHEJ, the Mre11 complex plays a structural role by bridging the DNA ends, thereby stimulating ligation (23). As a participant in HR, the Mre11 complex regulates end-resectioning that produces invasive ssDNA intermediates (28, 29).
In this study, we explore a molecular interaction between Srs2 and Nej1. We showed that Srs2 was recruited to a DSB and that efficient recruitment depended on Nej1. Further, we demonstrated that Nej1 supports DSB repair by an SSA-like mechanism. These observations seem to soften the distinction between DSB repair pathways and may reflect cross-talk between NHEJ and HR.
Results
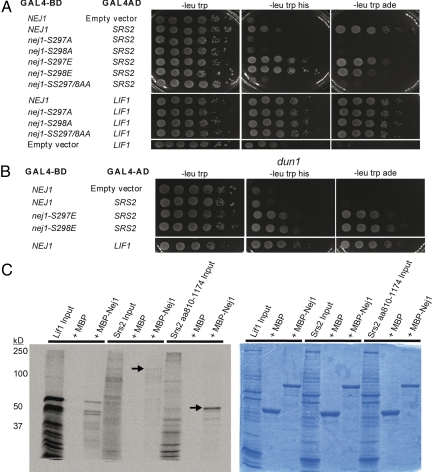
To identify proteins interacting with Nej1, we performed a 2-hybrid screen using a Gal4 DNA binding domain (DB)-Nej1 fusion protein as bait. A plasmid library consisting of a Gal4-activation domain (AD) fused to random genomic S. cerevisiae DNA was screened for interaction partners. The cotransformants were screened for adenine and histidine prototrophy, because the tester strain contained GAL-regulated ADE2 and HIS3 genes. Plasmid DNA was isolated from positive colonies and reintroduced into the tester strain. Among the library plasmids that passed this test were 2 independent isolates representing GAL4-AD-SRS2 fusion genes. In light of the common role of Nej1 and Srs2 in DNA repair, we found this interaction highly interesting. Both of the GAL4-AD-SRS2 fusion genes were joined in the 3′ part of the SRS2 gene, encoding fusion proteins containing amino acids 862 to 1,174 and 1,104 to 1,174 of Srs2, respectively. Hence, the last 70 amino acids of the Srs2 C terminus were sufficient to mediate the observed 2-hybrid interaction. Phenotypically, both truncated forms of Srs2 interacted equally well with Nej1. In subsequent 2-hybrid experiments we continued with the Srs2 construct encoding amino acids 862 to 1,174 (Fig. 1A).
Fig. 1.
Analysis of the Nej1-Srs2 interaction. (A) Shown is the 2-hybrid strain PJ69–4A containing pAS1-NEJ1 (Gal4BD-NEJ1) or pASI-nej1 substitution derivatives as bait. Prey constructs contain the indicated LIF1 or SRS2 gene fusions in pACTII (Gal4AD). Cells are spotted as 10-fold serial dilutions on 2-hybrid indicator medium lacking adenine or histidine. (B) Two-hybrid analysis using the indicated bait and prey constructs in a dun1 derivative of PJ69–4A. (C) Left, Radiograph of an MBP pull-down is shown. Lanes show input, pull-downs with MBP alone and pull-downs with MBP-Nej1, as indicated. The in vitro translated protein added was Lif1 (lanes 1–3), full-length Srs2 (lanes 4–6), and amino acids 810 to 1,174 of Srs2 (lanes 7–9). Arrows indicate the recovery of full-length Srs2 and amino acids 810 to 1,174 of Srs2. Higher mobility peptides in the input and pull-down lanes are interpreted to represent truncated forms of the respective in vitro translated proteins. Size markers are shown to the left. Right, Coomassie staining of the same gel used to generate the autoradiograph is shown.
