Abstract
Therapeutic proteins such as antibodies constitute the most rapidly growing class of pharmaceuticals for use in diverse clinical settings including cancer, chronic inflammatory diseases, kidney transplantation, cardiovascular medicine, and infectious diseases. Unfortunately, they tend to aggregate when stored under the concentrated conditions required in their usage. Aggregation leads to a decrease in antibody activity and could elicit an immunological response. Using full antibody atomistic molecular dynamics simulations, we identify the antibody regions prone to aggregation by using a technology that we developed called spatial aggregation propensity (SAP). SAP identifies the location and size of these aggregation prone regions, and allows us to perform target mutations of those regions to engineer antibodies for stability. We apply this method to therapeutic antibodies and demonstrate the significantly enhanced stability of our mutants compared with the wild type. The technology described here could be used to incorporate developability in a rational way during the screening of antibodies in the discovery phase for several diseases.
Keywords: aggregation, antibody, genetic engineering, molecular simulation
Therapeutic antibodies are glyco-proteins tailor-made to seek out and attach themselves to specific antigens, such as biomarkers on the surface of cancer cells. Because of their potential in the cure of various diseases, antibodies currently constitute the most rapidly growing class of human therapeutics (1). Since 2001, their market has been growing at an average yearly growth rate of 35%, the highest rate among all categories of biotech drugs (2). One of the major problems encountered in antibody-based therapies is that these antibodies tend to aggregate under the high concentration formulations required for disease treatment (3). Aggregation leads to a decrease in antibody activity and could elicit an immunological response (4, 5). It also has implications for regulatory approval and for delivery methods. Stabilization of therapeutic antibodies, however, is generally performed during the development phase using trial and error methods. These are both costly and time consuming. Thus, there is a need for a screening tool that will assess aggregation behavior. Such a tool could be used, for example, to evaluate the developability of a bio-pharmaceutical coming during the discovery phase. Protein aggregation in vivo is also shown to be directly responsible for many diseases such as type II diabetes and Alzheimer's (6, 7). Thus, an understanding of the fundamental mechanisms and regions involved in protein aggregation is of importance both for stabilizing protein therapeutics and for devising strategies to prevent in vivo aggregation.
Although there are many possible mechanisms for aggregation, hydrophobic interactions were shown to be the predominant interactions in extensive studies of protein folding and protein–protein binding (8–13). Our results below show that these interactions also play a key role in antibody aggregation. For antibodies in particular, Delano et al. (14) showed that the consensus binding region of the antibody (i.e., the hinge region between CH2 and CH3 domains where at least 4 different protein scaffolds bind) was distinguished by a high degree of solvent accessibility and a predominantly nonpolar character, suggesting that hydrophobic interactions are an important driving force behind binding at this site (14).
In contrast to the studies of aggregation of short peptides involved in amyloid fibril formation, there is relatively little work with regards to predicting the aggregation prone regions for antibodies. This can be attributed to the relatively huge size and thus complexity of antibody molecules (≈1,300 residues, 150 kDa). The prior work in predicting protein aggregation prone regions can be broadly divided into 2 categories, (i) phenomenological models and (ii) molecular simulation techniques. The phenomenological models are based on using physicochemical properties such as hydrophobicity, β-sheet propensity, etc. to attempt to identify aggregation prone regions for unstructured peptides or for globular proteins from their primary sequence (15–19). Molecular simulation techniques use the 3-dimensional structure and dynamics of proteins to locate the regions prone to aggregation (20–26). Applying an accurate fully atomistic model for the simulation of an antibody is very computationally demanding because of the huge size of an antibody. For this reason, although there have been simulations of small parts of the antibody such as the Fab fragment (27, 28), there is no known full antibody atomistic simulation in the literature.
In this current work, we perform atomistic simulation of a full antibody molecule with an explicit solvent. We use the simulations to study the dynamic fluctuations and to characterize the extent of aggregation prone hydrophobic patches exposed on the antibody surface. These hydrophobic patches could be either natively exposed, or exposed due to dynamic fluctuations or conformational changes, as observed in our simulations. Based on a molecular method that we develop to identify aggregation prone regions, we target particular residues for mutation, and using 2 therapeutic antibodies demonstrate that those mutations lead to substantial increases in antibody stability.
Results and Discussion
Prediction of Aggregation Prone Regions Using SAP.
One common way to find the exposure of different residues is through solvent accessible area (SAA). However, SAA by itself does not provide the correct estimate of hydrophobic patches because (i) SAA does not distinguish between hydrophobic and hydrophilic regions, (ii) SAA is not directly proportional to a residue's hydrophobicity (for example, MET has more surface area than LEU but is less hydrophobic), and (iii) SAA does not indicate whether several hydrophobic residues are in close proximity and thus could enhance the hydrophobicity of a certain region.
