Abstract
We describe a novel scanning optical microscope based on a polarization-modulated nulling ellipsometry. The new microscope employs a combination of scanning mirror and sample translation and thus enables high-throughput label-free detection of biomolecular microarrays with more than 10,000 protein or small molecule targets. For illustration, we show the image of a 2760-spot protein microarray on a functionalized glass slide obtained with such a microscope. The new scanning microscope is also capable of determining, in parallel, the real-time binding kinetics of multiple molecular species under aqueous conditions.
I. INTRODUCTION
Microarrays of surface-immobilized macromolecules or chemical ligands are powerful tools for in vitro analysis of multiple biochemical binding events in a single experiment [1,2]. They are particularly useful for proteomics and glycomics research as the number and variety of structures and functions of proteins, glycans, and their ligands are large. The cooperative effect in a probe-target binding reaction due to the presence of other molecules further increases the complexity of proteomic and glycomic studies. Microarray technology in various forms enables parallel detection of hundreds or thousands of biochemical binding processes, thus promises to shorten the discovery and analysis time. In most cases the microarray technology is associated with some form of fluorescence detection such that molecular probes are labeled with fluorescent tags either extrinsically (e.g. with Alexa 480) or “intrinsically” through genetic engineering (e.g., incorporation of green fluorescent protein or phytochrome proteins) [3–5]. The fluorescence detection is widely used for its superior sensitivity and low background. The choices of fluorescent tags are many, some being nearly free of photo-bleaching.
However fluorescent labeling has its shortcomings. High cost and variation in labeling efficiency are well known. For proteomics and glycomics studies, tagging molecules of interest with fluorescent agents inevitably changes the properties of host molecules. For example, the rate of incorporating deoxyribonucleotide triphosphates (dNTP such as dCTP, dATP, etc.) by high-fidelity polymerases is reduced by more than an order of magnitude when dNTP’s are fluorescently labeled with Cy3 or Cy5 molecules. The binding affinity of a protein to a small molecule or peptide ligand may also be significantly reduced when the protein is labeled by a fluorescent molecule such as Cy3. Often such an effect of fluorescent labeling is not known or characterized a priori due to lack of comparative label-free studies [6,7]. It is thus important and necessary to develop label-free optical methods for high-throughput detection of microarrays.
II. NOVEL SCANNING OPTICAL MICROSCOPE BASED ON DETECTION OF OBLIQUE-INCIDENCE REFELCTIVITY DIFFERENCE
At oblique incidence a surface-bound change such as binding of molecules in liquid phase to surface-immobilized molecular targets alters the ratio of reflection coefficients, rp/rs = tan ψ exp(iδ), for p-polarized (transverse magnetic mode) and s-polarized (transverse electric mode) components of a monochromatic light beam. Optical ellipsometry in one form or another measures Ψ and δ, and thus can be used to study the surface-bound process. Based on the detection of oblique-incidence optical reflectivity difference (OI-RD), - a special form of polarization-modulated nulling ellipsometry, we have developed an optical scanning microscope for label-free, high-throughput detection of microarrays over large areas (equivalent of the field of view). In this report, we describe the optical arrangement and performance of a scanning OI-RD microscope capable of in situ detecting protein, peptide, and small molecule microarrays with thousands of target spots immobilized on conventional microscope glass slides. The microscope readily scans over 10,000 distinct molecular target spots and is capable of real-time kinetic as well as end-point measurements of thousands of binding reactions.
Compared to imaging surface plasmon resonance (SPR) scanners [8–11], an OI-RD microscope does not depend on gold-coated substrates for detection and can have a “field of view” as large as 2-cm×5-cm. We should note that microarrays with up to 10,000 ∼ 15,000 spots on a single solid support (e.g., a 1-in.×3-in. glass slide) are suitable high-throughput platforms for proteomic and glycomic studies or for drug lead optimization. This is because varieties of proteins and glycans and small molecule ligand candidates are in thousands.
Compared to imaging ellipsometers based on the PCSA scheme (polarizer-compensator-sample-analyzer) [12–14], the OI-RD microscope is inherently more sensitive to surface-bound changes (e.g., thickness, density, etc.) by at least one order of magnitude. In a typical PCSA imaging ellipsometer, the phase compensator (C) is fixed while the polarizer (P) and the analyzer (A) are adjusted. The reflected light beam from an illuminated area on the sample surface (S) passes through the analyzer (A) and forms an image on a CCD camera. To yield ψ0 and δ0 maps of the illuminated region before for example a molecular binding reaction takes place on the sample surface, P and A are adjusted until the photocurrents are minimized over the image region on the CCD. During the subsequent binding reaction, the thickness of a surface-bound layer changes by Δd. This causes ψ and δ to deviate from ψ0 and δ0 by Δψ ∼ Δd/λ and Δδ ∼ Δd/λ. However the corresponding change in photocurrent under this off-null condition is proportional to (Δψ)2 and/or (Δδ)2. The quadratic dependence of the off-null photocurrent on already small quantities Δψ and Δδ sets the detection limit of this type of imaging ellipsometers to roughly Δψ ∼ 0.01° and Δδ ∼ 0.01° (i.e., ∼ 0.0002 radians).
