Abstract
The virion host shutoff protein product of the UL41 gene of herpes simplex virus 1 is an endoribonuclease that selectively degrades mRNAs during the first hours after infection. Specifically, in contrast to the events in uninfected cells or cells infected with a mutant lacking the RNase, in wild-type virus-infected cells mRNA of housekeeping genes exemplified by GAPDH is degraded rapidly, whereas mRNAs containing AU elements are cleaved and the 5′ cleavage product of these RNAs persists for many hours. We report that in wild-type virus-infected cells there was a rapid increase in the number and size of processing bodies (P-bodies). These P-bodies were also preset in cycloheximide (CHX)-treated cells but not in either treated or untreated uninfected cells or cells infected with the RNase minus mutant. Additional studies revealed that polyribosomes extracted from cytoplasm of wild-type virus-infected cells treated with CHX and displayed in sucrose gradients contained ribosome-loaded, truncated AU-rich mRNAs lacking the 3′ UTR and poly(A) tails. The results suggest that the virion RNase is bound to polyribosomes by virtue of the reported association with translation machinery and cleaves the RNAs 5′ to the AU elements. In contrast to the slow degradation of the of the residual 5′ domain, the 3′ UTR of the AU-rich mRNA and the GAPDH mRNA are rapidly degraded in wild-type virus-infected cells.
Keywords: AU-rich elements RNA, RNase
The UL41 gene of herpes simplex virus 1 (HSV-1) encodes an endoribonuclease with substrate specificity of RNase A and designated the virion host shutoff (VHS)-RNase (1, 2). It is packaged in virions and introduced into cells during virus entry. Its function is to degrade preexisting and newly transcribed mRNAs at early times after infection (reviewed in ref. 3). The outcome of degradation of cellular mRNAs after infection is sequence specific (4). Thus, in contrast to the events in uninfected or ΔUL41-infected cells, normally stable RNAs exemplified by GAPDH mRNA are rapidly degraded, whereas normally rapidly turning over AU-rich elements (ARE) containing mRNAs are cleaved and the 5′ UTR and the coding sequence but not the 3′ UTR persist for many hours (4, 5). This article focuses on the site of degradation of the mRNAs.
Relevant to this article are the following. The enzymes involved in mRNA decay and the complex regulation that determines the pathway and rates of mRNA degradation in noninfected cells have recently been reviewed (6). In eukaryotes, degradation of mRNAs is initiated by removal of the poly(A) tail by the deadenylases (7, 8). Deadenylation induces the transition of the mRNA from an active translational state maintained by the interaction of the translation initiation factors eIF4E/eIF4G with the poly(A)-binding protein (PABP) to an intermediate inactive state, where it becomes target for degradation either by exosomes [multimeric complexes of 3′-to-5′ exonucleases (9)] or decapping and 5′-to-3′ degradation by exoribonuclease Xrn1 (7, 8). In addition to exonuclease-mediated mRNA decay pathways, vertebrate cells can use endonucleases to catalyze mRNA degradation (10), such as the polysomal ribonuclease 1 (PMR1) (11–13).
The proteins involved in the 5′-to-3′ mRNA decay pathway are enriched in cytoplasmic processing bodies (P-bodies) (14). P-bodies become enlarged when the mRNA decay machinery is rendered limiting by an increase in the level of polyribosome-free cytoplasmic mRNAs after treatment with puromycin (15), whereas they disappear when mRNA substrates become limiting upon entrapment of translated mRNAs in polyribosomes by exposure to cycloheximide (CHX) (15, 16).
Soon after infection the host cell polyribosomes are dispersed, contributing to the global decline of host protein synthesis (17, 18). The HSV-induced RNA decay and polyribosome disassembly were not inhibited by exposure of the cells to CHX or Actinomycin D, suggesting that it might be caused by some constituent of the infecting viral particles (19, 20).
In this study, we report that P-bodies are abundant and enlarged in wild-type virus-infected cells but are small and uncommon in uninfected or ΔUL41-infected cells. The unexpected finding was that P-bodies were present in CHX-treated, wild-type virus-infected cells and not in uninfected or ΔUL41-infected cells. Following up on this observation, we discovered that polyribosomes extracted from CHX-treated, wild-type virus-infected cells displayed in sucrose gradients contained ribosome-loaded RNAs lacking the 3′ UTR. These truncated RNAs were not present in polyribosomes of drug-treated uninfected cells or ΔUL41-infected cells.
Results
P-Bodies Are Enlarged in Cells Infected by Wild-Type HSV-1.