We define a new parameter called spatial aggregation propensity (SAP), which gives the effective dynamically exposed hydrophobicity of a certain patch on the protein surface. This is illustrated in Fig. 1D and is defined as
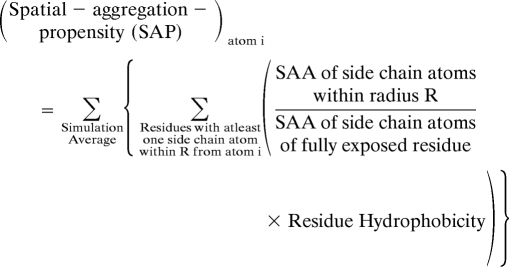 |
Here, (i) SAA of side chain atoms within radius R is computed at each simulation snapshot; (iii) SAA of side chain of fully exposed residue (e.g., for amino acid X) is obtained by calculating the SAA of side chains of the middle residue in the fully extended conformation of tripeptide Ala-X-Ala; and (iii) Residue Hydrophobicity is obtained from the hydrophobicity scale of Black and Mould (29). The scale is normalized such that Glycine has a hydrophobicity of zero. Therefore, amino acids that are more hydrophobic than Glycine are positive and less hydrophobic than Glycine are negative on the hydrophobic scale.
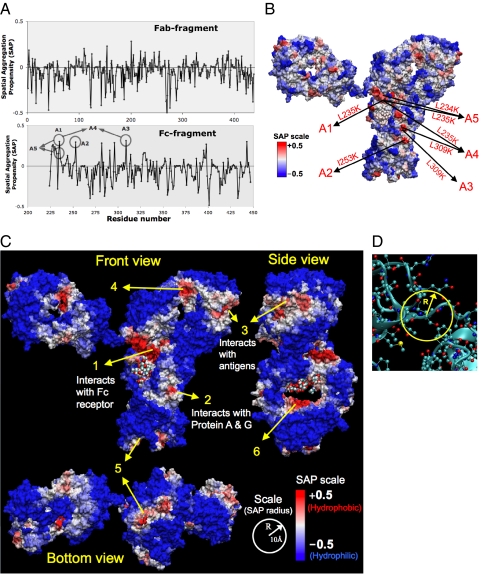
Fig. 1.
Spatial aggregation propensity (SAP) for the antibody A. (A) Values of SAP at R = 5 Å for the Fab and Fc fragments of antibody A, along with the peaks chosen for mutations, A1 through A5. (B) The SAP values at R = 5 Å values are mapped onto the antibody A structure where red regions represent positive peaks and blue regions are negative dips. Again the sites chosen for mutation are indicated A1 through A5. (C) The SAP values at R = 10 Å values are mapped onto the antibody A structure. (D) Definition of spatial aggregation propensity based on atoms within radius-R from a given atom.
Spatial aggregation propensity (SAP) is calculated for spherical regions centered on every atom in the antibody. This gives a unique SAP value for each atom. Then the SAP for a residue is obtained by averaging the SAP of all its constituent atoms. The values of SAP at R = 5 Å thus evaluated with a 30 ns simulation average for each residue in antibody A and antibody B are shown in Figs. 1A and 2A, respectively. For antibody A, the SAP values are obtained from the simulation of full antibody structure obtained by assembling the X-ray structures of Fc, Fab fragments. For antibody B, the X-ray structure of the Fab fragment was not available. Therefore, the structure was built from its sequence using the canonical structure method in combination with homology modeling. Thus the SAP values of antibody B are calculated from 30-ns simulation of the Fab fragment. The Fc fragment is the same for antibody A and antibody B. For both of these antibodies in Figs. 1 and 2 we notice that the majority of SAP values calculated are negative, indicating that most exposed regions are hydrophilic. This is as expected because most of the exposed protein surface is usually hydrophilic. We also observe that there are a few regions with positive peaks for SAP indicating high exposed hydrophobicity. These SAP values from simulation are mapped onto the antibody structure in Figs. 1B and 2B, respectively. In all these figures, the antibody surface is colored according to the values of SAP. Positive values of SAP (hydrophobic) are colored in red whereas negative values (hydrophilic) are in blue. The intensity of color is proportional to the magnitude of SAP. Therefore a highly exposed hydrophobic patch would be deep red, and similarly a highly exposed hydrophilic will be deep blue. For both antibodies A and B, we observe that the surface is predominantly blue indicating that the surface is mostly hydrophilic. Again, this is as expected because most of the protein surface is usually hydrophilic. However, we also notice a few red areas indicating exposed hydrophobic regions. The contrast between the red and blue regions is more prominent at the higher radii of patch (R = 10 Å) used in the calculation of SAP (Figs. 1C and 2C). These red (hydrophobic) regions also have excellent correlation with regions of the antibody known to interact with other proteins (Fig. 1C). The deep red region in the hinge region (region 1 in Fig. 1C) is where the Fc-receptor interacts, the red region in the Fc fragment (region 2) is where protein A and protein G interact, and the red patch at the end of Fab fragment (region 3) is where the antibody binds to antigens. Region-5 at the bottom of Fc is significantly hydrophobic, but it is somewhat buried inside, with hydrophilic region on its borders. Similarly regions 4 and 6 are hydrophobic and solvent exposed, but they are facing into the interior of the antibody. These regions 4 and 6 could still be potentially involved in interactions with other proteins if they are exposed due to significant conformational changes or unfolding of the antibody.