The oblique-incidence reflectivity difference (OI-RD) technique is a more sensitive form of ellipsometry [15–17]. It is a polarization-modulated nulling ellipsometry in which the measurable harmonics of modulated photocurrents are directly proportional Δψ and Δδ. This makes an imaging ellipsometer based on OI-RD signals at least an order of magnitude more sensitive than a typical PCSA imaging ellipsometer, namely, with the detection limit to Δψ ∼ 0.001° and Δδ ∼ 0.001°. Such sensitivity is needed for high-throughput affinity detection of low molecular weight analytes. The detection limit of Δδ ∼ 0.001° corresponds to 0.01 nm in detected protein thickness, similar to that of an imaging SPR microscope [8–11].
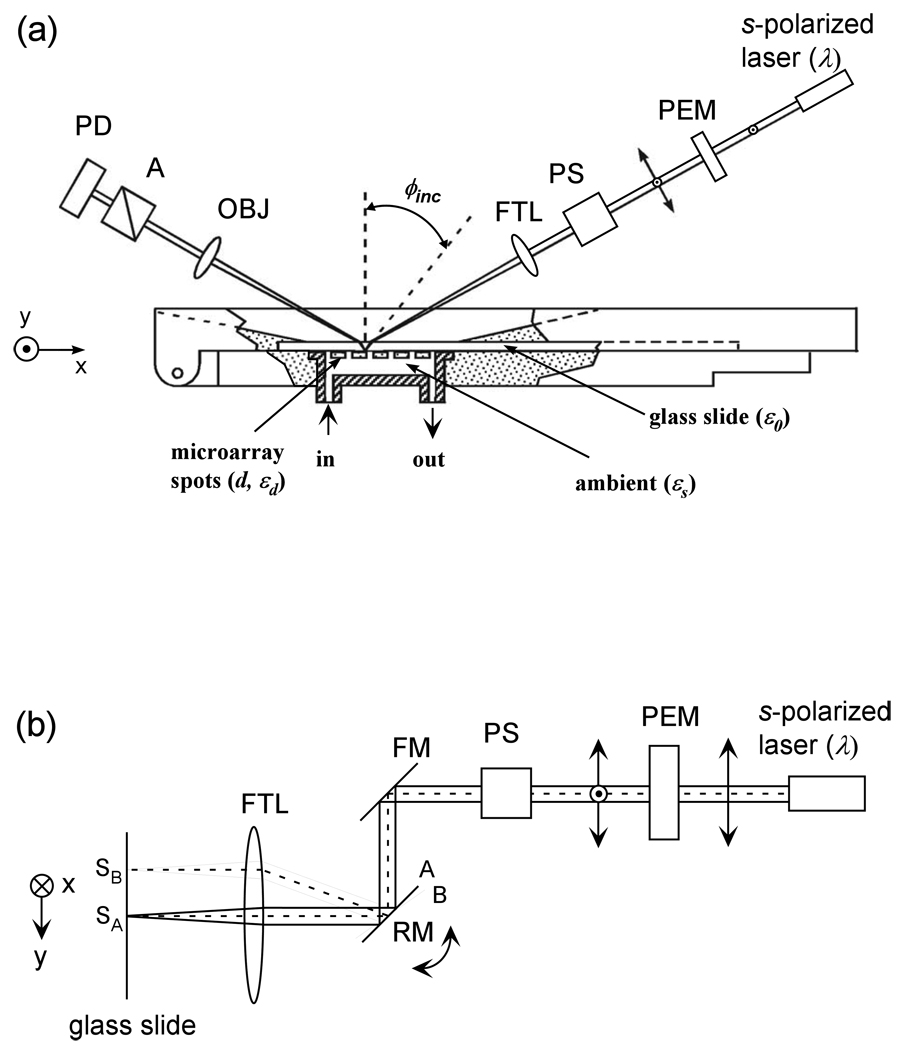
The optical arrangement of our new scanning OI-RD microscope is sketched in Fig. 1. It is a dual-axis mechanical scanning microscope. The key difference between this scanning microscope and our previous OI-RD microscopes is that the sample-holding stage in the present microscope only moves in one direction for x-scan and the y-scan is accomplished with a combination of a scan mirror and an f-theta lens. Rotating a low-mass scan mirror to achieve a y-scan of 20 mm on the focal plane of the f-theta lens takes much less time than moving the sample-holding stage. To illustrate the concept of such a hybrid scanning microscope and the performance in terms of “field of view” and image quality of protein microarrays, we mount a metal-coated mirror on a backlash-free rotation stage (from Physik Instruments, Germany) driven by a custom stepping motor.