A recent study showed that Nej1 was phosphorylated on either serine 297 or 298 or both. Moreover, the Dun1 protein kinase was necessary for this phosphorylation in vivo (30). Dun1 is a serine/threonine protein kinase that is activated by Rad53 in response to DNA damage (31, 32). Dun1 also appears to become activated during normal cell cycle progression (33). We generated mutant alleles of Nej1 in which serines 297 and 298 were replaced with nonphosphorylatable alanine residues either individually, or as a pair. In addition, individual glutamate substitutions of serines 297 and 298 of Nej1 were generated to mimic phosphorylated residues. We used a GAL4-AD-LIF1 fusion gene as a positive control because a direct interaction between Nej1 and Lif1 has been established by several laboratories (34–37). Lif1 interacted with all Nej1 substitution proteins showing that the constructs were expressed. The Srs2 construct, however, lost the interaction with both Nej1 alanine substitution proteins but retained the interaction with Nej1 proteins carrying glutamate substitutions (Fig. 1A). These results were consistent with the notion that the Nej1-Srs2 interaction was promoted by phosphorylation. To further test this idea, we generated a dun1 mutant derivative of the 2-hybrid strain. Interestingly, the Nej1-Srs2 interaction was lost in the dun1 background. In contrast, by selecting for isolates with high copy numbers of the respective NEJ1 allele, we found that both Nej1 glutamate derivatives retained the ability to interact with Srs2 in the dun1 strain (Fig. 1B).
To confirm the Nej1-Srs2 interaction and test whether it was direct, we performed pull-down experiments using in vitro translated Srs2 and a recombinant maltose-binding protein (MBP)-Nej1 fusion protein. In vitro translated Lif1 was used as a positive control. This experiment showed that Nej1 could interact with Srs2, both with full-length Srs2 and with its C terminus (amino acids 810–1,174). Moreover, comparing the input lanes to the pull-downs, the Srs2 interactions with Nej1 were comparable with the Nej1-Lif1 interaction (Fig. 1C).
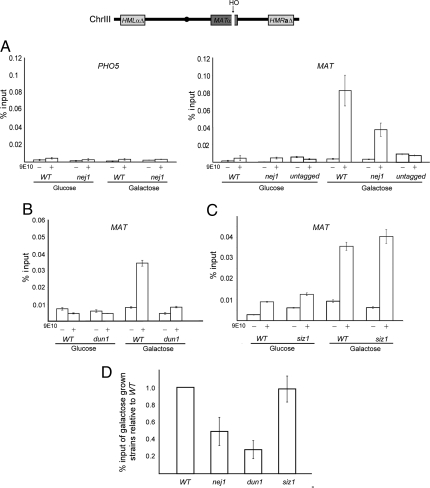
Given that Srs2 plays a role in several DSB repair pathways, we endeavored to determine whether we could detect Srs2 recruitment to an induced DSB in vivo. We generated an SRS2-13MYC fusion gene at the endogenous SRS2 locus. The strain also contained the HO endonuclease gene under the control of a galactose-inducible promoter and complete deletions of the cryptic mating-type loci, HMLα and HMRa. Galactose-induced expression of HO in this strain results in a single DSB at the MAT HO recognition site that cannot be repaired by gene conversion (38). We performed chromatin-immunoprecipitation (ChIP) followed by qPCR to quantify relative amounts of MAT-associated DNA in the SRS2-13XMYC WT strain and its derivatives. Relative to input DNA, galactose-induction in the SRS2-13XMYC WT strain resulted in the recovery of 0.082% of MAT-associated DNA vs. 0.004% in noninduced cells, or 0.003% of a control locus located immediately upstream of the PHO5 gene (Fig. 2A). In addition, the ChIP was specific for Srs2–13XMyc because MAT-associated DNA was not preferentially precipitated after break induction in a strain containing untagged SRS2. To test whether Nej1 may be targeting Srs2, we tested Srs2 recruitment to the DSB in a nej1 strain. Deletion of NEJ1 resulted in an approximate 2-fold reduction in Srs2 recruitment to the DSB (Fig. 2A). Consistently, we observed an almost complete loss of Srs2 recruitment to the DSB in a dun1 strain (Fig. 2B). Within the context of PRR, Srs2 is recruited to DNA replication forks partly through its interaction with PCNA. This interaction is regulated by sumoylation of PCNA at lysine 164, a modification carried out by the SUMO E3 ligase Siz1 (22). To determine whether Srs2 localization to the DSB depended on PCNA sumoylation, we tested Srs2 recruitment efficiency in a siz1 strain. Cells lacking Siz1 showed no appreciable defect in Srs2 recruitment to the DSB, as the enrichment levels of MAT-associated DNA were comparable between the siz1 strain and the WT control (Fig. 2C). This result suggests a mechanistic difference between Srs2 recruitment to DSBs and replication forks. The results presented in Fig. 2 A–C are single representative experiments. Reassuringly, the relative levels of Srs2–13XMyc recruitment for each experimental strain remained constant in several experiments (Fig. 2D). Importantly, differences in recruitment efficiency between strains were not because of differences in Srs2–13XMyc steady-state levels (Fig. S1) or different levels of DSB induction (Fig. S2).
