Abstract
We describe a general application of the nonsense suppression methodology for unnatural amino acid incorporation to probe drug–receptor interactions in functional G protein-coupled receptors (GPCRs), evaluating the binding sites of both the M2 muscarinic acetylcholine receptor and the D2 dopamine receptor. Receptors were expressed in Xenopus oocytes, and activation of a G protein-coupled, inward-rectifying K+ channel (GIRK) provided, after optimization of conditions, a quantitative readout of receptor function. A number of aromatic amino acids thought to be near the agonist-binding site were evaluated. Incorporation of a series of fluorinated tryptophan derivatives at W6.48 of the D2 receptor establishes a cation–π interaction between the agonist dopamine and W6.48, suggesting a reorientation of W6.48 on agonist binding, consistent with proposed “rotamer switch” models. Interestingly, no comparable cation–π interaction was found at the aligning residue in the M2 receptor.
Keywords: D2 receptor, fluorination, membrane protein
G protein-coupled receptors (GPCRs) represent the largest family of transmembrane receptor proteins in the human genome and constitute a prominent class of targets for the pharmaceutical industry (1–3). Accordingly, they have been studied extensively throughout academia and industry, by using the full range of chemical, biochemical, and biophysical techniques. In recent years, the field has been energized by several high-resolution crystal structures of mammalian GPCRs that build upon the earlier, highly informative structural studies of rhodopsin and bacteriorhodopsin (4–9).
The structural snapshots provided by crystallography greatly enhance our understanding of specific receptors but also raise many new issues. Key among these is the extent to which the information from available structures can be extrapolated to the hundreds of other GPCRs. In addition, a key goal in the study of GPCRs—and all receptors—is a description of the interconversions among several structural states that underlie the protein's biological function. It can be a challenging task to deduce a signaling mechanism from static images. As such, structure–function studies, guided by the new structural advances, will remain an important tool in evaluating GPCR function and the nature of drug–receptor interactions in this family.
In recent years, unnatural amino acid mutagenesis on ion channels and receptors expressed in Xenopus oocytes has provided a powerful tool for uncovering crucial drug–receptor interactions and signaling mechanisms (10, 11). GPCRs present an especially attractive target for unnatural amino acid mutagenesis, given the importance of the family, the significant pharmacological variations among closely related family members, and the central role of structural rearrangements in their biological function.
Incorporating unnatural amino acids into GPCRs, however, presents unique challenges. Most unnatural amino acid mutagenesis studies in eukaryotic cells have focused on ion channels. These studies exploit the exquisite sensitivity of electrophysiology, which allows for detailed characterization even when only small quantities of protein are produced, as is often the case with unnatural amino acid mutagenesis. Because GPCRs are not ion channels and instead produce downstream signals through second messenger systems, a direct readout of GPCR activation during an unnatural amino acid experiment is not possible.
In the present work, we describe a general strategy for chemical-scale studies of GPCRs using unnatural amino acid incorporation in a vertebrate cell. Electrophysiology again provides the functional readout, through downstream activation of a K+ channel. We also report studies of key aromatic amino acids in the agonist-binding region of the M2 muscarinic acetylcholine (ACh) receptor and the D2 dopamine receptor. We found that W6.48, a residue long postulated to play an important role in signaling, makes a cation–π interaction to dopamine in the active state of the D2 receptor. Interestingly, ACh does not make the same interaction to the conserved W6.48 of the M2 receptor.
Results
Optimization of a GPCR Readout System.
We have developed a robust assay for studying GPCRs containing unnatural amino acids expressed in Xenopus oocytes. Key issues are described here; full details can be found elsewhere (12).
We began with an established readout system based on a G protein-coupled, inward-rectifying K+ channel (GIRK). Upon activation of a Gi/o-coupled receptor, Gβγ subunits dissociate from the GPCR and then bind to and activate the GIRK channel; Gα subunits also alter channel activation (13–16). This is the most direct known pathway from a GPCR to a channel, providing, in principle, a straightforward electrophysiological assay for GPCR activation.
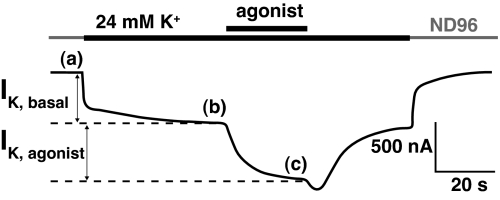
Fig. 1 illustrates the basic protocol. The basal K+ current (IK,Basal) results primarily from the presence of free intracellular Gβγ (17, 18). The agonist-induced current (IK,Agonist) is measured relative to the basal K+ current. Both basal and agonist-induced currents are measured in the presence of a high K+ concentration (24 mM), which provides appropriately large K+ currents at a modest holding potential (−60 mV).
Fig. 1.
Exemplar current traces during a GPCR voltage-clamp experiment on the D2 receptor. Agonist concentration was 10 μM dopamine. IK, Basal is defined as the current difference between b and a; subtraction of b from c yields IK, Agonist
The challenge in implementing this system was to ensure that dose–response relationships provided direct assays of GPCR activation. To yield reproducible data, we optimized the assay system, primarily in experiments with expressed M2 receptors. In previous studies, RGS proteins were shown to accelerate the deactivation kinetics of GIRK currents via Gα-mediated GTP hydrolysis, while also increasing the activation rates of GIRK currents (18–20). Coexpression of RGS4 did indeed result in faster activation and deactivation kinetics in IK,Agonist traces.