Fig. 1.
(a) Top-view of a hybrid scanning OI-RD microscope with a combination of y-scan by a combination of a rotating mirror (RM) and a f-theta lens (FTL) and x-scan by translation of the sample holding stage. The sample is a biomolecular microarray printed on a glass slide. The microarray-bearing surface is in contact with an aqueous solution as a part of a fluidic handling system. (b) Side view of the microscope that illustrates the y-scan. PEM: photoelastic modulator. PS: phase shifter. FM: fixed mirror. OBJ: objective lens. A: analyzer. PD: photodiode detector.
Changes in surface mass density and/or thickness of a thin non-absorbing layer of molecules on a transparent substrate only alter δ significantly. Our OI-RD microscope measures the change in δ as follows. For illumination, we use a 30-mW, linearly polarized, continuous-wave Nd:YAG laser operated at λ = 532 nm. The beam intensity Iinc is stable within ± 0.4% over at least two hours. The beam is initially s-polarized. As shown in Fig. 1(a) it passes through a photo-elastic modulator (PEM, Hinds Instruments, OR) so that the beam emerging after the modulator changes from being s-polarized to p-polarized at a frequency Ω. The PEM is made of a fused quartz slab driven by a transducer at Ω = 50 kHz (a mechanical frequency of the slab). The resultant beam then passes through a phase shifter (PS) that alters the relative phase between the s- and p-polarized components by a variable amount Φps for nulling ellipsometry. The phase-shifter here is a rotatable multi-order crystalline quartz half-wave plate (HWP) for λ = 532 nm. It is mounted on a precision rotation stage with the rotation axis coinciding with the fast axis (FA) of the waveplate. We align the fast axis of the waveplate parallel to the s-polarized component of the laser beam and the slow axis parallel to the p-polarized component of the beam. By rotating the waveplate about the fast axis, we variably change the path length of the laser beam and in turn the phase between the two polarization components. We then use an assembly of a rotating mirror (RM) and a f-theta lens (FTL) to focus the beam into a spot (∼ 10 µm in diameter) on the microarray-bearing surface of a glass slide (with optical dielectric constant ε0) at incident angle ϕinc = 34.7°. The microarray-bearing surface is in contact with an aqueous solution (characterized by optical dielectric constant εs). The reflected beam from the illuminated spot passes through an analyzer (A) with its transmission axis set at θA (45° in the present study) from p-polarization. The beam is then imaged with an objective lens onto a single-element Si PIN photodiode operated in photo-conductive mode. The photodiode converts the light intensity incident onto its surface, I(t), into a photoelectric current. Through a current-to-voltage converter with a gain resistor R = 5×104 Ohm, the photocurrent is detected by an SR830 Digital Lock-in Amplifier (Stanford Research Systems, CA). The light intensity I(t) consists of harmonics of polarization modulation frequency Ω. In a typical OI-RD experiment, we measure the first- and second-harmonics. The amplitude of the first-harmonic is given by I(Ω)= Iinc|rprs|sin 2θAsin(ηsys+δ+Φps) [16]. The second-harmonic is given by I(2Ω)=Iinc(|rp|2cos2θA–|rs|2sin2θA) [16]. ηsys is the phase difference between the p-polarized and s-polarized components of the light beam contributed by optical components in the beam path other than the sample surface. δ is the phase difference due to the reflection from the microarray-bearing glass surface alone.
Let δ0 be the value for the bare glass slide surface. We adjust Φps until ηsys+δ0+Φps=0 on the bare surface so that I(Ω)= 0. When a thin layer of molecules (characterized by thickness d and optical dielectric constant εd) is added to the bare surface or when the region of the surface covered with microarray features (again characterized by a thickness d and an optical dielectric constant εd) is moved under the illumination optics, the first-harmonic becomes finite, namely, I(Ω)= Iinc|rprs|sin 2θAsin(δ–δ0). We determine Iinc|rprs|sin 2θA≡IMAX(Ω) separately by adjusting Φps until I(Ω)= Iinc|rprs|sin 2θAsin(ηsys+δ+Φps) reaches its maximum. The ratio of I(Ω)=Iinc|rprs|sin 2θAsin(δ–δ0) to IMAX(Ω) yields sin(δ–δ0)≈δ–δ0 ≡ Δδ.