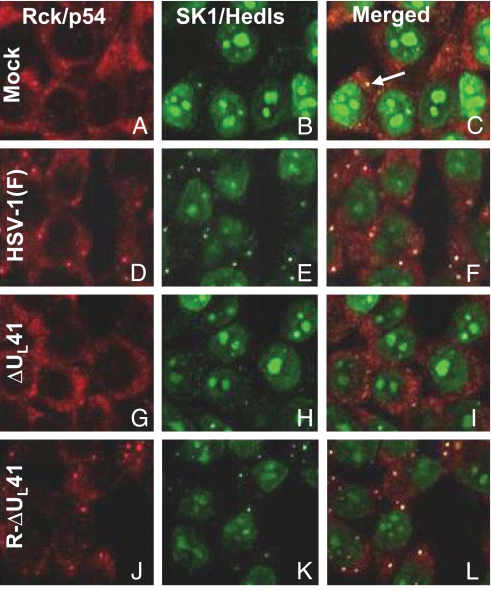
In this series of experiments HeLa cells grown in 4-well slides and either mock-infected or infected with 10 pfu of wild-type HSV-1(F), ΔUL41, or R-UL41 viruses per cell and incubated for 3 h in spent medium to reduce cell stress were fixed and reacted with antibodies to Rck/p54 and Hedls according to the recommendation reported in ref. 21. The results illustrated in Fig. 1 were as follows:
In mock-infected cells Rck/p54 appeared diffused throughout the cytoplasm (Fig. 1A), whereas the antibody directed against SK1-Hedls showed only a strong nuclear staining (Fig. 1B) caused by reactivity with its intended target, p70 S6 kinase or SK1. Small punctuate cytoplasmic structures in which the 2 P-bodies components colocalized were occasionally observed (Fig. 1C, white arrow). The percentage of cells exhibiting P-bodies (≈20%) in our studies was comparable with those reported earlier for actively growing cells (21).
As shown in Fig. 1 D–F, in the majority of the cells (>80%) exposed to HSV-1(F), Rck/p54 and Hedls/Ge-1 were both localized in discrete cytoplasmic foci defined elsewhere as P-bodies (21). P-bodies have been detected in the cytoplasm of infected HEp-2, U2OS, and mouse and human embryonic fibroblasts, indicating that the HSV-1-induced accumulation of P-bodies is not cell-type dependent.
P-bodies were not detectable in cells exposed to ΔUL41 mutant virus (Fig. 1 G–I). In these cells the antibody staining patterns were identical to those of mock-infected cells.
P-bodies were readily detected in cells infected with the mutant virus in which the UL41 gene was restored (R-ΔUL41; Fig. 1 J–L). The size, distribution, and number of cells exhibiting the P-bodies were similar to those of wild-type virus-infected cells.
We conclude that wild-type virus induces the formation of P-bodies and their presence correlates with the expression of the ΔUL41 gene.
Fig. 1.
P-bodies formation in response to HSV-1 infection. HeLa cells grown in 4-well slides were either mock infected (A–C) or exposed to10 pfu of HSV-1(F) (D–F), ΔUL41 mutant virus (G–I), or R-ΔUL41 virus (J–L) per cell. The cells were fixed and stained 3 h after exposure to virus as described in Materials and Methods. P-bodies are occasionally present in uninfected cells (white arrow) as explained in Results.
Enlarged P-Bodies Persist in Wild-Type Virus-Infected Cells Even Later in Infection.
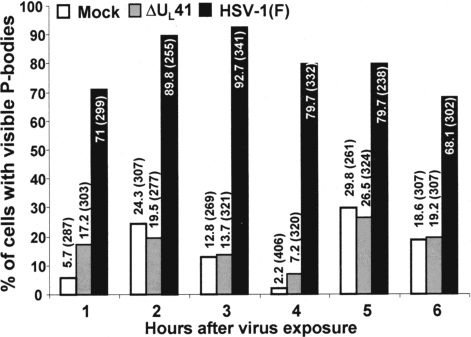
To exclude the possibility that the lack of P-body formation in ΔUL41-infected cells was a consequence of the slower replication kinetic of this virus, we monitored the formation of P-bodies at later time points and in cells infected at a higher multiplicity of infection. HeLa cells grown in 4-well slides were either mock-infected or infected with 20 pfu per cell of HSV-1(F) or ΔUL41 mutant virus and fixed at 1-h intervals for 6 h after infection. At least 250 cells per well were counted and the percentage showing visible P-bodies was plotted as a function of time. The results (Fig. 2) were as follows:
A large number (≈70%) of cells infected with HSV-1(F) showed microscopically visible P-bodies as early as 1 h after virus exposure. The percentage of cells with enlarged P-bodies increased up to 3 h postinfection (≈95%) and declined slightly thereafter.