We also experimented with coinjection of GαoA mRNA. The added GαoA could bind endogenous Gβγ, and thus minimize IK,Basal, another source of variability. Although this coinjection did suppress basal currents, it also produced aberrant shifts in EC50 (SI Text). We therefore abandoned GαoA mRNA coinjections.
The nonsense suppression methodology can yield low levels of expression for the protein of interest, and thus weak agonist-induced GIRK1/GIRK4 signals. To increase expression levels for the M2 receptor, we used 2 injections of suppressor tRNA. The first injection occurred 48 h before recording and included the aminoacyl-tRNA, along with the M2 receptor and GIRK mRNAs. The second tRNA injection, along with the RGS4 mRNA, occurred 24 h before the assay. We evaluated this protocol on 2 mutants in the M2 receptor: W6.48Trp and W7.40Trp. In nonsense suppression experiments, we list the wild-type residue in one-letter code, the location using the X.50 nomenclature system (Fig. S2) (21), and then the amino acid appended to the tRNA. These experiments are, thus, wild-type “recovery” by nonsense suppression. In both cases, a second injection of tRNA led to larger IK,Agonist: 67% and 89% increases for W6.48 and W7.40, respectively.
Considerable variability was seen from oocyte to oocyte in single-cell EC50 values (cEC50), as quantified by the coefficient of variation (CV) (22). On varying the ratio of mutant M2 receptor to GIRK1/GIRK4 mRNAs, by using ratios of 0.4, 1.0, 2.0, and 4.0, a strong correlation (R = 0.98) between the mRNA ratio and the CV was seen, with smaller ratios producing smaller CVs. Although the mRNA ratio of 0.4 gave the least variability, the expression efficiency was quite low and irregular, and other studies showed that EC50 variability increased when current was small (Fig. S1). We therefore chose an M2/GIRK mRNA ratio of 1.0 (10 ng of M2 receptor mRNA and 10 ng of GIRK mRNA). In 7 different nonsense suppression experiments, these conditions gave cEC50 CVs that ranged from 0.28 to 0.52, which are adequate for meaningfully interpreting variations in EC50 among mutants. The equivalent CV for conventional mutagenesis was 0.55.
The nonsense suppression studies of the M2 receptor were thus conducted as follows. Forty-eight hours before recording we injected 10 ng of the stop codon mutant M2 mRNA, 10 ng each of GIRK1 and GIRK4 mRNA, and 25 ng of the suppressor tRNA ligated to the amino acid of choice. Twenty-four hours later, we injected an additional 25 ng of aminoacyl-tRNA and 10 ng of RGS4 mRNA.
In the D2 receptor system, IK,Agonist levels were consistently lower than those for the M2 receptor, prompting the use of a different mRNA ratio. Adequate expression in the D2 system was achieved by increasing the amount of D2 receptor mRNA and suppressor tRNA 4-fold. Perhaps because of the lower expression levels, including RGS4 affected response waveforms only weakly. Thus, conditions used for D2 receptor experiments were: 40 ng of stop codon mutant D2 receptor mRNA, 10 ng each of GIRK1 and GIRK4 mRNA, and 100 ng of suppressor tRNA 48 h before recording. Twenty-four hours later, we injected an additional 100 ng of aminoacyl-tRNA. These conditions resulted in adequate GIRK currents and in cell-to-cell CVs ranging from 0.21 to 0.46.
Nonsense Expression Experiments.
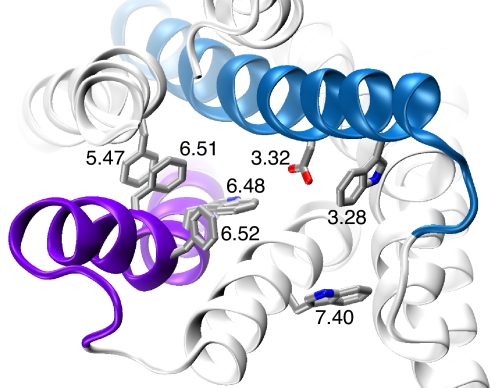
The binding region of class A GPCRs, such as the M2 and D2 receptors, is rich in aromatic amino acids (Fig. 2) (4–8, 23). Both ACh and dopamine have charged ammonium groups, suggesting the possibility of a cation–π interaction (24, 25). In the Cys-loop family of ligand-gated ion channels, ACh and serotonin (which bears structural similarities to dopamine) make cation–π interactions to a conserved Trp in the nicotinic (26, 27) and the 5-HT3 receptors (28), respectively.
Fig. 2.
An image of the β2 structure [Protein Data Bank (PDB) ID code 2RH1] with the residues considered here highlighted. Helix 3 is blue; helix 6 is purple.
