Abstract
Screening of Salmonella mutants for the ability to increase β-lactam resistance has led to the identification of a mutation in hns, which codes for the histone-like nucleoid structuring protein (H-NS). In this study, we report that H-NS modulates multidrug resistance through repression of the genes that encode the AcrEF multidrug efflux pump in Salmonella enterica serovar Typhimurium.
Bacterial multidrug efflux pumps confer resistance to a wide range of antibiotics, dyes, and biocides. In gram-negative bacteria, pumps belonging to the resistance-nodulation-cell division family are especially effective in conferring resistance (14). For example, high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204 has been shown to be largely attributable to multiple target gene mutations and active efflux by the AcrAB-TolC system (3, 4). Recent studies have shown that S. enterica has nine functional drug efflux pumps (15). Because many of these multidrug transporters have overlapping substrate spectra, it is intriguing that bacteria, with their economically organized genomes, harbor such large sets of multidrug efflux genes. The key to understanding how bacteria utilize these multiple efflux pumps lies in the regulation of pump expression. Currently available data show that multidrug efflux pumps are often expressed under precise and elaborate transcriptional control (16). Expression of acrAB, which encodes the major AcrAB efflux pump, is subject to multiple levels of regulation. In S. enterica, it was reported that mutation in the acrR repressor contributes to the overexpression of acrAB and increases resistance to multiple drugs (20). Recent reports showed that the RamA activator (2, 13, 22) and RamR (1) repressor are also involved in the regulation of acrAB.
However, few data are available on the regulation of S. enterica multidrug efflux genes other than acrAB. We therefore screened for S. enterica mutations that increase multidrug resistance levels in this organism. The S. enterica strains used in this study were derived from wild-type strain ATCC 14028s (9). We used a host strain (NKS175) lacking a functional acrB gene in the screening in order to identify regulatory elements involved in the expression of other multidrug resistance systems. NKS175 was subjected to transposon mutagenesis with the EZ-Tn5<R6Kγori/KAN-2>Tnp Transposome kit (Epicentre) according to the manufacturer's instructions. Briefly, the transposome complex was electroporated into NKS175 and then approximately 10,000 colonies were screened. Cells were plated on LB agar medium (21) containing 25 μg/ml kanamycin and inhibitory concentrations of various drugs. In one experiment, the medium contained 8 μg/ml oxacillin, which had an MIC of 2 μg/ml against NKS175. When a mutant colony that grew on this medium was purified and reexamined, we indeed found an eightfold increase in the oxacillin MIC (16 μg/ml) against the transposon insertion mutant (data not shown).
Sequencing determined that the transposon was inserted into the coding sequence of hns. H-NS belongs to a family of small, abundant, nucleoid-associated proteins of gram-negative bacteria that have the ability to bind DNA, and it has been shown to act as a transcriptional repressor (7). It seemed possible that deletion of hns might be causing the transcriptional activation of genes involved in oxacillin resistance. To test whether deletion of hns confers oxacillin resistance on the S. enterica NKS175 strain, a nonpolar deletion was made in the hns gene with the lambda Red system (5). The oxacillin MICs for cells lacking hns (NKS291) was eight times higher than that for NKS175 cells (16 versus 2 μg/ml) (Table 1), suggesting that the deletion of the H-NS regulator indeed conferred oxacillin resistance on S. enterica. We investigated the effect of hns deletion on the susceptibility of S. enterica to other toxic compounds. Various drugs were tested, including common substrates of multidrug efflux pumps, and we found that hns deletion increased the resistance of the NKS175 strain to cloxacillin, nafcillin, cefamandole, methylene blue, and rhodamine 6G (Table 1). These results indicate that the deletion of H-NS induces multidrug resistance in S. enterica. It has been reported that many multidrug efflux systems need the membrane channel TolC for their function (8, 10, 17). To determine whether H-NS-mediated multidrug resistance is attributable to the TolC-dependent drug efflux pump(s), we investigated the effect of tolC deletion on the drug resistance of the hns deletion-containing cells. MICs against NKS419 were the same as those against NKS174, indicating that the tolC deletion completely inhibited H-NS-mediated multidrug resistance (Table 1). This result indicates that H-NS-mediated multidrug resistance is attributable to increased functioning of a TolC-dependent drug efflux pump.
TABLE 1.