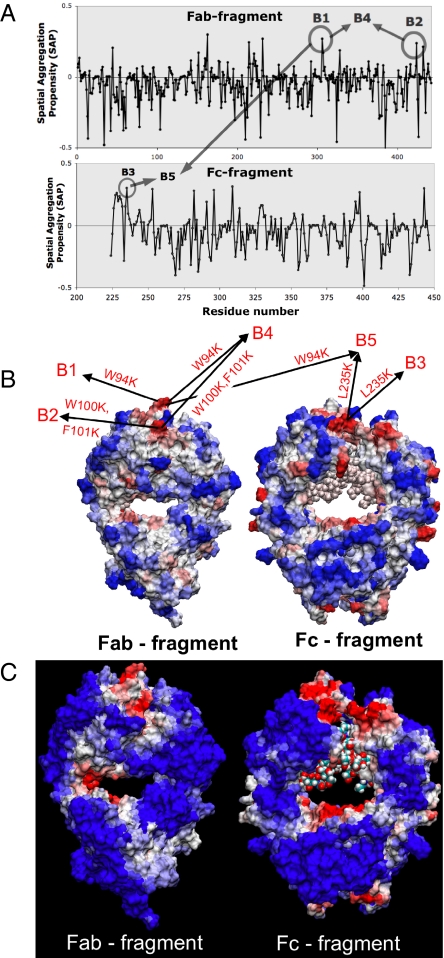
Fig. 2.
Spatial aggregation propensity (SAP) for the antibody B. (A) Values of SAP at R = 5 Å for the Fab and Fc fragment of antibody B, along with the peaks chosen for mutations, B1 through B5. (B) The SAP values at R = 5 Å are mapped onto the antibody B structure where red regions represent positive peaks and blue regions are negative dips. Again, the sites chosen for mutation are indicated B1 through B5. (C) The SAP values at R = 10 Å values are mapped onto the antibody B structure.
Selection of Mutation Sites for Protein Engineering.
The SAP tool was applied to 2 different therapeutic antibodies, A and B. The peaks in the plots of SAP and the corresponding aggregation prone regions (in red) are identified on the antibodies A and B in Figs. 1 and 2, respectively. Based on these SAP values at high resolution (R = 5 Å), we selected the sites to be engineered for enhanced antibody stability. These sites are represented as A1 through A5 for antibody A in Fig. 1 and B1–B5 for antibody B in Fig. 2. The hydrophobic residues that correspond to these positive peaks in SAP (A1 to A5, B1 to B5) were mutated to less hydrophobic (or more hydrophilic) residues as shown in Figs. 1 and 2. Whereas some of these mutants are single site mutants, others are double or triple mutants (such as A4, A5, B2, B4, and B5). The mutants are then tested for their aggregation behavior using accelerated aggregation experiments under heat stress. The resulting aggregates are characterized using size exclusion chromatography–high performance liquid chromatography (SEC-HPLC) and turbidity analysis.
SAP Selected Mutants Are More Stable than Wild Type.
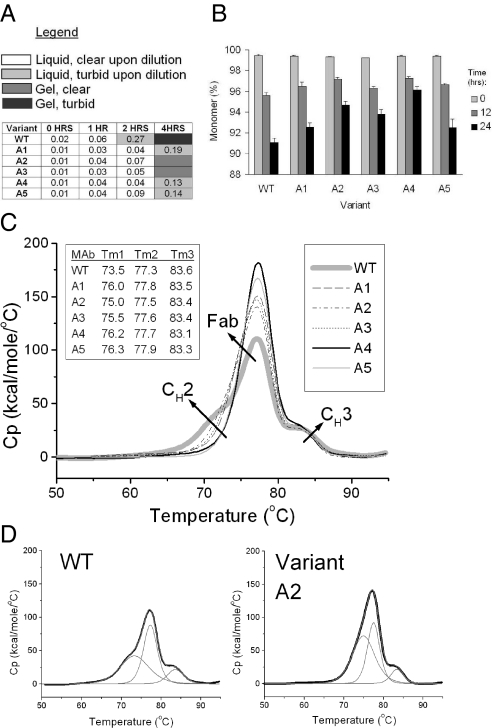
Expression and purification of stable, highly monomeric antibody variants was confirmed by SDS/PAGE (Fig. S1). Variant A1 was also compared with antibody A wild type by circular dichroism, which shows an intact secondary structure upon mutation (Fig. S1). The stability of engineered antibody A variants and wild type were compared using a turbidity assay and SEC-HPLC. The turbidity assay was carried out at 65 °C for up to 4 h with protein samples at 150 mg/mL. SEC-HPLC was used to determine monomer loss over time after heat stress at 150 mg/mL at 58 °C for up to 24 h. Both assays indicate improved stability of all variants of up to 50% compared with wild type (Fig. 3). The thermodynamic stability of antibody A wild type and variants were also compared by differential scanning calorimetry (DSC). A comparison of the thermograms shows an increase of the CH2 melting transition in the variants compared with wild type by 1 °C to 3 °C, with the difference most pronounced for the double variants A4 and A5 (Fig. 3). The results from turbidity, SEC-HPLC and DSC experiments of antibody A wild type and variants are also summarized in Table 1.