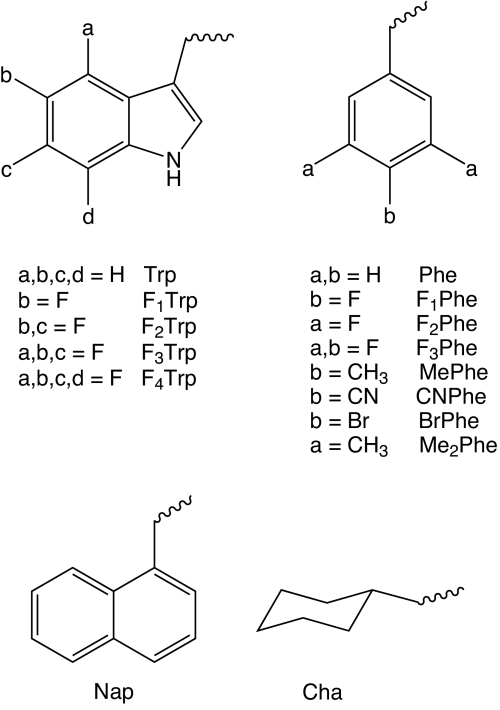
The nonsense suppression protocol for identifying a cation–π interaction is well-established. If a cation–π interaction between the agonist and the particular aromatic is essential, progressive fluorination of the aromatic amino acid steadily diminishes the affinity of the drug for the binding site. To probe a potential cation–π interaction at a Trp site, the appropriate unnatural amino acids are 5-F-Trp (F1Trp); 5,7-F2-Trp (F2Trp); 5,6,7-F3-Trp (F3Trp); 4,5,6,7-F4-Trp (F4Trp); and 1-napthylalanine (Nap) (Fig. 3). At a Phe site, the appropriate analogues are 4-F-Phe (F1Phe); 3,5-F-Phe (F2Phe); 3,4,5-F-Phe (F3Phe); 4-methyl-Phe (MePhe); 4-cyano-Phe (CNPhe); 4-bromo-Phe (BrPhe), 3,5-dimethyl-Phe (Me2Phe); and cyclohexylalanine (Cha).
Fig. 3.
Structures of unnatural amino acids used in this study.
In the M2 receptor, 3 Trp residues were studied: W3.28, W6.48, and W7.40 (Fig. 2). At W6.48 and W7.40, fluorination studies produced no consistent trends (Fig. 4A and Table 1). Most importantly, F3Trp produced an EC50 value very near that of wild-type at both W6.48 and W7.40. These data rule out any possibility of a strong cation–π interaction.
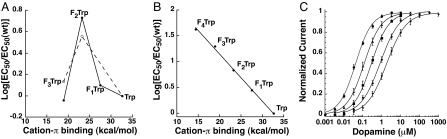
Fig. 4.
Fluorination plots and dose-response curves. (A) FnTrp data for the M2 receptor analyzed in terms of gas phase cation–π binding energies of fluorinated indole rings vs. the log of the ratio of the FnTrp EC50 and wild-type EC50. Dashed line, W7.40; solid line, W6.48. (B) Fluorination plot as in A for W6.48 of the D2 receptor. (C) Dose–response relations for W6.48 of the D2 receptor; EC50 values plotted in B.
Table 1.
EC50 values (μM), with SEM values in parentheses
| Site | Residue | EC50 |
|---|---|---|
| M2 receptor | ||
| W3.28 | –* | 1,900 (80) |
| W6.48 | Trp | 310 (6) |
| F2Trp | 1,100 (70) | |
| F3Trp | 420 (30) | |
| D2 receptor | ||
| F3.28 | Phe | 55 (1) |
| F1Phe | 140 (10) | |
| F2Phe | 36 (1) | |
| F3Phe | 140 (10) | |
| Cha | 97 (2) | |
| F5.47 | Cha | 78 (1) |
| F6.51 | Phe | 64 (4) |
| F1Phe | 76 (6) | |
| F2Phe | 4,200 (350) | |
| F3Phe | 6,200 (400) | |
| CNPhe | 1,340 (160) | |
| MePhe | 690 (40) | |
| Me2Phe | 75,000 (5,000) | |
| Cha | 55,000 (4,000) | |
| W7.40 | Trp | 190 (20) |
| F1Trp | 240 (9) | |
| F2Trp | 1000 (80) | |
| F3Trp | 170 (10) | |
| W6.48 | Trp | 42 (4) |
| F1Trp | 120 (10) | |
| F2Trp | 290 (30) | |
| F3Trp | 840 (60) | |
| F4Trp | 1,800 (300) | |
| Nap | 190 (20) | |
| F6.52 | Phe | 45 (3) |
| F1Phe | 41 (2) | |
| F2Phe | 1,700 (100) | |
| F3Phe | 5,500 (400) | |
| CNPhe | 240 (30) | |
| MePhe | 91 (6) | |
| Me2Phe | 33,000 (3,000) | |
| BrPhe | 1,500 (100) |
Hill coeffiecients generally range from 0.9 to 1.1; number of cells is generally >7. Full data are given in Table S1.
*Experiment with unacylated tRNA.
Studies of W3.28 in the M2 receptor were problematic. When we injected mutant mRNA and full-length tRNA without an appended amino acid, we observed IK,Agonist values that were substantially larger than what is typically seen in this essential control experiment. As such, W3.28 is presently an uninformative site for studies using nonsense suppression with the amber suppressor THG73 tRNA. We were able to determine an EC50 value of 1,900 nM for currents created in this experiment, a 10-fold increase over wild type.
Five different aromatic amino acids were evaluated in the D2 receptor (Fig. 2 and Table 1). F3.28 and F5.47 were quite tolerant of substitution. The largest structural perturbation introduced—Cha—gave essentially wild-type behavior. In sharp contrast, F6.51 and F6.52 were very sensitive to substitution. The primary factor appears to be sterics, with larger substituents producing larger effects.