Fig. 2.
Recruitment of Srs2–13XMyc to an induced DNA DSB. Chromatin immunoprecipitation (ChIP) was performed on strain SAY1110 (SRS2-13XMYC) and derivatives using an anti-myc antibody (9E10). Cell extracts were prepared from each strain following growth in glucose or galactose, with galactose resulting in the induction of a single DSB by the HO endonuclease at MAT, schematically shown on top. Following ChIP, associated DNA was quantified by qPCR. Mock immunoprecipitations lacking α-myc (9E10) are included. Shown are representative results of experiments repeated at least 3 times, except the experiment in which 9E10 antiserum was added to an untagged strain (SRS2 JKM115), in which a single experiment was performed. The standard deviations of triplicate qPCR reactions for each strain and condition are included. (A) Amplification of MAT associated DNA approximately 0.2 kb away from the induced DSB, and amplification of nonspecifc DNA immediately upstream of the PHO5 locus of chromosome II in a WT (SAY1110) and nej1 (SAY1126) strains. (B) Identical experiments performed in dun1 (SAY1180) or (C) siz1 (SAY1196) strains. (D) For each of the indicated strains, the amplification of MAT associated DNA following DSB induction from multiple experiments is shown as an average relative to WT. The averages represent 3 independent experiments for the nej1 and dun1 strains, and 2 independent experiments for the siz1 strain. The standard deviation between separate experiments is indicated.
We next explored the biological consequences of the interaction between Srs2 and Nej1. We began by investigating any potential cooperation between Srs2 and Nej1 in PRR using UV-sensitivity assays. Strains lacking NEJ1 displayed no sensitivity to UV radiation. Furthermore, deletion of NEJ1 in rad6, srs2 or rad6 srs2 backgrounds had no effect on the parental phenotypes (Fig. S3). Hence, it is unlikely that the Nej1-Srs2 interaction regulates PRR.
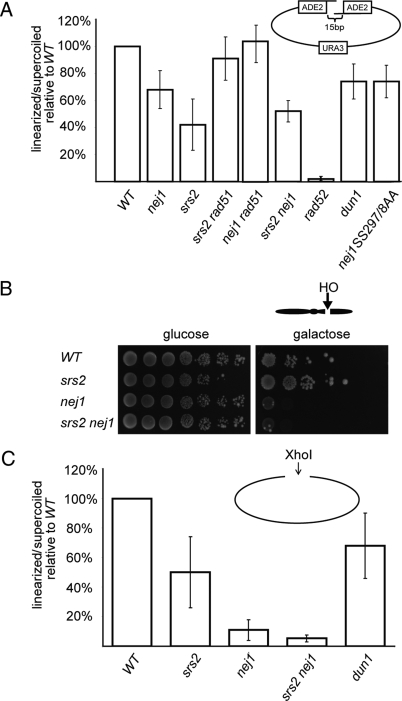
Srs2 has been shown to promote SSA by removing Rad51 from ssDNA overhangs, thus allowing Rad52-dependent strand-annealing and subsequent repair. Therefore, we next used a plasmid based rejoining assay using a linearized vector modified to contain 15bp 3′ overhangs flanking a disrupted ADE2 gene (39) (See Methods). Precise repair of the construct results in the restoration of ADE2, and can be screened for by white colony color and expressed as transformation efficiency. As expected, we found that repair of the construct was mediated by HR-dependent machinery, as deletion of RAD52 abolished nearly all repair events compared with WT (Fig. 3A). Furthermore, an srs2 strain transformed with less than half the efficiency of the WT control. In agreement with previous observations characterizing the biochemical activity of Srs2, deletion of RAD51 suppressed the srs2 phenotype and raised the transformation efficiency of an srs2 rad51 strain to nearly WT levels. Most surprisingly, a nej1 strain transformed with a reduced efficiency (Fig. 3A). This result suggests that a protein previously shown only to participate in NHEJ also contributes to a Rad52-dependent repair process. Importantly, deleting rad51 suppressed the nej1 defect, suggesting that the role of Nej1 in promoting SSA in this assay was similar to the role of Srs2. An srs2 nej1 double mutant transformed with an efficiency similar to that of the srs2 single mutant. The dun1 and nej1-SS297/8AA strains displayed a slightly less pronounced phenotype than the nej1 strain, with transformation efficiencies of 74% and 73% of WT, respectively.