The number of cells showing enlarged P-bodies after infection with ΔUL41 was almost indistinguishable from that observed in mock-infected cells, although the percentage in both groups varied within the range typically reported for actively growing cells (21). From these series of experiments, we conclude that enlarged P-bodies appear very rapidly upon virus exposure and persist for several hours during the infection.
Fig. 2.
Time course of appearance and maintenance of P-bodies in the cytoplasm of uninfected and infected cells. HeLa cells grown in 4-well slides were either mock-infected (white bars) or infected with 20 pfu per cell of HSV-1(F) (black bars), or ΔUL41 mutant virus (gray bars). The cells were fixed and stained at the times indicated. The cells were counted in 10 randomly chosen fields. The percentages of cells exhibiting visible P-bodies were counted and shown as a percentage of total cells counted as indicated in parentheses.
HSV-1-Dependent P-Bodies Do Not Disappear After Treatment with CHX.
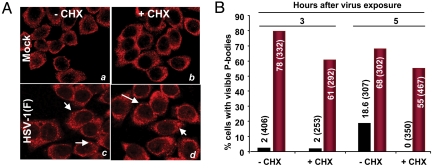
In uninfected cells the size of P-bodies directly correlates with the availability of mRNA substrates. Consequently, the entrapment of mRNAs in polyribosome after exposure to CHX results in rapid disassembly of P-bodies (15, 16). To determine whether wild-type virus-infected cells respond in a similar manner, HeLa cells grown in 4-well slides either mock-infected or infected with 20 pfu of HSV-1(F) per cell were incubated for 3 or 5 h and then exposed to medium containing CHX (50 μg/mL). After 1 additional h of incubation, the cells were fixed and reacted with antibodies. Treatment of mock-infected cells with CHX resulted in the complete disappearance of background levels of P-bodies (Fig. 3A, compare b with a) whereas they were still visible in drug-treated cells infected with wild-type virus (Fig. 3A, compare d with c), albeit at lower percentage compared with untreated infected cells as illustrated in Fig. 3B.
Fig. 3.
HSV-1(F)-induced P-bodies do not disappear after treatment with CHX. HeLa cells grown in 4-well slides were either mock-infected or exposed to 10 pfu of HSV-1(F) per cell for 3 or 5 h. At these times the inoculum was replaced with spent medium alone or medium containing 50 μg of CHX per mL of medium. After 1-h incubation at 37 °C, the cells were fixed and immunostained with antibody against Rck/p54 (red). The cells were counted in 10 randomly chosen fields. The percentages of cells exhibiting visible P bodies were counted and shown as a percentage of total cells counted as indicated in parentheses. (A) Representative images of mock- and HSV-1(F)-infected cells either untreated (a and c) or incubated for 1 h in medium containing 50 μg of CHX per mL 3 h after infection (b and d). (B) Histogram showing the percentage of cells containing visible P-bodies in mock- and HSV-1(F)-infected cells untreated or CHX-treated after either 3 or 5 h of exposure to the virus. Total cells counted are reported in parentheses.
VHS Plays a Role in the Polyribosome Disassembly Early After Infection.