Fig. 3.
Stability comparison of antibody A wild type and variants. (A) Turbidity Assay. Samples at 150 mg/mL were incubated at 65 °C for up to 4 h. Color-coding indicates the state of the solution upon 15-fold dilution, or if the sample had gelified. The numbers represent absorbance at 320 nm after further dilution of the samples to 1 mg/mL. (B) SEC-HPLC. Monomer loss of antibody A wild type and variant A1-A5 upon heat stress at 58 °C. Data are mean ± SD. (n = 3 experiments). (C) Overlay of DSC thermograms for antibody A wild-type and each of the 5 variants. Right Inset indicates the line coding for each sample. Wild type is in thick light-gray line. Left Inset summarizes the melting transition temperatures in degrees Celsius for wild type and each variant after peak deconvolution. (D) Examples of thermogram deconvolution: wild type and variant A2. The original thermogram is in black line, and all fitted peaks are in gray.
Table 1.
Summary of stability results for antibody A wild type and variants
| Variant | Mutation | Location | Relative stability based on |
||
|---|---|---|---|---|---|
| Turbidity | HPLC | DSC | |||
| WT | na | na | ++ | ++ | ++ |
| A1 | L235K | CH2 lower hinge | ++++ | +++ | +++ |
| A2 | I253K | CH2-CH3 junction | +++ | +++ | +++ |
| A3 | L309K | CH2 | +++ | +++ | +++ |
| A4 | L235K L309K | CH2 | ++++ | ++++ | ++++ |
| A5 | L234K L235K | CH2 | ++++ | +++ | ++++ |
+ least stable; ++ as stable as WT; +++ more stable than wild type; ++++ most stable; na, not applicable.
Each of the 3 single mutants A1, A2, and A3 show improved stability by each of the 3 analytical methods. In the turbidity assay, dilution of antibody A wild type sample stressed at 65 °C for 2 h results in clouding of the solution, whereas the solutions for all variants remain clear. SEC-HPLC results of samples stressed at 58 °C for 24 h indicate monomer increase from 91% for wild type to 93–95% for the variants. Because the initial monomer population is 99%, the nonmonomeric species in the variants decrease up to a half compared with wild type. DSC analysis shows an increase of the melting transition for CH2 (the domain with the lowest melting transition in antibody A) from 73.5 °C for wild type to 75.0–76.0 °C for the variants. Substituting additional high-SAP residues in variant A1 further improves stability, as evidenced by the turbidity results and the DSC thermograms for variants A4 and A5. The SEC-HPLC results show an improvement over variant A1 only for variant A4 (96% monomer after 24 h of stress) and not for variant A5 (93% monomer after 24-h stress, as variant A1).
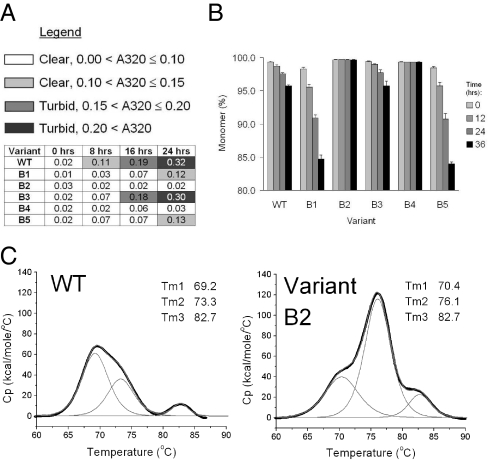
The stability of antibody B wild type and variants were compared in a similar fashion. The turbidity assay was carried out at 55 °C for up to 24 h at 60 mg/mL. SEC-HPLC analysis was carried out after heat stress of protein samples at 60 mg/mL at 52 °C for up to 36 h. Variants B2 and B4 feature a strongly enhanced stability (Fig. 4). The thermodynamic stability of antibody B wild type and variant B2 were also compared by DSC. A comparison of the thermograms of wild type and variant B2 shows an increase of melting transitions in variant B2 (Fig. 4). The results from turbidity and SEC-HPLC experiments of antibody B wild-type and variants are summarized in Table 2.
Fig. 4.