Fig. 3.
DSB repair by an SSA-like mechanism and NHEJ. (A) The indicated strains (YW465, SAY1105, SAY1103, SAY1193, SAY1362, SAY1198, SAY1124, SAY1104, and SAY1199) were transformed with a plasmid construct containing a DSB flanked by 15-bp complementary overhangs embedded within an ADE2 gene. Repair efficiency was assessed by counting white colonies (Ade+) growing on medium lacking uracil and normalized to parallel transformations with supercoiled pRS413 and growth on medium lacking histidine. Transformation efficiency of each mutant strain is shown as a percentage of WT, which was defined as 100%. The standard deviation of 3 separate experiments is included. (B) GAL-HO assay of 10-fold serial dilutions showing cell survival during continuous HO expression and DSB induction at MAT using the indicated strains (JKM115, SAY272, SAY264, and SAY274). (C) NHEJ efficiency of the indicated strains (YW465, SAY1103, SAY1105, SAY1198, and SAY1104) is shown in a plasmid-rejoining assay of XhoI-linearized pRS416. Values for individual strains are plotted as a relative percentage of WT and include the standard deviation of 3 separate experiments.
To determine whether the interaction between Srs2 and Nej1 affect NHEJ, we performed 2 separate assays designed to measure NHEJ efficiency. We introduced a plasmid carrying the HO endonuclease gene under the control of a galactose-inducible promoter into nej1, srs2 and srs2 nej1 strains. Since the cryptic mating type loci, HMLα and HMRa have been deleted in these strains, expression of the HO endonuclease results in the generation of a single DSB at MAT that cannot be repaired by gene conversion. Because an unrepaired DSB is lethal, cell survival following plating on medium containing galactose is interpreted as a relative measure of NHEJ efficiency. By this assay, no contribution to NHEJ by Srs2 was detectable, as survival of the srs2 strain on galactose containing medium was similar to that of WT (Fig. 3B). In contrast, the nej1 single or srs2 nej1 double-mutant strains performed NHEJ at a greatly reduced efficiency, as expected. In a more sensitive approach, we next performed plasmid-rejoining assays of XhoI-digested vector in srs2, nej1, srs2 nej1, and dun1 strains. Our results indicated that the srs2 strain was defective in performing plasmid repair by 50% relative to WT (Fig. 3C). The srs2 nej1 double-mutant strain transformed with an efficiency of 6% relative to WT, indicating a plasmid-rejoining defect similar to that of the nej1 single mutant. The dun1 strain transformed with an efficiency of 68% compared with WT, a result that confirms an initial finding that Dun1 contributes to efficient NHEJ (30).
Discussion
In conclusion, our data expands on the current view of DSB repair by describing an overlap between molecular components of the NHEJ and HR pathways. Our 2-hybrid results show an interaction between Nej1 and Srs2, and strongly suggest that Dun1-dependent phosphorylation of Nej1 promotes this interaction. A compelling indication of this notion was that the Nej1-Srs2 2-hybrid interaction was lost in a dun1 strain, but Nej1 containing the glutamate substitutions maintained their interaction with Srs2 in the absence of Dun1. A previously performed large scale 2-hybrid screen using Srs2 as bait identified 166 potential interacting proteins, but did not identify Nej1 (40). This data raised the possibility that the 2-hybrid interaction we observed was indirectly mediated through another protein. We reasoned that confirming the Srs2-Nej1 interaction by means of coimmunoprecipitation would not address whether the interaction was direct or not. Instead, we performed in vitro interaction assays in which both full-length Srs2 and the C terminus of Srs2 interacted with recombinant Nej1. Hence, the Nej1-Srs2 interaction appears to be direct. Although within this context phosphorylation of Nej1 is highly unlikely, we suggest the pull-down assay might enable the detection of an interaction too weak or transient to result in the activation of reporter genes necessary for observation by 2-hybrid.