When the thickness d of the interrogated microarray feature or the added molecular layer is much less than the wavelength λ, Δδ is related to the properties of the surface-bound layer by [15–17]
| (1) |
Here Θ is the fractional surface coverage of the molecules, defined as the area covered by the molecules divided by the total area available. Θ can be determined experimentally by exposing the microarray-covered glass surface to a solution of bovine serum albumin (BSA). The additional change in Δδ from the originally Θ-covered region is proportional to 1 - Θ while the signal from the originally uncovered region is proportional to 1. From the two values of Δδ, one extracts Θ. With our current scan microscope, we can detect a submonolayer of protein molecules such as bovine serum albumin at a level as small as Θ = 0.005 or 1/200 of a monolayer.
From Eq. (1) it is clear that the first-harmonic I(Ω), after the initial nulling step, increases linearly with d/λ. In contrast, when the ambient, the substrate, and the molecular layer are all transparent to the laser beam (as in our present study), I(2Ω) changes quadratically with d/λ and is thus negligible [17].
An image in Δδ of a biomolecular microarray on a glass slide is obtained by scanning the illumination beam in y-direction with the rotating mirror as shown in Fig. 1(b) and moving the sample stage in x-direction. The incremental angle for the rotating mirror driven by a stepping motor is as small as 42.5 μ-radians in our present microscope. Together with the f-theta lens, this gives a rise to a linear step of 4.68 µm in y-direction on the sample surface (on the focal plane of the f-theta lens). In actual scan, we use a step size of 18.7 µm in order to reduce the scan time and to match the beam diameter on the sample surface (∼ 10 µm).
III. EXPERIMENTAL RESULTS
For target microarrays, we use rabbit immunoglobulin-G (IgG), human IgG, mouse IgG, bovine serum albumin (BSA) (all purchased from Jackson ImmunoResearch Laboratories), and biotin-BSA conjugates that we have synthesized in house [18]. The choice of biotin–BSA conjugates as a target is motivated by our intent to apply the OI-RD microscope to high-throughput screening small-molecule libraries for protein ligands. Our strategy is to conjugate small molecules to carrier proteins such as BSA through a flexible linker and then print small-molecule-BSA conjugate microarrays on an epoxy-functionalized glass surface. The conjugate binds to the epoxy groups on the glass surface through amine residues on the surface of BSA, leaving the small-molecule target accessible to protein probes in subsequent analysis. In our case more than one biotin molecule is attached to one BSA molecule. All the target molecules, in 1×PBS (phosphate buffered solution) at same concentration of 7.6 µM, are printed on an epoxy-functionalized glass slide (CEL Associates, TX) with a contact-printing robot (OmniGrid 100, Genomic Solutions, MI). The center-to-center separation between neighboring spots is 200 µm. The spots are more or less circular with an average diameter of ∼100 µm. After printing, the microarray-bearing slide is washed with 1×PBS (pH = 7.4) to remove unbound protein targets. The slide is then assembled into a fluid-handling system filled with 1×PBS so that the microarray is examined in situ in either the buffer solution or a probe solution during subsequent reactions with the scanning OI-RD microscope. After the excess printed target materials are washed away, the microarray spots are optically flat on the glass substrate. There are no observable edge effects when these spots are imaged with our scanning microscope.