Susceptibility of Salmonella strains to β-lactams and toxic compounds
| Strain | Genotype | MIC (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| OXA | CLOX | NAF | FAM | MB | R6G | ||
| ATCC 14028s | Wild type | 512 | 512 | >512 | 1 | >512 | >512 |
| NKS175 | ΔacrB | 2 | 4 | 8 | 0.063 | 16 | 8 |
| NKS291 | ΔacrB hns | 16 | 128 | 64 | 0.25 | 512 | 32 |
| NKS174 | ΔtolC | 0.5 | 0.5 | 1 | 0.063 | 16 | 8 |
| NKS419 | ΔtolC hns | 0.5 | 0.5 | 1 | 0.063 | 16 | 8 |
| NKS176 | ΔacrEF | 512 | 512 | >512 | 0.5 | >512 | >512 |
| NKS422 | ΔacrB acrEF | 1 | 4 | 4 | 0.063 | 16 | 8 |
| NKS416 | ΔacrB acrEF hns | 1 | 4 | 4 | 0.063 | 16 | 8 |
OXA, oxacillin; CLOX, cloxacillin; NAF, nafcillin; FAM, cefamandole; MB, methylene blue; R6G, rhodamine 6G. Values in bold are larger than those of the ΔacrB strain. MIC determinations were repeated at least three times.
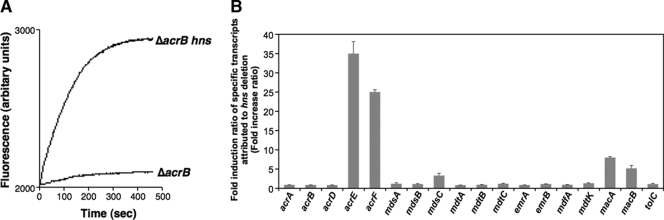
We investigated drug efflux activity in the hns-deficient mutant. The rhodamine 6G efflux activities of S. enterica NKS175 (ΔacrB) and NKS291 (ΔacrB hns) were measured as described previously (18). Briefly, exponential-phase cultures were harvested and washed with 100 mM potassium phosphate buffer (pH 7.5) and then the cells were incubated with 1 μM rhodamine 6G and 40 μM carbonyl cyanide m-chlorophenylhydrazone at 37°C for 1 h. These cells were then resuspended in the same buffer containing 25 mM glucose and subjected to fluorescence measurement. As shown in Fig. 1A, rapid efflux of rhodamine 6G from ΔacrB hns cells was observed as an increase in fluorescence intensity. On the other hand, no significant efflux was observed in ΔacrB mutant cells. This result clearly indicates that hns deletion induces an active efflux system.
FIG. 1.
Effect of hns deletion on efflux activity and of drug efflux gene expression levels. (A) Rhodamine 6G efflux activity in S. enterica cells. Energy-starved cells of S. enterica NKS175 (ΔacrB) and NKS291 (ΔacrB hns) were loaded with rhodamine 6G, and efflux activities were monitored with an F-2000 fluorescence spectrophotometer (Hitachi, Japan). Rhodamine 6G efflux was measured with excitation at 529 nm and emission at 553 nm. (B) Changes in drug efflux and outer membrane channel gene expression. The mRNA transcript level was determined by quantitative real-time PCR. The n-fold change ratio was calculated by dividing the expression level of the gene in the NKS285 (Δhns mutant) strain by that in the ATCC 14028s (wild-type) strain. The data correspond to mean values of three independent experiments. Error bars represent the standard deviation.
It has been reported that S. enterica has nine drug efflux systems, including AcrAB, AcrD, AcrEF, EmrAB, MacAB, MdfA, MdsABC, MdtABC, and MdtK (15). In order to determine which drug efflux pump shows increased expression when hns is deleted, we used quantitative reverse transcription-PCR to investigate changes in the levels of drug efflux gene mRNAs dependent on hns deletion. Total RNAs were isolated from exponential-phase cultures of the wild-type and Δhns mutant strains, and cDNA samples were synthesized with TaqMan reverse transcription reagents (PE Applied Biosystems) with random hexamers as primers. Real-time PCR of the cDNAs was performed with each specific primer pair and SYBR green PCR Master Mix (PE Applied Biosystems) as described previously (15). The expression levels of drug efflux pump genes and tolC in the Δhns mutant were compared with those in the wild-type strain. The results are shown in Fig. 1B. Expression of acrE and acrF was significantly increased (more than 10-fold). The expression of mdsC, macA, and macB was slightly increased in the Δhns mutant.