Incorporation of fluorinated tryptophans at W6.48 resulted in systematic increases in EC50, with 2.8-, 6.9-, 20-, and 43-fold shifts in the series from 1 to 4 fluorines. The standard plot of the calculated gas-phase cation–π binding energies against log EC50 gave the hallmark linear relationship of a cation–π interaction. (Fig. 4 B and C)
The electron-withdrawing ability of a fluorine attached to an indole ring would also be expected to diminish the hydrogen-bonding ability of the indole NH. If a hydrogen bond to this indole NH were essential to receptor function, a linear fluorination plot could also arise. To test for a hydrogen-binding effect, we removed any possibility of such a hydrogen bond by incorporating the unnatural amino acid Nap, which is sterically very similar to Trp but lacks the NH. The modest shift caused by the Nap mutation (Table 1) rules out an essential hydrogen-bonding role for the indole NH of W6.48, especially in contrast with the much larger 43-fold shift for F4Trp, which has the indole NH. Note that Nap is a weaker cation–π donor than Trp (26), consistent with the modest rise in EC50.
Discussion
In the present work, we have developed a general protocol to prepare and functionally characterize GPCRs containing unnatural amino acids. We have applied the methodology to both the M2 ACh receptor and the D2 dopamine receptor. In this initial study, we have identified a distinctive cation–π interaction between dopamine and W6.48, a residue that has been proposed to play a key role in receptor function.
Unnatural Amino Acid Mutagenesis at GPCRs.
Given the broad range of structures and activities for GPCRs, as well as their undeniable pharmaceutical importance, it has been appreciated for some time that unnatural amino acids could provide a valuable probe of this essential class of membrane receptors. An early study incorporated the fluorescent unnatural amino acid NBD-Dap into the NK2 receptor and showed that exposure to tachykinin did produce measurable electrophysiological currents in Xenopus oocytes (due to opening of Ca2+-activated Cl− channels that are endogenous to the oocyte) (29). A very recent study used an orthogonal tRNA–synthetase pair to incorporate a benzophenone-containing unnatural amino acid into the Ste2p GPCR (30) and the CCR5 receptor (31). Certainly, extensive conventional mutagenesis studies on GPCRs have provided a wealth of valuable information about which residues are important to receptor function (32). However, the more subtle variations that are possible with the unnatural amino acid methodology can provide additional insights into the precise role of a given residue.
Here, we describe a general application of nonsense suppression methodology to GPCRs, incorporating 13 different unnatural amino acids and developing a reliable readout system that can be used in chemical-scale studies of many GPCRs. Our initial focus has been on the M2 muscarinic ACh receptor and the D2 dopamine receptor. These are class A GPCRs, a group that also includes adrenergic, serotonin, odorant, peptide, and glycoprotein hormone receptors. The binding site in this class lies within a crevice formed by several of the transmembrane helices and includes the highly conserved Asp 3.32 (Fig. 2). In addition, it has long been appreciated that a cluster of aromatic amino acids shapes much of the binding crevice, and recent structural studies position many of them in locations that could be expected to contribute to agonist and antagonist binding.
We chose the M2 and D2 receptors for this initial study partly because they both couple to the Gi/o pathway, which gates GIRK channels, along with inhibiting adenylate cyclase. GIRK channels provide a sensitive readout of GPCR activation, a critical feature given the often small quantities of protein made by nonsense suppression. However, the use of a downstream signal added significant complications to the process, compared with our previous nonsense suppression studies on ion channels. We can readily control the expression levels of some of these proteins, such as GIRK, but it is less straightforward to control others, such as the G protein and the GPCR itself, when incorporating unnatural amino acids. In addition, other cellular pathways can intersect with the desired signaling pathway in unanticipated ways. Fortunately, we found conditions to minimize this variability; variations in EC50 reported in this study are meaningful to a confidence level of >99% (12).
After controlling for adequate expression efficiencies and consistent dose–response relationships, we arrived at optimum conditions for the M2 and D2 receptor systems. RSG4 expression was used in the M2 system to provide more uniform, faster electrophysiological responses. The delayed injection of RGS4 mRNA produced more consistent expression of the RGS protein, as observed through changes to trace kinetics. We believe that this delay in injection provides the cell's translational and membrane-trafficking machinery a chance to process the M2 and GIRK mRNAs before expressing the RGS4 protein. In addition, relatively low ratios of M2 to GIRK1/GIRK4 mRNA were necessary. We consider these ratios low because the typical expression efficiency of the nonsense suppression methodology is roughly 10% that of conventional expression. Thus, a 1:1 ratio of M2 to GIRK1/GIRK4 mRNA could be considered to produce a ≈0.1:1 ratio of proteins. Presumably, the rather low GPCR expression levels minimize the possibility that receptor activation saturates G proteins, GIRK channels, or other downstream elements in the signaling pathway, which would distort the dose–response relations. Injecting cells with wild-type recovery conditions alongside cells with mutant conditions provided an additional means to assess the variability of a given batch of cells.
Interactions at GPCR-Binding Sites.