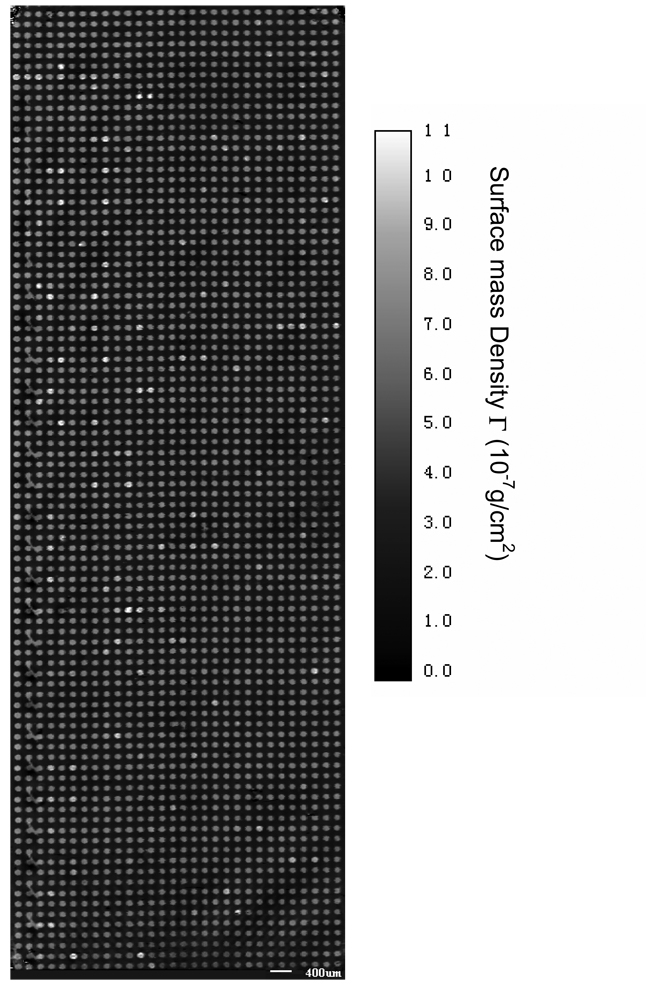
We first demonstrate that the new scanning microscope is capable of imaging large microarrays under aqueous conditions. Fig. 2 shows the image of a 2760-spot bovine serum albumin (BSA) microarray immersed in a 1×PBS buffer solution after the washing step. The microarray covers an area of 1.83 cm×0.64 cm, total of ∼ 1cm2. The scan step is 17.8 µm in x-direction and 18.7 µm in y-direction, the dimension of one image pixel is 17.8 µm × 18.7 µm. If we reduce the step size by a factor of 2, this OI-RD microscope can scan a protein microarray 4 times as dense over the same area, with over 11,000 spots. In Fig. 2 we have converted the optical signal Δδ to the surface mass density Γ using the following procedure. Using the fact that ΓBSA/ρBSA= ΘBSAdBSA, we rewrite Eq. (1) into
| (2) |
By rearranging Eq. (2) and putting in the values of ε0=2.307 for the glass slide, εs=1.788 for the aqueous buffer solution, εd= εBSA=2.031 for BSA in solution, and ρBSA = 1.35 gm/cm3, we arrive at
| (3) |
From Eq. (3) we covert the experimental Δδ to the surface mass density of the BSA layer. If we model a bovine serum albumin molecule as an ellipsoid with major axes being 4 nm × 4 nm × 14 nm [19], a fully packed monolayer of BSA molecules with their long axes parallel to the surface (side-on geometry) will have a surface mass density of 1.9 × 10−7 g/cm2. A full monolayer of BSA molecules with the long axes perpendicular to the surface (end-on geometry) will have a surface mass density of 6.8 × 10−7 g/cm2. Fig. 2 shows that after the excess protein targets are washed off, nearly all printed spots consist of a full monolayer of BSA in end-on geometry.
Fig. 2.
Image of a 2760-spot BSA microarray acquired with the hybrid scanning OI-RD microscope as illustrated in Fig. 1. The image area is 18.3 mm×6.4 mm and the pixel dimension is 17.8 µm × 18.7 µm. The image contrast has been converted from the optical signal to the surface mass density of surface-bound BSA molecules.
The image of a nearly 3000-spot BSA microarray takes 69 minutes to acquire. Out of the total image acquisition time, only 10 minutes are used for signal averaging at 2 ms per “pixel”, while the remaining 59 minutes are spent to rotate the scan mirror that currently is driven by a stepping motor. By using a galvanometer-based scan mirror, the time needed to rotate the mirror in order to acquire the same image can be reduced to less than 3 minutes. This means that the scan time for an image as shown in Fig. 2 can be acquired in less than 15 minutes.