Stability comparison of antibody B wild type and variants. (A) Turbidity assay. Antibody-B wild type and variants were incubated at 60 mg/mL at 55 °C for up to 24 h. Color-coding indicates the state of the solution upon 6-fold dilution. The numbers represent absorbance at 320 nm after further dilution of the samples to 1 mg/mL. (B) SEC-HPLC. Monomer loss of antibody B wild type and variants B1-B5 upon heat stress at 52 °C. Data are mean ± SD. (n = 3 experiments). (C) DSC thermograms of antibody B wild type and variant B2. Fitted peaks are in gray, and the original thermograms are in black. The figure also includes the melting transition temperatures after deconvolution of the thermograms.
Table 2.
Summary of stability results for antibody B wild type and variants
| Variant | Mutation | Location | Relative stability based on* |
|
|---|---|---|---|---|
| Turbidity | HPLC | |||
| WT | na | na | ++ | ++ |
| B1 | W94K | LC CDR | +++ | + |
| B2 | W100K F101K | HC CDR | ++++ | ++++ |
| B3 | L235K | CH2lower hinge | ++ | ++ |
| B4 | W94K W100K F101K | LC CDR HC CDR | ++++ | ++++ |
| B5 | W94K L235K | LC CDR CH2 | +++ | + |
+, least stable; ++ stable as WT; +++ more stable; ++++ most stable; na, not applicable.
In case of antibody B, the replacement of 2 hydrophobic residues in the CDR of VH dramatically improves its stability. Variant B2 has adjacent tryptophan and phenylalanine residues in the heavy chain CDRs substituted with lysines. These 2 mutations lead to a remarkable stabilization compared with wild type. In the turbidity assay, stress at 55 °C for 24 h results in no perturbation of the variant B2 solution, whereas wild-type turns very cloudy; stress at 52 °C for up to 36 h does not lead to any monomer loss in variant B2, whereas the wild-type sample experiences 4% monomer loss. The DSC thermogram of variant B2 also indicates the stabilization effect of the CDR mutations (Fig. 4C). Variant B1 recapitulates some but not all features of variant B2. For instance, variant B1 does not meet the quality control criteria because unstressed variant B1 sample shows only 97% monomer as opposed to the standard 99% or above for wild type and the other variants (SEC-HPLC). Although variant B1 forms significantly more aggregates than wild type after stress at 52 °C, the increase is proportional to the initial amount of nonmonomeric species (3% for variant B1 and 1% for wild type). Nevertheless, variant B1 shows improvement in the turbidity assay compared with wild type. This apparently conflicting data may indicate a destabilizing effect of the mutation, but the lysine mutation may prevent protein denaturation and precipitation that most likely account for solution clouding of wild type in the turbidity assay.
The lower hinge region has little if any role in antibody B aggregation. Variant B3 has a leucine in the lower hinge region substituted with lysine. This variant shows modest stability improvement compared with wild type in the 8-h time point in the turbidity assay. In the longer incubations of the assay, and in the SEC-HPLC experiment, the stability profile of the variant is nearly identical to wild type. The stabilizing effect of the mutations in VH CDR is dominant over the mutation in VL CDR. Variant B4 is a combination of the mutations for variant B1 and B2. According to Turbidity Assay results and SEC-HPLC results, variant B4 shows a stability profile very similar to B2. The stability effects of the mutation in VL CDR are dominant over the mutation in the lower hinge region of CH2. Variant B5 is a combination of the mutations for variant B1 and B3. Both Turbidity Assay results and SEC-HPLC results indicate that variant B5 has the stability features of B1 and not of B3. Thus we observe that the variant in Fc region of antibody B (variant B3) has less effect compared with the variants in the Fab region. The strong stabilizing effect of mutations in variant B2 in the Fab region suggest that amino acids W100 and F101 are the major contributors to aggregation in antibody B.
In addition to the stability analysis, we also performed functional analysis of antibody A and antibody B variants to test if they preserved their antigen binding activity (shown in Tables S1 and S2). The analysis for antibody A showed that all of the variants preserved their function. For antibody B, the variants with mutations in the CDR regions lost their function, whereas the mutation elsewhere preserved its function. This is expected because the antigen binds in the CDR regions, and therefore a mutation there can disrupt binding to antigen. Therefore, in such cases where CDR regions have high SAP, these regions should be carefully engineered such that the stability is improved without losing antigen activity. As shown in the case of antibody A, the stability can be increased without losing activity if the mutations are performed far away from the antigen binding region. In summary, except for mutants that are presumably in the antigen-binding region, most of our mutants showed stable function in addition to improvement in stability.
We have so far used the SAP tool to predict the aggregation prone regions of antibodies successfully. To validate our SAP tool further on other proteins as well, we perform SAP analysis on sickle cell hemoglobin, which is a well-established aggregation model. It was shown that the sixth position of the β-chain in hemoglobin S, which is mutated from glutamic acid in hemoglobin A to valine in hemoglobin S, is the aggregating region leading to sickle cell disease (30). We perform SAP on the X-ray structure of hemoglobin S, PDB entry 2HBS (31). The resulting SAP analysis (Fig. S2) shows excellent correlation between the highest SAP region and the known aggregating region involving the valine residue at the sixth position of the β-chain, thereby validating our SAP tool further. Therefore, in addition to antibodies, the SAP tool could be used to predict aggregation prone regions on other proteins as well.