This initial study focused on several aromatic amino acids in or near the agonist-binding site. W(F)3.28 was chosen because of its position 4 amino acids—approximately 1 turn of an α-helix—above the highly conserved D3.32. If the cationic moiety of the agonist makes an electrostatic (ion pair) interaction with D3.32, then W(F)3.28 could be well positioned to augment the binding. W6.48 is highly conserved throughout the class A GPCRs and has been proposed to be in close proximity to the agonist-binding site and to play a central role in receptor activation (9, 33). In particular, binding-induced changes in the rotameric state of W6.48 are thought to act as part of a “switch” that is critical to receptor function (4, 9, 33–37). W7.40 is the next most highly conserved residue associated with the aminergic class of GPCRs (38). F6.51 and F6.52 were chosen because the rhodopsin and β2AR structures place them in close proximity to the agonist. Previous studies on the D2 receptor and other aminergic GPCRs have shown that mutations to these helix 6 residues have substantial effects on agonist affinity (32, 39).
The most compelling results are seen for W6.48 of the D2 receptor. A clear linear correlation is seen in the “fluorination plot” (Fig. 4B), establishing a cation–π interaction. By using the β2 structure as a guide (5, 7), one finds no cationic residues (Lys/Arg) within 8 Å of W6.48. Thus, we propose that dopamine contains the cationic moiety forming the cation–π interaction with W6.48. This establishes an energetically significant cation–π interaction between dopamine and W6.48 of the D2 receptor.
The fluorination strategy used here has been used previously to identify cation–π interactions in 8 different Cys-loop receptors for 4 different monoamine neurotransmitters (10). These studies have led to the conclusion that the slope of a fluorination plot is related to the energy of the cation–π interaction. Primary ammonium ions (RNH3+), such as those in serotonin and GABA, produce larger slopes than the quaternary ammonium ion [RN(CH3)3+] of ACh. Studies have established that a cation–π interaction is intrinsically stronger for the smaller, higher-density charge of primary ammonium ions vs. quaternary ammonium ions (40, 41).
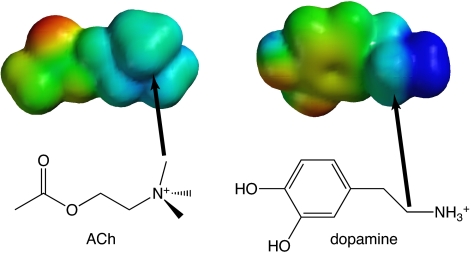
In the D2 receptor, the fluorination plot for W6.48 has a slope of 0.092, which is smaller than would be expected for an agonist with a primary ammonium group. For example, the primary ammonium of serotonin interacts with Trp-183 in the 5-HT3 receptor with a fluorination slope of 0.17 (28). The value for dopamine is much closer to that measured for the interaction between the quaternary ammonium of ACh and a Trp of the nAChR (0.096) (26). This suggested an alternative type of cation–π interaction for dopamine. Despite the typically used symbolism (R4N+), the positive charge of an alkylated ammonium group is not focused on the nitrogen, but rather on the directly attached hydrogens or alkyl groups. The CH2 group adjacent to the ammonium of dopamine (the β-methylene carbon) carries a significant positive charge, one that is similar to that of the methyl groups on the quaternary ammonium of ACh (Fig. 5). Based on the similarity of the slopes for the fluorination plots for dopamine and ACh (in the nAChR), we propose that it is the β-methylene group rather than the ammonium group on dopamine that forms a cation–π interaction with W6.48. Such a cation–π interaction is, in fact, quite common. In a previous survey of cation–π interactions that stabilize protein secondary structure, for energetically significant cation–π interactions involving lysine [i.e., Lys···(Phe,Tyr,Trp)], most structures involved the ε carbon of lysine contacting the face of the aromatic ring (42).
Fig. 5.
Calculated electrostatic potential surfaces (eps) for ACh and dopamine. Color represents relative electrostatic potential, with blue as most positive (limit, +150 kcal/mol), and red as least positive (limit, +13 kcal/mol). Also shown are the structures of each molecule, as well as arrows from a carbon attached to the ammonium N to the corresponding carbon in the eps, showing the similarities in potential.
A binding orientation in which the β-methylene carbon of dopamine forms a cation–π interaction would leave the ammonium group free to hydrogen bond/ion pair with the highly conserved D3.32. A geometry such as that of Fig. 6 seems quite plausible. A prominent role in binding for D3.32 is well-established in several GPCRs (32, 43), and in the present work we found that D3.32E and D3.32N produced 1,000- and 3,000-fold shifts in dopamine EC50, respectively. Our data thus suggest that helices 3 and 6 jointly interact with the  CH2
CH2 NH3+ group of dopamine.
NH3+ group of dopamine.
Fig. 6.
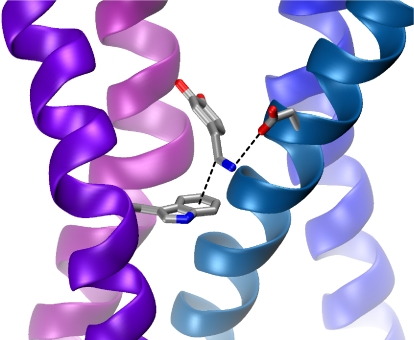
A hypothetical docking mode for dopamine in the D2 receptor. Shown are W6.48 (Lower Left), dopamine (Center), and D3.32 (Upper Right). The side chain of W6.48 has been rotated from that seen in PDB ID code 2RH1, as discussed in text. In the orientation, the dopamine hydroxyls are positioned to enable hydrogen bonding to serine residues on helix 5 (pink).