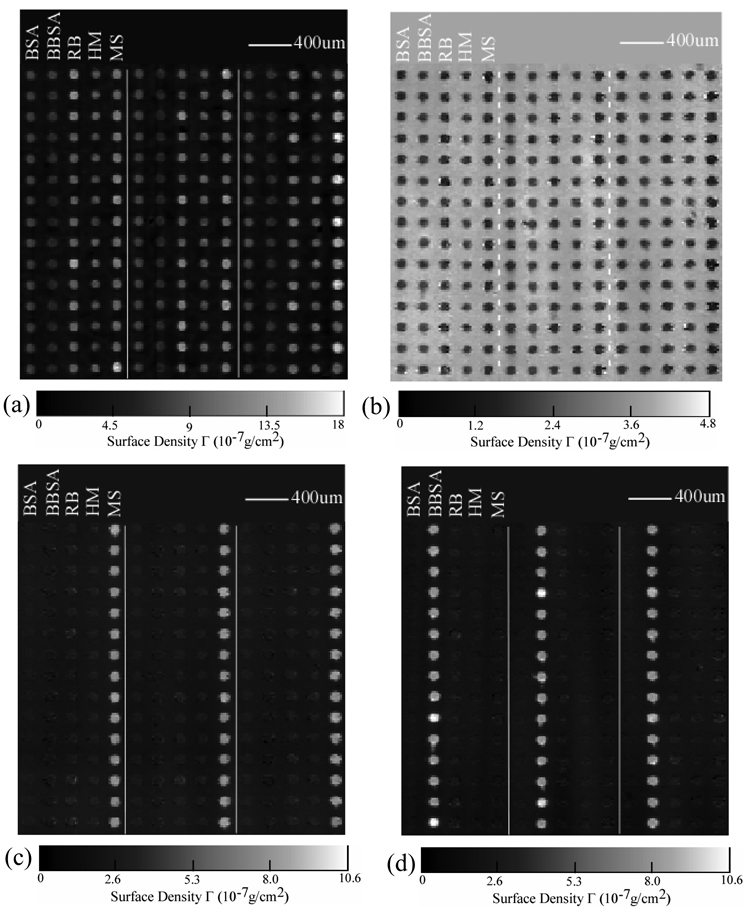
We subsequently use the new hybrid scanning OI-RD microscope to detect two known protein-protein and protein-small molecule binding reactions on a separately prepared microarray. The microarray consists of rabbit IgG (RB), human IgG (HM), and mouse IgG (MS) as antigen targets; biotin-BSA conjugate as a small-molecule target, and unmodified BSA as a control target. The microarray is printed with each target in 1×PBS solution at a concentration of 7.6 µM. Before the optical scan, the printed microarray is washed to remove the excess targets. In Fig. 3(a) we show the image of the target microarray obtained with the microarray immersed in 1×PBS. Each column consists of one target and has 90 spots (only 15 spots are shown). The sections in the middle and on the right are duplicates. The spot size and the surface mass density are uniform for each target; they do vary for different targets presumably due to the detailed make-up of surface residues of the printed proteins. The average surface mass densities of the immobilized BSA and biotin-BSA targets is ΓBSA= 5.8×10−7 g/cm2 (calculated using Eq. (3) from the optical signals Δδ). The surface mass densities of the immobilized IgG targets (as shown in Fig, 3(a)) are also converted from the respective optical signals using Eq. (3) since the optical dielectric constant and volume mass density for IgG are roughly the same as BSA [20]. The average surface mass density of the IgG targets is ΓIgG= 11.5×10−7 g/cm2. Again if we model an IgG molecule as an ellipsoid with three major axes measuring 4.4 nm × 4.4 nm × 23.5 nm , a full monolayer of IgG molecules with their long axes perpendicular to the surface will a surface mass density of 13 × 10−7 g/cm2. This suggests that both IgG and BSA targets bind to epoxy-functionalized surface in end-on geometry. It is reasonable to expect that half of the immobilized IgG targets have the Fc fragments pointing away from the surface and thus accessible to antibodies against these fragments.
Fig. 3.
Images of a protein microarray after a sequence of post-printing treatment. The microarray consists of mouse IgG (MS), human IgG (HM), rabbit IgG (RB), biotin-BSA (BBSA), and BSA as targets printed in columns. Dashed lines separate three microarray replicates. (a) Image in surface mass density after washing; (b) mass density change due to BSA-blocking treatment; (c) mass density after reaction with goat polyclonal antibody against the Fc domain of mouse IgG;. (d) mass density change after reaction with streptavidin.
Before reactions, we flow 0.5mg/ml BSA (i.e., 7.6 µM) in 1×PBS into the flow cell so that free epoxy groups on the microarray-bearing surface are reacted with the BSA molecules. This “blocking” step prevents non-specific binding during subsequent reactions. We take an image after the blocking step (not shown here). The differential image (as shown in Fig. 3(b)) taken by subtracting the image after blocking from the image shown in Fig. 3(a) reveals the coverage Θ of the printed targets is uniform and full. It means that the variation in Δδ or in surface mass density from target to target in Fig. 3(a) results from the variation in mean thickness of the targets. The mean thickness depends on the orientation of the targets.