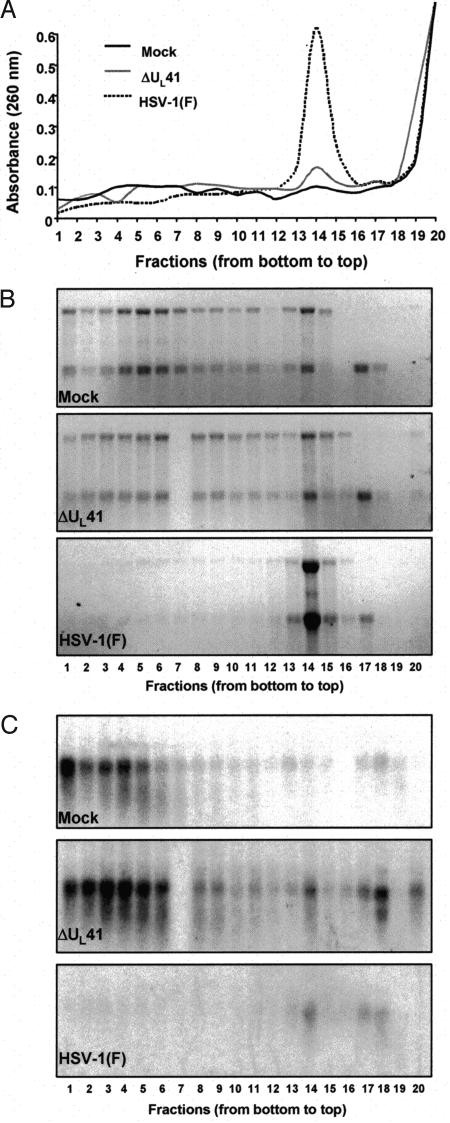
The objective of these experiments was to determine whether VHS-RNase attacks mRNA in polyribosomes as could be inferred from the presence of enlarged P-bodies in CHX-treated cells described above and earlier studies that showed that the host shutoff was unaffected by inhibitors of protein translation (20). HeLa cell monolayers were either mock-infected or exposed to 30 pfu of HSV-1(F) or ΔUL41 viruses per cell at 37 °C for 1 h, then incubated in medium containing CHX (50 μg/mL) for 2 h at 37 °C. Cell extracts, prepared as described in Materials and Methods, were subjected to centrifugation through 10–45% sucrose gradients. The fractions was assayed for absorbance of 260 nm and the total RNA in each fraction was purified and analyzed as described in Materials and Methods. The results were as follows: The UV absorbance pattern (Fig. 4A) of fractions derived from HSV-1(F)-infected cells was consistent with those previously observed, in which most ribosomal subunits were dissociated and only a minor fraction engaged in protein synthesis. In contrast, the UV absorbance profiles of mock-infected or those of ΔUL41-infected cells were similar and consistent with the expectation that the vast majority of ribosomes were sequestered in polyribosomes. The RNA distribution (Fig. 4B) among the fractions mirrored the UV absorbance profiles. Indeed ribosomal RNA was evenly distributed in the fractions collected from the gradient loaded with cell lysate from either mock- or ΔUL41-infected cells (Fig. 4B Top and Middle), whereas they accumulated at the top of the gradient loaded with HSV-1(F)-infected cell lysate (Fig. 4B Lower). Taken together, these results associate the dissociation of polyribosome immediately after wild-type virus infection with the VHS-RNase.
Fig. 4.
VHS plays a role in the polyribosomes' disassembly early after infection. HeLa cells were either mock-infected or infected with HSV-1(F) or ΔUL41 mutant virus at a multiplicity of 30 pfu per cell for 1 h. The inoculum was then replaced with medium containing CHX (50 μg/mL). After 2-h incubation at 37 °C, the cells were lysed and fractionated on a 10–45% sucrose gradient as described in Materials and Methods. The fractions were collected from the bottom of the gradients. (A) Graph of A260 versus fraction number for sucrose density gradient fractionation of cytoplasmic extract from mock-infected (black continuous line) or ΔUL41 mutant virus (gray continuous line) or wild-type HSV-1(F) (dotted line) HeLa cells. One-third of each fraction was used for determination of absorbance. (B) Visualization by ethidium bromide staining of RNA extracted from each fraction and resolved on a denaturing formaldehyde gel as described in Materials and Methods. (C) Northern blotting analysis for GAPDH mRNA.
The patterns of degradation of cellular mRNAs change after HSV-1 infection. Thus, mRNAs that in uninfected cells are relatively stable, exemplified by GAPDH, are rapidly degraded in infected cells. In contrast, mRNas that are relatively unstable, such as the AU-rich mRNA IEX-1, are only partially degraded and tend to persist for many hours after infection (4, 5). Therefore, it was of interest to analyze the distribution of these 2 mRNAs in the sucrose gradient-fractionated cell lysates from either infected or uninfected cells.
The results, presented in Figs. 4C and 5were as follows:
In mock- or ΔUL41-infected cells the majority of GAPDH transcript was recovered within the polyribosomal fractions at the bottom of the gradient (Fig. 4C), indicating that in these cells the mRNAs were loaded with ribosomes and actively engaged in protein synthesis. Conversely, no GAPDH mRNA was detected in the fractions containing polyribosomes and isolated from HSV-1(F)-infected cells (fractions 1–7), whereas a barely visible amount remained associated with the single ribosomal subunits (fractions 14–18). The results suggest that VHS-RNase is able to target ribosome-bound mRNAs for degradation.