Implications for the Rotamer Switch Mechanism at W6.48.
The joint interaction of the  CH2
CH2 NH3+ group with helices 3 and 6 agrees with the contemporary model for receptor activation that has the extracellular portion of helix 6 moving toward helix 3. This movement of helix 6 is part of a rotamer switch activation mechanism (4, 9, 33–37). A key component of this model is a reorientation of the side chain of indole of W6.48 from a perpendicular to a parallel orientation, relative to the plane of the membrane, as the receptor transitions from the inactive to active state. This conformational change is associated with straightening of a proline kink in helix 6 and movement of the extracellular portion of helix 6 toward helix 3.
NH3+ group with helices 3 and 6 agrees with the contemporary model for receptor activation that has the extracellular portion of helix 6 moving toward helix 3. This movement of helix 6 is part of a rotamer switch activation mechanism (4, 9, 33–37). A key component of this model is a reorientation of the side chain of indole of W6.48 from a perpendicular to a parallel orientation, relative to the plane of the membrane, as the receptor transitions from the inactive to active state. This conformational change is associated with straightening of a proline kink in helix 6 and movement of the extracellular portion of helix 6 toward helix 3.
In a simple docking of dopamine into the β2 receptor structure, it is not possible to make the ion pair interaction to D3.32 and the cation–π interaction to W6.48 simultaneously. In this structure, a presumed inactive form of the receptor (7), the indole side chain is in the perpendicular orientation (Fig. 2). We found that rotation of the indole side chain with no further relaxation of the structure does allow formation of both the ion pair and the cation–π interaction, as shown in Fig. 6. Thus, we propose that a key component of the rotamer switch mechanism in the D2 receptor is a reorientation of the side chain of W6.48 so that a cation–π interaction can form to the β-carbon on dopamine. This is consistent with the fact that dose–response relationships for receptor function are rightward-shifted with successive fluorination (Fig. 4C), suggesting that dopamine and W6.48 interact more strongly while the receptor is in the active, functional state.
Other Residues.
The present data show that 2 other aromatic amino acids of the D2 receptor—F6.51 and F6.52—do not make a cation–π interaction but are very sensitive to substitution. Data from the monosubstituted Phe derivatives suggest the sensitivity is at least in part due to a steric interaction more than an electronic effect. For F6.51, F1Phe is essentially wild type, but MePhe is significantly perturbed. Methyl is sterically larger than fluorine but has essentially no electronic impact when compared to the electron-withdrawing fluorine. For F6.52, an electronic effect can be ruled out, because the magnitude of the EC50 shift for the Br analog is larger than that of the cyano analog, which is the reverse of their electron-withdrawing effects. Recent structural data for the β-adrenergic receptor are consistent with the steric sensitivity of F6.51 and F6.52; these residues contact each other with a specific geometry (Fig. 2).
A completely different pattern is seen with F3.28 and F5.47 in the D2 receptor. At both sites, Cha is only minimally different from wild type. Clearly, aromaticity is not a critical feature of the side chains at these sites. Instead, hydrophobicity is probably the key determinant.
Given that a large number of ACh-binding and R-N(CH3)3+-binding sites employ cation–π interactions (24, 25, 44), and the fact that position 6.48 of the M2 receptor is also a Trp, one might have expected to find a cation–π interaction at W6.48 of the M2 receptor. However, that is clearly not the case. The data of Table 1 and Fig. 4A do not support a straightforward cation–π interaction at this site. Mutations at W7.40, another aromatic that is thought to be near the agonist-binding site, produced similar data. Given the unusual nature of the fluorination plots of Fig. 4A, we hesitate to ascribe a specific role to these residues.
The third site considered for the M2 receptor, W3.28, was especially susceptible to distorted results from misacylated suppressor tRNA, as evidenced by the large current observed in experiments in which we injected tRNA without a chemically appended amino acid. As such, we cannot state with confidence what role this residue might play. We note that previous experiments have shown that the amino acid incorporated by misacylation of the THG73 tRNA is most likely Gln (45). If that is the case here, the mere 10-fold shift in EC50 for what is effectively a W3.28Gln mutation is too small to be consistent with a cation–π interaction. We also note that more recently developed tRNAs that are less prone to acylation may allow study of the W3.28 site (45, 46).
Materials and Methods
Molecular Biology.
In these experiments, the cDNA for GαoA was in a pCI plasmid, GIRK1 and GIRK4 were in pBSMXT plasmids, D2 receptor (human long form) and RGS4 were in the pcDNA3.1 plasmid, and the human M2 receptor was in the pGEM3 plasmid. Plasmids were linearized with the appropriate restriction enzymes (GαoA with ClaI, the GIRK plasmids with SalI, D2DR with XhoI, RGS4 with StuI, and the M2AChR with HindIII). The mRNA was prepared by in vitro runoff transcription using the Ambion T7 mMessage mMachine kit for all of the constructs except for GIRK1 and GIRK4, which required the T3 kits. For unnatural amino acid mutants, the site of interest was mutated to the TAG stop codon by standard means, verified by sequencing through both strands.
Oocyte Preparation and RNA Injection.