Conclusions
Aggregation is a major degradation pathway for therapeutic proteins such as antibodies during their storage for disease treatment. Analysis of molecular simulations using SAP can be used to predict the antibody aggregation prone regions. The mutants based on these predicted regions yielded stable variants for both antibodies. The SAP simulation tool developed could be used to improve the stability of potentially all therapeutic antibodies against aggregation. Application of SAP could thus have a huge impact considering that antibody based therapies are growing at the highest pace among all classes of human therapeutics. The same simulation tool could also be used to identify the aggregation prone regions on other proteins and peptides, as we successfully demonstrated using the example of sickle cell hemoglobin. With the mounting number of protein therapeutics, this technology could greatly improve the developability screening of candidate bio-pharmaceuticals for many diseases such as cancer, rheumatoid arthritis and other chronic inflammatory and infectious diseases.
Materials and Methods
Molecular Simulations.
Molecular dynamics simulations are performed for the full antibody A using an all atom model with explicit solvent. The starting structure for simulation was obtained from the X-ray structures of individual Fab and Fc fragments; Fab fragment structure from Novartis Pharma AG and Fc structure from that of another IgG1 antibody 1HZH (32). The structure of a full antibody is then obtained by aligning the Fab and Fc fragments using 1HZH structure as a model template. This structure is then used to perform explicit atom simulations for 30 ns. The CYS residues in the resulting antibody A were all involved in disulphide bonds, including the ones in the hinge region. We have used a G0 glycosylation pattern for our simulations because this is the most common glycosylation pattern observed in antibodies.
We used the CHARMM simulation package (33) for set-up and analysis, and the NAMD package (34) for performing simulations. The CHARMM fully atomistic force field (35) was used for the protein and TIP3P (36) solvent model for water. The simulations are performed at 298 K and 1 atm in the NPT ensemble. The parameters for the sugar groups involved in glycosylation of the Fc fragment were derived in consistence with the CHARMM force field, following from the CSFF force field (37). The protonation states of Histidine residues at pH-7 were decided based on the spatial proximity of electro-negative groups. The full antibody was solvated in an orthorhombic box with periodic boundary conditions in all 3 directions, and a water solvation shell of 8 Å. The resulting total system size was 202,130 atoms. Sufficient ions were added to neutralize the total charge of the system as required by the Ewald summation technique used to calculate the electrostatic contribution. After the antibody was solvated, the energy was initially minimized with steepest descents (SD) by fixing the protein to allow the water to relax around the protein. Then the restraints are removed and the structure is further minimized with SD and adopted basis Newton-Raphson (ABNR). The system was then slowly heated to room temperature with 5 °C increment every 0.5 ps using a 1-fs time step. The system is then equilibrated for 1 ns before we start computing the various properties from simulation. The configurations are saved every 0.1 ps during the simulation for further statistical analysis.
We also performed simulation of a second antibody, the sequence of which was obtained from Novartis Pharma AG. We call this antibody B. Because the X-ray structure of this antibody was not available, we built its structure from its sequence using the canonical structure method (38–40) in combination with homology modeling. This involved identifying the respective canonical structures for each of the CDR loops and modeling the rest of antibody framework through homology modeling. We used the modeling tool WAM (41) to identify the canonical structures. The Fab fragment of the resulting antibody B structure was solvated in a cubic box with a water solvation shell of 8 Å in the longest dimension. The rest of the setup is similar to that of antibody A.
Protein Expression.
Vectors that carry the light chain or the heavy chain genes of the IgG1 Antibodies antibody A and antibody B were obtained by subcloning the genes from proprietary vectors (Novartis) into a gWIZ vector (Genlantis), optimized for high expression from transiently transfected mammalian cells. Antibody variants were generated using site-directed mutagenesis by PCR. All constructs were confirmed by DNA sequencing. Plasmid DNA at the mg scale was purified from bacterial cultures with DNA Maxi Prep columns (Invitrogen). Tissue culture and transient transfection of FreeStyle HEK 293 cells were carried out following the manufacturer's protocols (Invitrogen), except that polyethyleneimine (Polysciences) was used as the transfection reagent. Transfected cells were incubated in a CO2 incubator at 37 °C for 7–9 days.
Protein Purification.
Antibody wild type and variants were purified from the tissue culture supernatant on a Protein A column (GE Healthcare). Antibodies were eluted from the column with 50 mM citrate buffer pH 3.5, and equilibrated to pH 6.6–7.0 with 1 M Tris·HCl pH 9.0, and further purified on a Q Sepharose column (GE Healthcare). Solutions of antibody A wild type and variants were further concentrated with 30 K MWCO filters and buffer exchanged with 20 mM His buffer pH 6.5 to a final concentration of 150 mg/mL. Solutions of antibody B wild type and variants were also concentrated with 30 K MWCO filters and buffer exchanged with 10 mM His buffer pH 6.0 to a final concentration of 60 mg/mL. As a quality control, aliquots of the purified and concentrated samples were analyzed by nonreducing and reducing SDS/PAGE, and by circular dichroism.