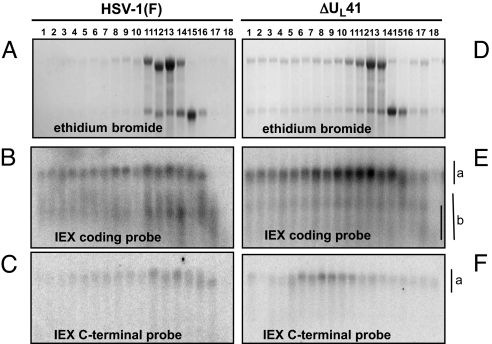
IEX-1 mRNA was readily detected in sucrose gradient containing polyribosomes of cells infected with the ΔUL41 mutant virus. On the basis of its electrophoretic mobility, the mRNA appears to be full-length (Fig. 5 E and F). The RNAs derived from wild-type virus-infected cells and extracted from sucrose density gradient fractions formed 2 bands. The electrophoretic mobility of the slow migrating and identified by the vertical “a” line in Fig. 5 was similar to the band formed by the IEX-1 RNA in ΔUL41-infected cells. This band hybridized with both the probe representing the 5′ UTR and coding sequence of IEX-1 mRNA (Fig. 5B) and the 3′ UTR of IEX-1 RNA (Fig. 5C). The faster migrating band identified by the vertical line “b” in Fig. 5 was heterogeneous in size, and although it was present in most fractions of the sucrose gradient it hybridized with the probe representing the 5′ UTR and coding sequence (Fig. 5B) but not with the probe representing the 3′ UTR of IEX-1 mRNA (Fig. 5C).
We conclude from these studies the following:
IEX-1 mRNA is cleaved in polyribosomes. The 5′ portion of IEX-1 mRNA is heterogeneous in size and is present in all fractions of the sucrose density gradients bound to ribosome.
The 3′ cleavage products containing the 3′ UTR and linked poly(A) tail are found in minute amounts at the top of the gradient and given the inequality of the amounts of 5′ and 3′ product of the cleavage reaction, they may be presumed to be rapidly degraded.
The normally stable mRNAs lacking AREs, exemplified by GAPDH, rapidly disappear from CHX-treated, wild-type virus-infected cells. In this respect their fate appears to be similar to that of the 3′ UTR of IEX-1 mRNA.
Fig. 5.
Pattern of IEX-1 degradation in HSV-1(F) and ΔUL41-infected cells. HeLa cells were either infected with HSV-1(F) (A–C) or ΔUL41 mutant virus (D–F) at a multiplicity of 30 pfu per cell for 1 h. The inoculum was then replaced with medium containing CHX (50 μg/mL). After 2-h incubation at 37 °C, the cells were collected and lysates were fractionated on a sucrose gradient as in Fig. 4. Total RNA, extracted from each fraction, was loaded on a denaturing formaldehyde gel, visualized by ethidium bromide staining (A and D), and analyzed by Northern blotting for IEX-1 mRNA with either a probe specific for the IEX-1 coding sequence (B and E) or an single-stranded oligoprobe complementary to a region of the 3′ UTR downstream of AREs (C and F). The vertical line designated “a” indicates the migration of full-length mRNA, and the line labeled “b” shows the distribution of truncated RNAs.
Discussion
In this article we show the following: (i) P-bodies accumulate in the cytoplasm of cells infected with wild-type HSV-1 at early times after virus exposure. (ii) The accumulation of P-bodies correlates with the presence of virion-bound VHS-RNase inasmuch as they are not visible in cells infected with a ΔUL41 mutant virus. (iii) P-bodies are present in wild-type virus-infected cells treated with CHX. (iv) The appearance of cleaved mRNA in polyribosomes of wild-type virus-infected cells indicates that VHS-RNase can cleave polyribosome-bound RNA. The significance of our results stems from the following considerations.
Two hypotheses may account for the appearance and persistence of P-bodies throughout infection with wild-type virus. The first, as yet unproven, is that VHS-RNase specifically interacts with components of P-bodies and fosters their formation. The second, based largely on current literature, is that infected cells accumulate a large amount of untranslatable RNA. When the decay enzymes are limiting, RNAs are stored in enlarged P-bodies. P-bodies store these RNAs until the process of degradation catches up with the supply of RNA substrates destined for degradation (14–16). This view is tempered somewhat by 2 considerations. Foremost, although in wild-type-infected cells the degradation of AU-rich mRNAs, e.g., IEX-1, is slower than in uninfected cells, that of housekeeping mRNAs, e.g., GAPDH, is faster than in uninfected cells (4, 5). The simplistic hypothesis that in infected cells the appearance of P-bodies results from oversupply of unselected mRNAs destined for degradation may not fully account for the appearance and persistence of P-bodies in wild-type infected cells.