In order to determine whether multidrug resistance mediated by hns deletion is due to increased expression of acrEF, we investigated the effects of deleting these genes on H-NS-mediated multidrug resistance. In the ΔacrB acrEF mutant strain, deletion of hns conferred no drug resistance (Table 1), whereas it conferred multidrug resistance on the ΔacrB mutant strain. Together, these data indicate that the multidrug resistance conferred by H-NS deletion is due to derepression of the acrEF multidrug efflux genes.
In this study, we performed a genome-wide search for a regulator of multidrug resistance in S. enterica by random insertion and found that H-NS downregulates the expression of acrEF. We initially found by random insertion that the mutation in hns conferred oxacillin resistance on the NKS175 strain. Then we investigated the susceptibility of the strain lacking hns to various drugs, including common substrates of multidrug efflux pumps, and found that H-NS modulates S. enterica resistance to oxacillin, cloxacillin, nafcillin, cefamandole, methylene blue, and rhodamine 6G (Table 1). The DNA-binding protein H-NS is a global transcriptional regulator that specifically silences horizontally acquired virulence genes in Salmonella (12). This process is achieved through interactions between H-NS and AT-rich DNA regions with low specificity (6). For the intergenic region located upstream of acrE in which H-NS was found to repress expression to 1/35 of the original level, the average GC content is 31.7%, whereas the average GC content of the entire S. enterica serovar Typhimurium LT2 genome is 52.2% (11). It should be noted that this intergenic region harbors a 100-bp AT-rich DNA region located 295 to 196 bp upstream of the start codon of acrE, which has a GC content of 16.0%. The AcrF efflux pump is highly homologous to AcrB in S. enterica with an 81.4% amino acid identity and similar substrate specificities (15). We found that acrEF is repressed by H-NS, whereas acrAB is not and is constitutively expressed. Because overexpression of acrEF confers an effect similar to that of acrAB (15), H-NS represses acrEF and might confer a fitness advantage on this organism. AcrEF might be required for drug resistance in S. enterica when AcrAB does not function. Indeed, in a fluoroquinolone-resistant mutant selected from S. enterica serovar Typhimurium DT204 acrB mutant cells, an insertion sequence (IS1 or IS10) was found integrated upstream of the acrEF operon, which increased the expression level of acrEF (19). Further investigation of the regulation of multidrug efflux systems in several natural environments such as those inside hosts is needed to elucidate the biological significance of their regulatory networks.
Acknowledgments
This research was supported by the Astellas Foundation for Research on Metabolic Disorders; the Kanae Foundation for the Promotion of Medical Science; the Mochida Memorial Foundation for Medical and Pharmaceutical Research; the Uehara Memorial Foundation; the Asahi Glass Foundation; the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation; a Grant-in-Aid for Scientific Research on Priority Areas, Matrix of Infection Phenomena, from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; a Grant-in-Aid for Young Scientists (S) from the Japan Society for the Promotion of Science; and PRESTO, Japan Science and Technology Agency, Japan.
Footnotes
Published ahead of print on 8 June 2009.
REFERENCES
- 1.Abouzeed, Y. M., S. Baucheron, and A. Cloeckaert. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52:2428-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, A. M., I. T. Paulsen, and L. J. Piddock. 2008. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob. Agents Chemother. 52:3604-3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baucheron, S., E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of TolC and parC mutation in high-level fluoroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J. Antimicrob. Chemother. 53:657-659. [DOI] [PubMed] [Google Scholar]
- 4.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorman, C. J. 2007. H-NS, the genome sentinel. Nat. Rev. Microbiol. 5:157-161. [DOI] [PubMed] [Google Scholar]
- 7.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 8.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 12.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 13.Nikaido, E., A. Yamaguchi, and K. Nishino. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 283:24245-24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishino, K., T. Latifi, and E. A. Groisman. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:126-141. [DOI] [PubMed] [Google Scholar]
- 16.Nishino, K., E. Nikaido, and A. Yamaguchi. 2009. Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 1794:834-843. [DOI] [PubMed] [Google Scholar]
- 17.Nishino, K., J. Yamada, H. Hirakawa, T. Hirata, and A. Yamaguchi. 2003. Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to β-lactams. Antimicrob. Agents Chemother. 47:3030-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olliver, A., M. Valle, E. Chaslus-Dancla, and A. Cloeckaert. 2005. Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar Typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob. Agents Chemother. 49:289-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olliver, A., M. Valle, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 238:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.van der Straaten, T., R. Janssen, D. J. Mevius, and J. T. van Dissel. 2004. Salmonella gene rma (ramA) and multiple-drug-resistant Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 48:2292-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]