Further, using ChIP we provide evidence that Srs2 is recruited to an induced DSB, and that Nej1 promotes full recruitment efficiency. However, the residual levels of Srs2 detectable at the DSB in the absence of Nej1 clearly indicate additional recruitment means. Under these circumstances, the almost complete loss of Srs2 recruitment in the dun1 strain was unexpected. We speculate that this strong phenotype reflects the pleiotropic nature of Dun1. Srs2 itself is phosphorylated, and DNA damage induced phosphorylation of Srs2 is lost in dun1 strains (41). We also demonstrate that the mechanistic basis of Srs2 recruitment to a DSB appears to be distinctly different from mechanisms governing Srs2 recruitment to replication forks. This claim is based on the observation that the PCNA SUMO ligase Siz1 was not required for Srs2 recruitment to the HO-induced DSB, whereas Siz1 is required for efficient recruitment of Srs2 to replication forks (21, 22).
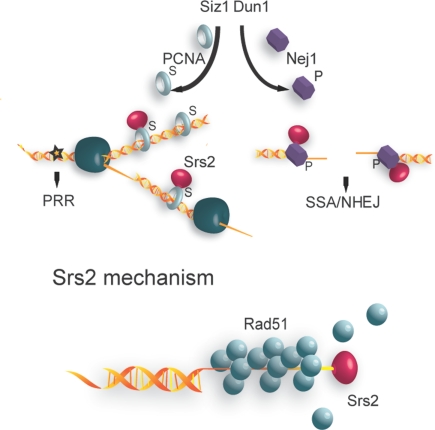
We present data that suggests a contribution of Nej1 to a Rad52-dependent SSA-like DSB repair mechanism. Both Srs2 and Nej1 are required for efficient repair in this 15-bp overhang assay. Significantly, absence of Rad51 suppresses the defect in both strains. This observation strengthens the notion that repair was mediated by an SSA-like mechanism. It further argues that Rad51 nucleation on the ssDNA overhangs was most likely the primary reason for the repair defect of the srs2 and nej1 strains. In addition to DSB repair by SSA, Srs2 has been shown to be necessary for cell recovery and adaptation following activation of the DNA damage response (20). In contrast to our 15-bp overhang assay, this study monitored much longer overhangs (25kb), making the interpretation of a role of Srs2 in adaptation vs. repair difficult. We also found a NHEJ-defect in srs2 strains. This result is in agreement with prior studies showing that Srs2 contributes to efficient NHEJ, although to a lesser degree than the core NHEJ components (35, 42). Furthermore, the less severe NHEJ defect of the srs2 strain compared with the srs2 nej1 double-mutant strain makes clear that the essential contribution of Nej1 to NHEJ remains a distinct and separate function from its role in recruiting Srs2 to DSBs. Based upon this study, we propose a model in which Srs2 is recruited through different mechanisms to replication forks or genomic DSBs (Fig. 4). During DNA replication, Srs2 is recruited to replication forks through an interaction with sumoylated PCNA, where it prevents deleterious HR events and thus shuttles encountered DNA lesions into the PRR pathway (21, 22). Outside of DNA replication, Srs2 recruitment to DSBs is supported through binding to Nej1. We surmise that Dun1-dependent phosphorylation of Nej1 functions to stabilize or modulate its interaction with Srs2. Studies have shown that Rad51 can bind to as few as 4 bp of ssDNA (43). Therefore, it is feasible that Rad51 binding to short overhangs could interfere with the repair of DSBs by NHEJ/SSA. We propose that the cooperation between Srs2 and Nej1 may counteract improper Rad51 binding to short overhangs, and thus support DSB repair by NHEJ/SSA.
Fig. 4.
Model of differential modes of Srs2 recruitment to replication forks and DNA DSBs. Srs2 is recruited to replication forks through an interaction with PCNA that is enhanced by Siz1-dependent sumoylation of PCNA. The star represents a DNA lesion. At DSBs, Srs2 recruitment is supported through an interaction with Nej1. Dun1-dependent phosphorylation of Nej1 might stabilize or modulate the interaction. Within both contexts, the biochemical ability of Srs2 to remove Rad51 from ssDNA is central to the outcome. At replication forks, preventing Rad51 nucleation on ssDNA antagonizes HR and shuttles DNA lesions into PRR. At DSBs, the same activity keeps shorter ssDNA tracts Rad51-free, and available for SSA/NHEJ.
Methods
Strains and Plasmids.