A fundamental observation reported in several studies is that in uninfected cells P-bodies disappear or are grossly diminished in size in cells treated with CHX (15, 16). The fundamental hypothesis underlying this observation is that in the presence of the drug the degradation machinery catches up with the supply and because ribosomes remain bound to the mRNA, little or no new RNA becomes available for degradation. In our studies P-bodies were abundant in infected, translation inhibitor-treated cells.
Of the various explanations for our results, 2 nonexclusive hypotheses are of interest. The first is that the rate of degradation of RNAs stored in P-bodies of wild-type virus-infected cells is RNA type rather than substrate concentration dependent. The second hypothesis, supported by the data presented here, is that in infected cells mRNA is cleaved in polyribosomes. We cannot exclude the first hypothesis and in fact comparison of the rates of degradation or AU-rich mRNAs vs. housekeeping RNAs sustain it. Indeed, the results shown in Fig. 5 indicate that drug-treated wild-type virus-infected cells have fully loaded polyribosomes containing the coding domains up to the AREs, but not the 3′ UTR and poly(A) portions of IEX-1 mRNA and that the accumulation of these polyribosomes is VHS-RNase dependent. The implication of this finding is that VHS-RNase is a component of the polyribosomes, a conclusion that is consistent with reports that VHS-RNase interacts with a translation initiation factor eIF4H, a component of the translation initiation complex eIF4F, and that in the context of infected cells RNA degradation by VHS-RNA is reduced in the absence of eIF4H (22–25).
The finding presented in this article raise questions concerning (i) the fate of the 3′ UTR and linked poly(A) tail, (ii) the impact of the absence of the 3′ UTR on the degradation of the 5′ domain of the AU-rich mRNAs, and (iii) the difference in the rates of degradation of AU-rich vs. stable mRNAs of housekeeping genes. Relevant to these issues are the following.
As noted in the Introduction, the consensus of numerous current studies supports the model that activation of translation involves circularization of mRNA resulting from the interaction of the eIF4F complex (eIF4E/eIF4A/eIF4G/eIF4H) at the cap with PABP at the poly(A) tail (for recent review see ref. 26). The decay of mRNAs is commonly preceded by removal of the poly(A) tail by the deadenylases such as CCR4-NOT or PARN (7, 8). This step is thought to induce degradation by either exosomes or multimeric complexes of 3′-to-5′ exonucleases (9) or decapping that enables the mRNA body to be degraded in 5′-to-3′ direction by exoribonuclease Xrn1 (7, 8). Our studies indicate that in the case of wild-type virus-infected cells, VHS-RNase cleaves in polyribosomes AU-rich mRNAs immediately upstream of AREs. In this instance, the 3′ UTR and linked poly(A) sequences disappear very rapidly, suggesting that exonucleolytic cleavage 5′-to-3′ rapidly degrades this portion of the RNA. In contrast, the truncated RNAs comprising the 5′ UTR and coding sequences linger longer, suggesting that the degradation of this RNA occurs by a different mechanism, most likely by exosome digesting the RNAs in 3′-to-5′ direction.
In light of these considerations, the rapid degradation of housekeeping RNAs resembles that of 3′ UTR and linked poly(A) tails of AU-rich mRNAs. One hypothesis that could explain their rapid disappearance is that VHS-RNase cleaves mRNAs lacking AREs close to the cap. The uncapped RNA would then be rapidly degraded most likely 5′-to-3′ in a manner similar to that of the 3′ UTR and linked poly(A)-tail of AU-rich mRNAs (Fig. 5).
The evidence that VHS-mRNA cleaves mRNAs in polyribosomes presumably by binding to a translation initiation factor raises 2 interesting questions for which there are no satisfactory answers. The first is whether VHS-mRNA effectively degrades RNA that is not bound to eIF4F and ribosomes. Purified VHS-RNase is active on synthetic RNAs, only in the presence of relatively high concentrations of the enzyme (1, 2). In the context of infected cells, VHS-mRNA was reported to be inefficient in degrading mRNA in cells lacking eIF4H (25).
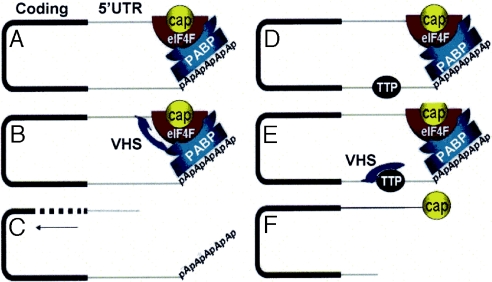
The second question is the mechanism by which VHS specifically targets AREs in the case of AU-rich RNAs and presumably sequences close to the cap in the case of RNAs lacking AREs. A possible explanation is that proteins bound to the AREs guide the VHS-RNase to that site. Interestingly, among the mRNAs induced after infection and that escape degradation is that of tristetrapolin, a protein that binds AREs and sequesters the RNAs in exosomes (27). Tristetrapolin induced in infected cells has been shown to bind VHS RNase (27). It is conceivable, as suggested in the model presented in Fig. 6 that tristetraprolin competes with eIF4F for binding VHS-RNase with the net result that translation is terminated by the cleavage 5′ to ARE rather than the cap structure. This hypothesis, however, remains to be tested.