Stage V-VI oocytes of Xenopus laevis were harvested and injected with RNAs as described previously (47). Typical oocyte injection volumes were 50 nL per cell for M2 receptor and 100 nL for D2 receptor experiments; doubly injected oocytes received 50-nL and 100-nL injections, respectively, at each injection session. Synthetic amino acids, which were conjugated to the dinucleotide dCA and ligated to truncated 74-nt tRNA as described previously, were deprotected via a 1-kW xenon lamp for 5 min by using WG-335 and UG-11 filters to remove the 6-nitroveratryloxycarbonyl protecting group. Injection mixture concentrations were typically made such that a 1:1 combination of an mRNA mixture solution and a volume of deprotected tRNA yielded the appropriate concentrations. Wild-type recovery conditions (injecting tRNA with the native amino acid) were injected alongside mutant conditions to control for data variability. Misacylation was assessed at every site of unnatural amino acid incorporation through the injection of 74-nt THG73 ligated to dCA (THG73-dCA).
Electrophysiology.
Oocyte recordings were made in 2-electrode voltage clamp mode by using the OpusXpress 6000A (Axon Instruments). Recording buffers were ND96 (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM Hepes, and 1.8 mM CaCl2, pH 7.5) and high-K ringer (96 mM NaCl, 24 mM KCl, 1 mM MgCl2, 5 mM Hepes, and 1.8 mM CaCl2, pH 7.5). Solution flow rates were 2 mL/min; drug application flow rates were 4 mL/min for the M2 receptor and 2.5 mL/min for the D2 receptor experiments. Initial holding potential was −60 mV. Data were sampled at 125 Hz and filtered at 50 Hz. The ND96 prewash lasted 10 s; the high-K application for basal currents lasted 50 s; drug applications were 15 s in duration for the M2 receptor and 25 s for the D2 receptor; the high-K and ND96 washings were 45 s and 90 s in duration, respectively. Acetylcholine chloride and dopamine were purchased from Sigma–Aldrich/RBI. All drugs were prepared in sterile distilled, de-ionized water for dilution into high-K ringer. Dose–response relations were fitted to the Hill equation, INorm = 1/[1+EC50/A)]nH, where INorm is the normalized current peak at [agonist] = A; EC50 is the concentration of agonist that elicits a half-maximum response; and nH is the Hill coefficient. The cEC50 values were obtained by fitting a single cell's INorm data to the Hill equation, whereas EC50 values were obtained by averaging the INorm values for each cell at a given dose and fitting those average INorm data to the Hill equation. Statistical calculations were performed by using Origin 7.0 (Origin Lab), MiniTab (MiniTab), or Excel (Microsoft).
Supplementary Material
Acknowledgments.
We thank C. Doupnik for advice and plasmids and A. Kovoor for helpful discussions. This work was supported by National Institutes of Health Grants GM 081662 and NS011756.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903260106/DCSupplemental.
References
- 1.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. erratum 542. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into 2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SGF, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherezov V, et al. High-resolution crystal structure of an engineered human 2-adrenergic G protein coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 10.Dougherty DA. Cys-loop neuroreceptors: Structure to the rescue? Chem Rev. 2008;108:1642–1653. doi: 10.1021/cr078207z. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty DA. Physical organic chemistry on the brain. J Org Chem. 2008;73:3667–3673. doi: 10.1021/jo8001722. [DOI] [PubMed] [Google Scholar]
- 12.Torrice MM. Pasadena, CA: California Institute of Technology; 2009. Chemical-scale studies of the nicotinic and muscarinic acetylcholine receptors. PhD Thesis. [Google Scholar]
- 13.Ivanina T, et al. Mapping the Gbetagamma-binding sites in GIRK1 and GIRK2 subunits of the G protein-activated K+ channel. J Biol Chem. 2003;278:29174–29183. doi: 10.1074/jbc.M304518200. [DOI] [PubMed] [Google Scholar]
- 14.Kofuji P, Davidson N, Lester HA. Evidence that neuronal G-protein-gated inwardly rectifying K+ channels are activated by Gbetagamma subunits and function as heteromultimers. Proc Natl Acad Sci USA. 1995;92:6542–6546. doi: 10.1073/pnas.92.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krapivinsky G, Krapivinsky L, Wickman K, Clapham DE. Gbetagamma binds directly to the G protein-gated K+ channel, IKACh. J Biol Chem. 1995;270:29059–29062. doi: 10.1074/jbc.270.49.29059. [DOI] [PubMed] [Google Scholar]
- 16.Mark MD, Herlitze S. G-protein mediated gating of inward-rectifier K+ channels. Eur J Biochem. 2000;267:5830–5836. doi: 10.1046/j.1432-1327.2000.01670.x. [DOI] [PubMed] [Google Scholar]
- 17.Rishal I, Porozov Y, Yakubovich D, Varon D, Dascal N. Gβγ-dependent and Gβγ-independent basal activity of G protein-activated K+ channels. J Biol Chem. 2005;280:16685–16694. doi: 10.1074/jbc.M412196200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang QL, Pacheco MA, Doupnik CA. Gating properties of GIRK channels activated by Gαo- and Gαi-coupled muscarinic m2 receptors in Xenopus oocytes: The role of receptor precoupling in RGS modulation. J Physiol Lond. 2002;545:355–373. doi: 10.1113/jphysiol.2002.032151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doupnik CA, Davidson N, Lester HA, Kofuji P. RGS proteins reconstitute the rapid gating kinetics of Gbeta gamma-activated inwardly rectifying K+ channels. Proc Natl Acad Sci USA. 1997;94:10461–10466. doi: 10.1073/pnas.94.19.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadja R, Alagem N, Reuveny E. Gating of GIRK channels: Details of an intricate, membrane-delimited signaling complex. Neuron. 2003;39:9–12. doi: 10.1016/s0896-6273(03)00402-1. [DOI] [PubMed] [Google Scholar]