Turbidity Assay.
A Turbidity Assay was carried out on samples stressed at a temperature for which we noticed clouding of a concentrated wild-type sample (150 mg/mL in 20 mM His, pH 6.5, for antibody A and 60 mg/mL in 10 mM His, pH 6.0, for antibody B) after several hours of incubation upon dilution to 10 mg/mL in 15 mM potassium phosphate buffer, pH 6.5. In addition to the qualitative observations, turbidity was quantified after further diluting the samples to 1 mg/mL and recording the absorbance values at 320 nm.
SEC-HPLC.
SEC-HPLC was used to determine monomer loss over time in accelerated aggregation experiments. Antibody-A wild type and variants were incubated at 58 °C at 150 mg/mL, and antibody B wild type and variants at 52 °C at 60 mg/mL. Stressed samples were diluted in 15 mM potassium phosphate buffer pH 6.5 to 10 mg/mL. Monomers were resolved from nonmonomeric species on a TSKgel Super SW3000 column (TOSOH Bioscience). Percent monomer was calculated as the area of the monomeric peak divided by the total area of all peaks.
Differential Scanning Calorimetry.
The thermodynamic stability of wild types and variants was also compared by DSC (Microcal). MAbs have characteristic DSC thermograms with 3 melting transitions, if not overlapping: Fab, CH2, and CH3 (42–45). Antibody-A wild type and variants A1-A5 were analyzed at a concentration of 2 mg/mL in 20 mM His pH 6.5 buffer at a 1.0 °C per minute scan rate. Antibody-B wild type and variant B2 were analyzed at a concentration of 1 mg/mL in 10 mM His pH 6.0 buffer at 1.0 °C per minute scan rate. The samples data were analyzed by subtraction of the reference data, normalization to the protein concentration and DSC cell volume, and interpolation of a cubic baseline. The peaks were deconvoluted by non-2-state fit.
Supplementary Material
Acknowledgments.
We thank Dr. Holger Heine from Novartis for his expert advice and help with setting up the antibody expression and purification systems; Dr. Burkhard Wilms from Novartis for providing the DNA vectors for antibody A and antibody B; Drs. Christoph Baechler, Thomas Millward, and Hans-Joachim Wallny from Novartis for performing functional analysis of our variants; Dr. Jean-Michel Rondeau from Novartis for providing X-ray structures of the Fab and Fc fragments of antibody A; and Prof. Dane Wittrup (MIT, Cambridge, MA) for helpful discussions and for use of laboratory equipment. DSC and CD training and experiments were performed at the Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems at Massachusetts Institute of Technology, which is supported by National Science Foundation Grant 0070319 and National Institutes of Health Grant GM68762. This work was supported by Novartis Pharma AG and the National Center for Supercomputing Applications under National Science Foundation Grant MCB060103 and used the National Center for Supercomputing Applications tungsten cluster.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904191106/DCSupplemental.
References
- 1.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal S. What's fueling the biotech engine? Nat Biotechnol. 2007;25:1097–1104. doi: 10.1038/nbt1007-1097. [DOI] [PubMed] [Google Scholar]
- 3.Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci. 2004;93:1390–1402. doi: 10.1002/jps.20079. [DOI] [PubMed] [Google Scholar]
- 4.Hermeling S, Crommelin DJA, Schellekens H, Jiskoot W. Structure-immunogenicity relationships of therapeutic proteins. Pharm Res. 2004;21:897–903. doi: 10.1023/b:pham.0000029275.41323.a6. [DOI] [PubMed] [Google Scholar]
- 5.Braun A, Kwee L, Labow MA, Alsenz J. Protein aggregates seem to play a key role among the parameters influencing the antigenicity of interferon alpha (IFN-α) in normal and transgenic mice. Pharm Res. 1997;14:1472–1478. doi: 10.1023/a:1012193326789. [DOI] [PubMed] [Google Scholar]
- 6.Selkoe DJ. Alzheimer's disease: Genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 7.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 8.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;31:7134–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 9.Guharoy M, Chakrabarti P. Conservation and relative importance of residues across protein–protein interfaces. Proc Natl Acad Sci. 2005;43:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S, Thornton JM. Principles of protein–protein interactions. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young L, Jernigan RL, Covell DG. A role for surface hydrophobicity in protein–protein recognition. Prot Sci. 1994;3:717–729. doi: 10.1002/pro.5560030501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai C, Lin SL, Wolfson HJ, Nussinov R. Studies of protein–protein interfaces: A statistical analysis of the hydrophobic effect. Prot Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conte LL, Chothia C, Janin J. The atomic structure of protein–protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 14.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein–protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 15.Caflisch A. Computational models for the prediction of polypeptide aggregation propensity. Curr Opin Chem Biol. 2006;10:437–444. doi: 10.1016/j.cbpa.