Fig. 6.
Model for VHS-dependent mRNA degradation in infected cells. (A and D) Current models of actively translated mRNAs show that they are maintained in a closed-loop conformation by the interaction of the translation initiation complex eIF4F bound to the 5′ cap with the PABP at the 3′ end of the RNA. (B and C) VHS-RNase bound to eIF4H, a component of the eIF4F complex, decaps the mRNA (B), thus initiating the degradation of mRNA by the 5′ exonuclease Xrn1 (C). (E) VHS RNase binds preferentially to tristetraprolin (TTP) and cleaves the ARE mRNA 5′ to the AU elements. (F) The product of cleavage inside the 3′ UTR is equivalent to a deadenylated mRNA accessible to the multimeric complex of 3′-to-5′ exonucleases known as the exosome.
Materials and Methods
Cells and Viruses.
The origin and propagation of HeLa cells and the properties of wild-type HSV-1, the ΔUL41 mutant virus R2621, and R2626 in which the gene was restored (R-ΔUL41) have been reported (28).
Immunofluorescence Staining.
HeLa cells grown on 4-well confocal slides (Erie Scientific) were either mock-infected or exposed to 10 or 20 pfu of wild-type or mutant viruses per cell and incubated at 37 °C. The procedures for fixation in 4% paraformaldehyde, blocking with 10% goat serum, and immunostaining has been reported (21). The antibodies used in these studies were the DDX6 (or Rck/p54) rabbit polyclonal antibody (Bethyl Laboratories) and the p70 S6 kinase α mouse monoclonal antibody (H-9; Santa Cruz Biotechnology), both used in a dilution of 1:1,000. The latter was shown (21) to react strongly with Hedls/Ge-1, a robust P-bodies marker (29). Although this antibody also reacts with its intended target p70 S6 kinase, the non-Hedls signal is nuclear and does not interfere with the antibody's ability to clearly recognize P-bodies. The secondary antibodies were Alexa Fluor 594-conjugated goat anti-rabbit and Alexa Fluor 488-conjugated goat anti-mouse (Molecular Probes), diluted 1:1,000 and 1:750 in PBS plus 5% goat serum, respectively. The immunostained cells were examined in a Zeiss confocal microscope equipped with software provided by Zeiss.
Sucrose Gradient Fractionation, RNA Purification, and Northern Analysis.
HeLa cell monolayers (60% confluent) were either mock-infected or infected with HSV-1(F) or ΔUL41 mutant viruses at a multiplicity of 30 pfu per cell. At the end of the incubation time, the inoculum was replaced with fresh medium containing CHX (50 μg/mL; Sigma), and the cells were incubated at 37 °C for 2 additional h. Cells were harvested by trypsinization, and the cell pellets were rinsed twice with ice-cold PBS containing CHX (50 μg/mL) and resuspended in gradient buffer [50 mM Tris·HCl (pH 7.6), 80 mM KCl, 5 mM MgOAc, 2 mM DTT, 1% sucrose] containing 1 unit/μL of RNase inhibitor (RNaseOUT; Invitrogen). Cells were lysed by sonication and lysates were cleared by centrifugation in a microcentrifuge at top speed for 10 min at 4 °C. Protein concentration was assayed with the aid of Bio-Rad protein assay, and the volume of cell extract corresponding to 750 μg of protein was layered on top of an 11-mL linear 10–45% sucrose gradient. Samples were centrifuged at 27,500 rpm in a Beckman SW41 ultracentrifuge rotor for 4 h. Gradients were fractionated from the bottom and RNA was purified with the aid of TRIzol LS reagent (Life Technologies) according to the manufacturer's instructions followed by phenol-chloroform extraction and ethanol precipitation. The entire amount of RNA recovered from each fraction was loaded into a denaturing formaldehyde gel and probed with random hexanucleotide-primed 32P-labeled specific probe after transfer onto a nylon membrane. The probes were the IEX-1 coding region generated as described (30) and a 40-nt oligoprobe (IEX-1 3′ UTR) complementary to a region downstream of the ARE present at the 3′UTR of IEX-1 mRNA that was labeled at the 5′ end with the aid of Kinase Max 5′-end labeling Ambion kit. Probe template for GAPDH was purchased from Ambion. Prehybridization and hybridization steps were performed as reported (30).