- 21.Ballesteros JA, Shi L, Javitch JA. Structural mimicry in G protein-coupled receptors: Implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Mol Pharmacol. 2001;60:1–19. [PubMed] [Google Scholar]
- 22.Harris DD. Quantitative Chemical Analysis. 6th Ed. New York: W. H. Freeman and Company; 2003. [Google Scholar]
- 23.Hibert MF, Trumpp-Kallmeyer S, Bruinvels A, Hoflack J. Three-dimensional models of neurotransmitter G-binding protein-coupled receptors. Mol Pharmacol. 1991;40:8–15. [PubMed] [Google Scholar]
- 24.Dougherty DA. Cation-pi interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 25.Ma JC, Dougherty DA. The cation-π Interaction. Chem Rev. 1997;97:1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 26.Zhong WG, et al. From ab initio quantum mechanics to molecular neurobiology: A cation-pi binding site in the nicotinic receptor. Proc Natl Acad Sci USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiu X, Puskar NL, Shanata JAP, Lester HA, Dougherty DA. Nicotine binding to brain receptors requires a strong cation-π interaction. Nature. 2009;458:534–537. doi: 10.1038/nature07768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beene DL, et al. Cation-pi interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: The anomalous binding properties of nicotine. Biochemistry. 2002;41:10262–10269. doi: 10.1021/bi020266d. [DOI] [PubMed] [Google Scholar]
- 29.Turcatti G, et al. Probing the structure and function of the tachykinin neurokinin-2 receptor through biosynthetic incorporation of fluorescent amino acids at specific sites. J Biol Chem. 1996;271:19991–19998. doi: 10.1074/jbc.271.33.19991. [DOI] [PubMed] [Google Scholar]
- 30.Huang LY, et al. Unnatural amino acid replacement in a yeast G protein-coupled receptor in its native environment. Biochemistry. 2008;47:5638–5648. doi: 10.1021/bi701866e. [DOI] [PubMed] [Google Scholar]
- 31.Ye SX, et al. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem. 2008;283:1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 32.Shi L, Javitch JA. The binding site of aminergic G protein-coupled receptors: The transmembrane segments and second extracellular loop. Annu Rev Pharmacol Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 33.Lu ZL, Saldanha JW, Hulme EC. Seven-transmembrane receptors: Crystals clarify. Trends Pharmacol Sci. 2002;23:140–146. doi: 10.1016/S0165-6147(00)01973-8. [DOI] [PubMed] [Google Scholar]
- 34.Shi L, et al. Beta2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J Biol Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz TW, Rosenkilde MM. Is there a ‘lock’ for all agonist ‘keys’ in 7TM receptors? Trends Pharmacol Sci. 1996;17:213–216. doi: 10.1016/0165-6147(96)10017-1. [DOI] [PubMed] [Google Scholar]
- 36.Lin SW, Sakmar TP. Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry. 1996;35:11149–11159. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Molecular mechanism of 7TM receptor activation–a global toggle switch model. Annu Rev Pharmacol Toxicol. 2006;46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 38.Huang ES. Construction of a sequence motif characteristic of aminergic G protein-coupled receptors. Protein Sci. 2003;12:1360–1367. doi: 10.1110/ps.0305603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho W, Taylor LP, Mansour A, Akil H. Hydrophobic residues of the D2 dopamine receptor are important for binding and signal transduction. J Neurochem. 1995;65:2105–2115. doi: 10.1046/j.1471-4159.1995.65052105.x. [DOI] [PubMed] [Google Scholar]
- 40.Deakyne CA, Meot-Ner Mautne M. Unconventional ionic hydrogen bonds. 2. NH+ …π complexes of onium ions with olefins and benzene derivatives. J Am Chem Soc. 1985;107:474–479. [Google Scholar]
- 41.Meot-Ner Mautner M, Deakyne CA. Unconventional ionic hydrogen bonds. 1. CHδ+…X complexes of quaternary ions with n- and π-donors. J Am Chem Soc. 1985;107:469–474. [Google Scholar]
- 42.Gallivan JP, Dougherty DA. Cation-π interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu ZL, Hulme EC. The functional topography of transmembrane domain 3 of the M1 muscarinic acetylcholine receptor, revealed by scanning mutagenesis. J Biol Chem. 1999;274:7309–7315. doi: 10.1074/jbc.274.11.7309. [DOI] [PubMed] [Google Scholar]
- 44.Zacharias N, Dougherty DA. Cation-pi interactions in ligand recognition and catalysis. Trends Pharmacol Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez EA, Lester HA, Dougherty DA. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: Minimizing misacylation. RNA. 2007;13:1703–1714. doi: 10.1261/rna.666807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez EA, Lester HA, Dougherty DA. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proc Natl Acad Sci USA. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowak MW, et al. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods Enzymol. 1998;293:504–529. doi: 10.1016/s0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.