The complete genotype of all strains used in this study is listed in Table S1. Unless noted otherwise, all gene deletions were carried out using a 1-step gene disruption procedure (44) using a kanMX or NAT PCR fragment amplified from pFA6a-KANMX (45) or pAG25 (46), respectively, and verified by a PCR-based strategy. Two-hybrid experiments were performed in PJ69–4A (47). A 3.2-kb fragment containing GAL-HO was excised from pBS283 (48) by SalI-EcoRI digestion, and cloned into XhoI-EcoRI digested pRS406, generating p490. p490 was introduced into JKM115 (38) by integrative transformation following digestion with NcoI, generating SAY282. SAY1110 (SRS2-13XMYC) was generated using a PCR-based procedure (49) by integrative transformation at the genomic SRS2 locus in SAY282. SAY1126 (nej1∷NAT), SAY1180 (dun1∷NAT), and SAY1196 (siz1∷NAT) were generated in SAY1110.
Wild-type strain UMY2060 and UMY2107 (rad6∷URA3) were gifts from Anders Byström. SAY1024 (srs2∷NAT) and SAY1030 (nej1∷kanMX) were generated in UMY2060. SAY1026 (srs2∷NAT), SAY1028 (nej1∷KanMX srs2∷NAT), and SAY1032 (nej1∷kanMX) were generated in UMY2107. YW465 (ade2Δ), was a gift from Dr. Thomas E. Wilson. SAY1103 (srs2∷NAT), SAY1104 (dun1∷NAT), and SAY1105 (nej1∷NAT) were generated in YW465. SAY1193 (rad51∷kanMX) and SAY1198 (nej1∷kanMX) were generated in SAY1103. SAY1362 (rad51∷kanMX) was generated in SAY1105. SAY230 was a gift from Lorraine Symington. SAY1124 (rad52∷TRP1) was generated by transforming YW465 with a PCR fragment amplified with primers flanking the rad52∷TRP1 allele in SAY230. In a 3-factor in vivo cloning technique, the nej1-SS297/8AA ORF was PCR amplified from the 2-hybrid vector pAS1-nej1-SS297/8AA with a 5′ primer containing 20 bp of sequence immediately upstream of the NEJ1 ORF and a 3′ primer containing 37 bp of sequence corresponding to nucleotides 2,124–2,161 in the MCS of pRS413. A 400-bp fragment containing the NEJ1 promoter was PCR amplified from genomic DNA using a 5′ primer containing 40-bp sequence corresponding to nucleotides 2,097–2,057 in the MCS of pRS413 and a 3′ primer containing 20 bp of sequence immediately downstream of the NEJ1 start codon. Both fragments were cotransformed with XhoI-EcoRI digested pRS413 into SAY215 (nej1∷kanMX). Plasmid rescue was performed by a standard protocol to recover pSC210 (pRS413-nej1-SS297/8AA). pSC210 was subsequently used as template to PCR amplify nej1-SS297/8AA with the endogenous NEJ1 promoter with primers introducing 5′ XhoI and 3′ SacI sites. The fragment was digested with XhoI and SacI and cloned into XhoI-SacI digested pRS404 generating pSC212. By digestion with HindIII, pSC212 was integrated into SAY1105, generating SAY1199 (nej1-SS297/8AA). SAY272 (srs2∷hisG) was generated in JKM115 using pM690 as described in ref. 50. SAY274 (nej1∷kanMX) was generated in SAY272. pSB283 (48) was used for GAL-HO assays. pTW423 (URA3 5′-ade2 polyterminator 3′-ade2) was a gift from Dr. Thomas E. Wilson. pTW423 was modified as previously described (39) with the following exceptions. All purification steps were performed using the QIAquick PCR purifcation kit (Qiagen), and oligonucleotide primers SDC274 (5′- GATCTGTTAACGGTTTAGTGTTTTCTTACCCAATTGTAGAGACTATCCACAAGGA), SDC275 (5′-TCTACAATTGGGTAAGAAAACACTAAACCGTTAACA), SDC276 (5- CAATATTTGTGACTTATGTTATGCGCCTGC) and SDC277 (5′- TCGAGCAGGCGCATAACATAAGTCACAAATATTGTCCTTGTGGATAGTC) were used to generate 15-bp overhangs and reconstitute the ADE2 ORF.
Other Methods.
Site-directed mutagenesis of pAS1-NEJ1 was performed according to the protocol outlined in the Stratagene QuikChange procedure.
MBP pulldown assay.