Acknowledgments.
This work was supported by National Cancer Institute Grant CA115662.
Footnotes
The authors declare no conflict of interest.
References
- 1.Taddeo B, Zhang W, Roizman B. The UL41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc Natl Acad Sci USA. 2006;103:2827–2832. doi: 10.1073/pnas.0510712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taddeo B, Roizman B. The virion host shutoff protein UL41 of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J Virol. 2006;80:9341–9345. doi: 10.1128/JVI.01008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smiley JR. Herpes simplex virus virion host shutoff protein: Immune evasion mediated by a viral RNase? J Virol. 2004;78:1063–1068. doi: 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esclatine A, Taddeo B, Evans L, Roizman B. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc Natl Acad Sci USA. 2004;101:3603–3608. doi: 10.1073/pnas.0400354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esclatine A, Taddeo B, Roizman B. The UL41 protein of herpes simplex virus mediates selective stabilization or degradation of cellular mRNAs. Proc Natl Acad Sci USA. 2004;101:18165–18170. doi: 10.1073/pnas.0408272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 7.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 8.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 10.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 11.Dompenciel RE, Garnepudi VR, Schoenberg DR. Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in selective destabilization of albumin mRNA. J Biol Chem. 1995;278:6108–6118. doi: 10.1074/jbc.270.11.6108. [DOI] [PubMed] [Google Scholar]
- 12.Yang F, Schoenberg DR. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol Cell. 2004;14:435–445. doi: 10.1016/j.molcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Peng Y, Schoenberg DR. c-Src activates endonuclease-mediated mRNA decay. Mol Cell. 2007;25:779–787. doi: 10.1016/j.molcel.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teixeira D, Sheth U, Vlencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sydiskis RJ, Roizman B. Polysomes and protein synthesis in cells infected with a DNA virus. Science. 1966;153:76–78. doi: 10.1126/science.153.3731.76. [DOI] [PubMed] [Google Scholar]
- 18.Sydiskis RJ, Roizman B. The disaggregation of host polyribosomes in productive and abortive infection with herpes simplex virus. Virology. 1967;32:678–686. doi: 10.1016/0042-6822(67)90043-8. [DOI] [PubMed] [Google Scholar]
- 19.Fenwick L, Walker MJ. Suppression of the synthesis of cellular macromolecules by herpes simplex virus. J Gen Virol. 1978;41:37–51. doi: 10.1099/0022-1317-41-1-37. [DOI] [PubMed] [Google Scholar]
- 20.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J Virol. 1978;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 22.Everly DN, Jr, Feng P, Mian IS, Read GS. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: Genetic and biochemical evidence that Vhs is a nuclease. J Virol. 2002;76:8560–8571. doi: 10.1128/JVI.76.17.8560-8571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng P, Everly DN, Jr, Read GS. mRNA decay during herpesvirus infections: Interaction between a putative viral nuclease and a cellular translation factor. J Virol. 2001;75:10272–10280. doi: 10.1128/JVI.75.21.10272-10280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng P, Everly DN, Jr, Read GS. mRNA decay during herpes simplex virus (HSV) infections: Protein–protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J Virol. 2005;79:9651–9664. doi: 10.1128/JVI.79.15.9651-9664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarma N, Agarwal D, Shiflett LA, Read GS. Small interfering RNAs that deplete the cellular translation factor eIF4H impede mRNA degradation by the virion host shutoff protein of herpes simplex virus. J Virol. 2008;82:6600–6609. doi: 10.1128/JVI.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in Eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esclatine A, Taddeo B, Roizman B. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J Virol. 2004;78:8582–8592. doi: 10.1128/JVI.78.16.8582-8592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon AP, Roizman B. Differentiation of the shutoff of protein synthesis by virion host shutoff and mutant γ134.5 genes of herpes simplex virus 1. Virology. 1997;229:98–105. doi: 10.1006/viro.1996.8425. [DOI] [PubMed] [Google Scholar]
- 29.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Taddeo B, Esclatine A, Zhang W, Roizman B. The stress-inducible immediate-early responsive gene IEX-1 is activated in cells infected with herpes simplex virus 1, but several viral mechanisms, including 3′ degradation of its RNA, preclude expression of the gene. J Virol. 2003;77:6178–6187. doi: 10.1128/JVI.77.11.6178-6187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]