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:336–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 17.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 18.Tartaglia GG, Cavalli A, Pellarin R, Caflisch A. Prediction of aggregation rate and aggregation-prone segments in polypeptide sequences. Prot Sci. 2005;14:2723–2734. doi: 10.1110/ps.051471205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 20.Cellmer T, Bratko D, Prausnitz JM, Blanch HW. Protein aggregation in silico. Trends in Biotech. 2007;25:254–261. doi: 10.1016/j.tibtech.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen HD, Hall CK. Molecular dynamics simulations of spontaneous fibril formation by random-coil peptides. Proc Natl Acad Sci USA. 2004;101:16180–16185. doi: 10.1073/pnas.0407273101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gsponer J, Haberthur U, Caflisch A. The role of side-chain interactions in the early steps of aggregation: Molecular dynamics simulations of an amyloid-forming peptide from the yeast prion Sup35. Proc Natl Acad Sci USA. 2003;100:5154–5159. doi: 10.1073/pnas.0835307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimov DK, Thirumalai D. Dissecting the assembly of Aβ16–22 amyloid peptides into antiparallel beta-sheets. Structure. 2003;11:295–307. doi: 10.1016/s0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 24.Harrison PM, Chan HS, Prusiner SB, Cohen FEJ. Thermodynamics of model prions and its implications for the problem of prion protein folding. Mol Biol. 1999;286:593–606. doi: 10.1006/jmbi.1998.2497. [DOI] [PubMed] [Google Scholar]
- 25.Dima RI, Thirumalai D. Exploring protein aggregation and self-propagation using lattice models: Phase diagram and kinetics. Protein Sci. 2002;11:1036–1049. doi: 10.1110/ps.4220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broglia RA, Tiana G, Pasqali S, Roman HE, Vigezzi E. Folding and aggregation of designed proteins. Proc Natl Acad Sci USA. 1998;95:12930–12933. doi: 10.1073/pnas.95.22.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noon WH, Kong Y, Ma J. Molecular dynamics analysis of a buckyball-antibody complex. Proc Natl Acad Sci USA. 2002;99:6466–6470. doi: 10.1073/pnas.022532599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha N, Smith-Gill SJ. Molecular dynamics simulation of a high-affinity antibody protein complex: The binding site is a mosaic of locally flexible and preorganized rigid regions. Cell Biochem Biophys. 2005;43:253–273. doi: 10.1385/CBB:43:2:253. [DOI] [PubMed] [Google Scholar]
- 29.Black SD, Mould DR. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal Biochem. 1991;193:72–82. doi: 10.1016/0003-2697(91)90045-u. [DOI] [PubMed] [Google Scholar]
- 30.Ingram VM. Gene mutations in human hæmoglobin: The chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 31.Harrington DJ, Adachi K, Royer WE., Jr The high resolution crystal structure of deoxyhemoglobin S. J Mol Biol. 1997;272:398–407. doi: 10.1006/jmbi.1997.1253. [DOI] [PubMed] [Google Scholar]
- 32.Saphire EO, et al. Crystal structure of a neutralizing human IGG against HIV-1: A template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 33.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. Comput Chem. 1983;4:187–217. [Google Scholar]
- 34.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of protein. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 37.Kuttel M, Brady JW, Naidoo KJ. Carbohydrate solution simulations: Producing a force field with experimentally consistent primary alcohol rotational frequencies and populations. J Comput Chem. 2002;23:1236–1243. doi: 10.1002/jcc.10119. [DOI] [PubMed] [Google Scholar]
- 38.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 39.Chothia C, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 40.Al-Lazikani B, Lesk A, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 41.Whitelegg NR, Rees AR. WAM: An improved algorithm for modeling antibodies on the WEB. Protein Eng. 2000;13:819–824. doi: 10.1093/protein/13.12.819. [DOI] [PubMed] [Google Scholar]
- 42.Tischenko VM, Abramov VM, Zav'yalov VP. Investigation of the cooperative structure of Fc fragments from myeloma immunoglobulin G. Biochemistry. 1998;37:5576–5581. doi: 10.1021/bi972647a. [DOI] [PubMed] [Google Scholar]
- 43.Mimura Y, et al. The influence of glycosylation on the thermal stability and effector function expression of human IgG1-Fc: Properties of a series of truncated glycoforms. Mol Immunol. 2000;37(12–13):697–706. doi: 10.1016/s0161-5890(00)00105-x. [DOI] [PubMed] [Google Scholar]
- 44.Ionescu RM, Vlasak J, Price C, Kirchmeier M. Contribution of variable domains to the stability of humanized IgG1 monoclonal antibodies. J Pharm Sci. 2008;97:1414–1426. doi: 10.1002/jps.21104. [DOI] [PubMed] [Google Scholar]
- 45.Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun. 2007;355:751–757. doi: 10.1016/j.bbrc.2007.02.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.