We expose the microarray first with unlabelled goat antibody against the Fc fragments of the mouse IgG at a probe concentration of 0.01mg/ml (i.e., 0.067 µM.) In Fig. 3(c) we show the difference between the image taken after the reaction and the image taken after the BSA blocking step. Only the mouse IgG targets show the strong evidence of binding with the goat anti-mouse IgG probe. The average change in surface mass density on the mouse IgG targets is ΔΓIgG= 5.3×10−7 g/cm2. This is one half of a full monolayer of goat IgG probe in end-on geometry. Since roughly one half of the mouse IgG targets in end-on geometry has the Fc fragments accessible to the probe, this means that all accessible mouse IgG targets have reacted with the probe. Fig. 3(c) also reveals noticeable cross-reactivity of goat anti-mouse IgG with surface-immobilized rabbit IgG and human IgG targets. The net change in surface mass density due to the cross-reacted goat IgG is 5 times smaller than on the mouse IgG targets. There is no evidence of binding for goat anti-mouse IgG probe on either biotin-BSA conjugate or BSA control targets.
We next subject the microarray to reaction with streptavidin at a probe concentration of 0.17 µM. Fig. 3(d) shows the differential image by subtracting the image taken before the reaction from the image after the reaction. Only on biotin-BSA conjugates we observe uniform changes in Δδ or in surface mass density while no changes at all on other targets. This indicates, not surprisingly, that the streptavidin molecules bind specifically to biotin targets that are conjugated to BSA through linker molecules. The surface mass density of captured streptavidin molecules by biotin-BSA conjugates is ΔΓ = 6.9×10−7 g/cm2. It is more than the surface mass density expected of a fully packed monolayer of streptavidin, which is ∼ Γstreptavidin= 4×10−7 g/cm2. This can be understood as follows: the area number density of biotin molecules is higher than that of a full monolayer of streptavidin; the linker molecules make the “excess” biotin targets accessible to streptavidin molecules beyond one fully packed monolayer.
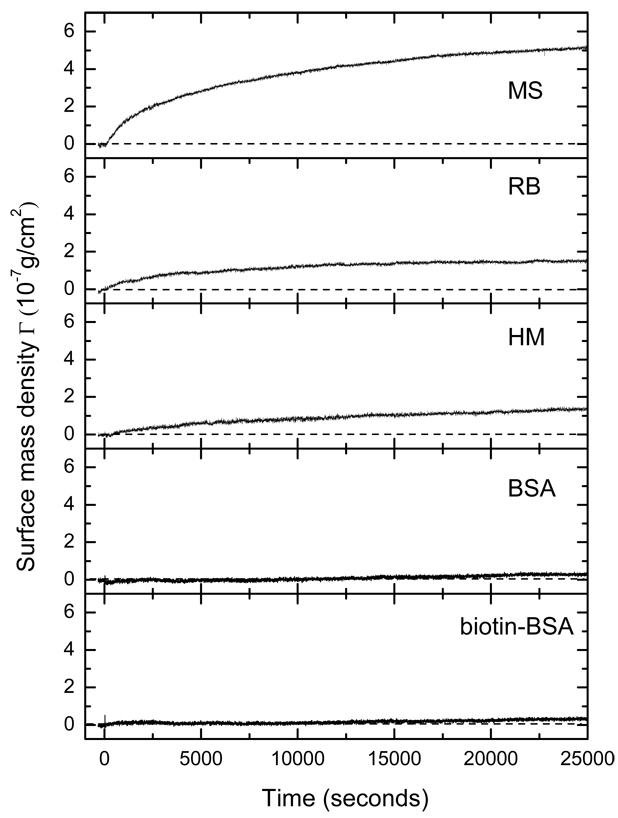
In addition to endpoint measurements, the present high-throughput OI-RD microscope is capable of monitoring reaction kinetics at multiple targets. In this mode of operation, we choose one pixel from each of the five targets and one pixel from the unprinted region as the reference “channel” and only measure Δδ from these pixels in sequence. By repeating the measurement sequence, we obtain the surface mass density change in real time on the selected targets. We minimize the effect of any drift in ηsys in the microscope by subtracting Δδ of the reference channel from Δδ of the targets. Fig. 4 shows the surface mass density change vs. time on five targets measured at 1/3 Hz (one time point every 3 seconds). The solution of goat anti-mouse IgG at a concentration of 0.067 µM is quickly introduced into the sample fluidic system at t = 0. The signal on the mouse IgG target increases until it reaches the equilibrium value ΔΓ =5.3×10−7 g/cm2 as also displayed in Fig. 3(c). The signal on biotin-BSA conjugate and BSA control targets remain unchanged. On human IgG (HM) and rabbit IgG (RB) targets the signals change noticeably but by a significantly smaller amount, ΔΓ =1.5×10−7 g/cm2, as also shown in Fig. 3(c). Other than confirming the end-point measurement shown in Fig. 3(c), the real-time data reveal the kinetics of the observed binding reactions.