MBP-Nej1 was purified from Rosetta Competent cells (Novagen) and immobilized on amylose resin beads (51). Lif1, Srs2, and Srs2aa810–1174 were in vitro translated with the TNT Quick Coupled Transcription/Translation System (Promega) in the presence of l-[35S] methionine (Amersham Pharmacia Biotech). Equal amounts of in vitro translated protein and MBP or MBP-Nej1 were incubated at room temperature for 1 h. The beads were washed 3 times in NTEN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40), followed by SDS/PAGE and autoradiography using an FLA3000 phosphoimager (Fuji).
Chromatin immunoprecipitation and quantitative PCR.
Fifty mL overnight cell cultures grown in YEPD were spun down, washed once with ddH2O, and split into 2 separate cultures at an OD600 of 0.2 in either YEPD or YEP plus 2% galactose. Following 6 h of growth, formaldehyde cross-linking and immunoprecipiations were performed as described in ref. 52. Input DNA was recovered before immunoprecipitation. The precipitating antibody was anti-c-Myc (9E10) (Santa Cruz Biotechnology). Quantitative PCR reactions were performed in triplicate, using SYBR Green PCR Master Mix (Applied Biosystems), and conditions recommended by the manufacturer on an ABI Prism 7000 Sequence Detection System instrument. Data analysis was performed using 7000 System SDS software.
DNA repair assays.
Plasmid rejoining and galactose survival assays were performed as described (24, 38, 39). For the plasmid rejoining assay employing 15-bp overhangs, 100% (144/144) of white colonies resulting from the transformation of the WT strain (YW465) were determined to be Ade+ by replica plating on synthetic complete medium lacking adenine.
Supplementary Material
Acknowledgments.
We thank Thomas Wilson, Satya Prakash, Anders Byström, and Lorraine Symington for strains and plasmids, Thomas Helleday and Christos Samakovlis for critical reading of the manuscript, and L. Åström for help preparing Fig. 4. This work was supported by grants from the Swedish Research Council and Swedish Cancer Society (S.U.Å).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903869106/DCSupplemental.
References
- 1.Macris MA, Sung P. Multifaceted role of the Saccharomyces cerevisiae Srs2 helicase in homologous recombination regulation. Biochem Soc Trans. 2005;33:1447–1450. doi: 10.1042/BST0331447. [DOI] [PubMed] [Google Scholar]
- 2.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence CW, Christensen RB. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J Bacteriol. 1979;139:866–876. doi: 10.1128/jb.139.3.866-876.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 6.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3, and rad52 mutations. Mol Gen Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 7.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rong L, Palladino F, Aguilera A, Klein HL. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krejci L, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 10.Veaute X, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 11.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 13.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupaigne P, et al. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 17.Wilson TE. A genomics-based screen for yeast mutants with an altered recombination/end-joining repair ratio. Genetics. 2002;162:677–688. doi: 10.1093/genetics/162.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaze MB, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 21.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 24.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne GT, Jin S, Shannon KB, Weaver DT. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 27.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Mimitou EP, Symington LS. Sae2, Exo1, and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahnesorg P, Jackson SP. The non-homologous end-joining protein Nej1p is a target of the DNA damage checkpoint. DNA Repair (Amst) 2007;6:190–201. doi: 10.1016/j.dnarep.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- 32.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valencia M, et al. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 35.Ooi SL, Shoemaker DD, Boeke JD. A DNA microarray-based genetic screen for nonhomologous end-joining mutants in Saccharomyces cerevisiae. Science. 2001;294:2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- 36.Frank-Vaillant M, Marcand S. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 2001;15:3005–3012. doi: 10.1101/gad.206801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kegel A, Sjöstrand JO, Åström SU. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol. 2001;11:1611–1617. doi: 10.1016/s0960-9822(01)00488-2. [DOI] [PubMed] [Google Scholar]
- 38.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiolo I, et al. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberi G, et al. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hegde V, Klein H. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 2000;28:2779–2783. doi: 10.1093/nar/28.14.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 44.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 45.Bahler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 47.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berlin V, Brill JA, Trueheart J, Boeke JD, Fink GR. Genetic screens and selections for cell and nuclear fusion mutants. Methods Enzymol. 1991;194:774–792. doi: 10.1016/0076-6879(91)94058-k. [DOI] [PubMed] [Google Scholar]
- 49.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 50.Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 51.Riggs P. Expression and purification of maltose-binding protein fusions. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb1606s28. Chapter 16:Unit16 16. [DOI] [PubMed] [Google Scholar]
- 52.Aparicio O, et al. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol. 2005 doi: 10.1002/0471142727.mb2103s69. Chapter 21:Unit 21 23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.