Fig. 4.
Simultaneous measurement of the binding reaction in real time of goat polyclonal antibody against mouse IgG (the Fc domain) with a protein microarray consisting of mouse IgG, human IgG, rabbit IgG, biotin-BSA conjugate, and BSA as surface-immobilized target using the hybrid OI-RD microscope as illustrated in Fig. 1.
IV. CONCLUSION
In summary, we have developed a new high-speed oblique-incidence reflectivity difference (OI-RD) microscope that can detect biomolecular microarrays with 2760 features in 1-cm2 area. This scanning microscope uses a combination of a rotating scan mirror for y-scan and a single-axis translation for x-scan. By increasing the scan field of view or the density of microarray spots by a factor of 4, this microscope should be able to detect biomolecular microarrays with over 10,000 features. For microarray spots with 100-µm diameters, this new microscope can acquire an end-point image of a 2760-spot microarray in less than 15 minutes with a pixel dimension of 20 µm × 20 µm. The new scanning OI-RD microscope is also capable of following the kinetics of molecular interactions on biomolecular microarrays in a high-throughput fashion.
ACKNOWLEDGEMENTS
This work was supported by NIH under NIH-R01-HG003827-02.
References
- 1.Kodadek T. Chem. & Biol. 2001;8:105. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 2.MacBeath G. Nature Genet. 2002;32:526. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 3.Schena M. Microarray Analysis. Hoboken: Wiley; 2003. [Google Scholar]
- 4.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Synder M. Science. 2000;293:2101. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P. Nature Biotech. 2002;20:225. [Google Scholar]
- 6.Shliom O, Huang M, Sachais B, Kuo A, Weisel JW, Nagaswami C, Nassar T, Bdeir K, Hiss E, Gawlak S, Harris S, Mazar A, Higazi AA. J. Biol. Chem. 2000;275:24304. doi: 10.1074/jbc.M002024200. [DOI] [PubMed] [Google Scholar]
- 7.Karlsson R. J. Mol. Recognit. 2004;17:151. doi: 10.1002/jmr.660. [DOI] [PubMed] [Google Scholar]
- 8.Singh BK, Hillier AC. Anal. Chem. 2006;78:2009. doi: 10.1021/ac0519209. [DOI] [PubMed] [Google Scholar]
- 9.Boozer C, Kim G, Cong S, Guan H, Londergan T. Curr. Opin. Biotechnol. 2006;17:400. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Shumaker-Parry JS, Campbell CT. Anal. Chem. 2004;76:907. doi: 10.1021/ac034962a. [DOI] [PubMed] [Google Scholar]
- 11.Usui-Aoki K, Shimada K, Nagano M, Kawai M, Koga H. Proteomics. 2005;5:2396. doi: 10.1002/pmic.200401171. [DOI] [PubMed] [Google Scholar]
- 12.Azzam RMA, Bashara NM. Ellipsometry and Polarized Light. 1987 Elsevier Science. [Google Scholar]
- 13.Jin G, Jansson R, Arwin H. Rev. Sci. Instrum. 1996;67:2930. [Google Scholar]
- 14.Wang ZH, Jin G. Anal. Chem. 2003;75:6119. doi: 10.1021/ac0347258. [DOI] [PubMed] [Google Scholar]
- 15.Landry JP, Zhu XD, Gregg JP. Opt. Lett. 2004;29:581. doi: 10.1364/ol.29.000581. [DOI] [PubMed] [Google Scholar]
- 16.Thomas P, Nabighian E, Bartelt MC, Fong CY, Zhu XD. Appl. Phys. A. 2004;79:131. [Google Scholar]
- 17.Zhu XD, Landry JP, Shun YS, Gregg JP, Lam KS, Guo XW. Appl. Opt. 2007;46:1890. doi: 10.1364/ao.46.001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Q, Miyamoto S, Lam KS. Mol. Diversity. 2004;8:301. doi: 10.1023/b:modi.0000036238.42200.3d. [DOI] [PubMed] [Google Scholar]
- 19.Wright AK, Thompson MR. Biophys. J. 1975;15:137. doi: 10.1016/s0006-3495(75)85797-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oncley JL, Scatchard G, Brown A. J. Phys. Colloid Chem. 1947;51:184. doi: 10.1021/j150451a014. [DOI] [PubMed] [Google Scholar]