Abstract
Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae have become more common in the United States and throughout the world. We used pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) to examine the molecular epidemiology of KPC-producing K. pneumoniae isolates sent to the Centers for Disease Control and Prevention (CDC) for reference testing from 1996 to 2008. A dominant strain, sequence type 258 (ST 258), was found and likely accounts for 70% of the CDC's K. pneumoniae PFGE database. Isolates with PFGE patterns related to ST 258 were identified in 10 of the 19 U.S. states currently reporting KPC-producing K. pneumoniae, in addition to one isolate from Israel. KPC subtyping and analysis of the surrounding genetic environment were subsequently performed on 23 representative isolates. Thirteen isolates identified as ST 258 possessed either blaKPC-2 or blaKPC-3 and some variability in the Tn4401 element upstream of the blaKPC gene. Escherichia coli DH10B was successfully transformed by electroporation with KPC-encoding plasmid DNA from 20 of the 23 isolates. Restriction analysis of plasmid DNA prepared from transformants revealed a diversity of band patterns, suggesting the presence of different plasmids harboring the blaKPC gene, even among isolates of the same ST.
Klebsiella pneumoniae carbapenemase (KPC) is an Ambler class A β-lactamase that confers resistance to all β-lactam agents, including carbapenems, cephalosporins, penicillins, and the monobactam aztreonam (20). KPC was first reported in a K. pneumoniae isolate from North Carolina in 1996 (32). Although this enzyme has been found primarily in K. pneumoniae, KPC has been identified in several other gram-negative bacilli, such as Citrobacter freundii (9, 21), Enterobacter aerogenes (4), Enterobacter cloacae (4, 9), Enterobacter gergoviae (9), Escherichia coli (3, 18), Klebsiella oxytoca (34), Pseudomonas aeruginosa (26), Salmonella enterica (14), and Serratia marcescens (9). Because the blaKPC gene is plasmid mediated and is carried in a Tn3-based transposon, Tn4401 (16), the potential ease of mobility of this resistance mechanism is a major concern.
Although the first health care-associated outbreaks of KPC-producing bacteria occurred in New York City (4, 31), recent reports indicate that these bacteria are globally prevalent and not limited to the northeastern United States. KPC-producing Enterobacteriaceae isolates have been identified in at least 33 U.S. states and have been reported in Brazil, China, Colombia, France, Greece, India (CDC, unpublished data), Israel, Norway, Scotland, and Sweden (6, 7, 15, 17, 19, 22, 23, 25, 27, 29). To better characterize the dissemination of this resistance mechanism, we used pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), sequencing of the blaKPC gene, PCR mapping of the Tn4401 flanking regions, and plasmid analysis to examine the molecular epidemiology of KPC-producing K. pneumoniae isolates sent to the CDC for reference testing from 1996 to 2008. The isolates we examined in this study were from 16 U.S. states, India, and Israel.
(This information was presented at the 48th annual Interscience Conference on Antimicrobial Agents and Chemotherapy and at the 46th annual meeting of the Infectious Diseases Society of America, Washington, DC, 28 October 2008.)
MATERIALS AND METHODS
Bacterial strains.
K. pneumoniae isolates sent to the CDC for reference antimicrobial susceptibility testing were analyzed by PCR for blaKPC if the MIC was ≥2 mg/liter for any of the carbapenems (imipenem, meropenem, or ertapenem) or if the isolate tested positive by the modified Hodge test (1). Limited clinical information was available for most isolates including the specimen type (e.g., blood, urine, sputum, or wound swab) used for sample isolation. No recognized epidemiologic connections between isolates from different geographic locations were known, although it has recently been suggested that there may be an association between a subset of K. pneumoniae isolates from Israel and isolates from the United States (19).
PFGE.
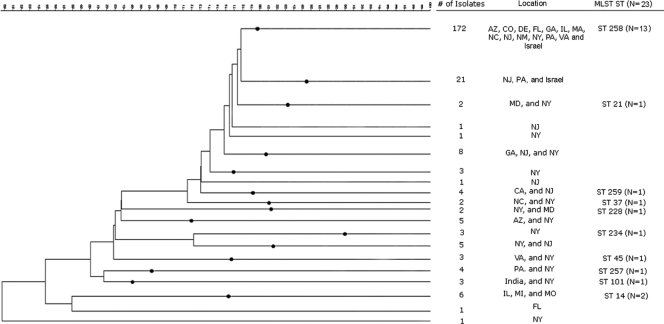
KPC-positive K. pneumoniae isolates representing diverse geographic locations were subsequently typed by PFGE of XbaI-digested total DNA as described for E. coli (http://www.cdc.gov/PULSENET/protocols.htm), resulting in a database of 248 PFGE patterns (Fig. 1). These isolates were originally received from New Jersey (26%), New York (25%), Arizona (15%), and 15 other states that represent each geographic region of the continental United States and account for 0.3% to 6.5% of the KPC-positive K. pneumoniae PFGE database. This database also included representative isolates from an outbreak in Israel (2.8%) and isolates from India (0.8%).
FIG. 1.
Representative dendrogram of the CDC's KPC-producing Klebsiella pneumoniae PFGE database (n = 248). Black markers have been placed on the major branches of the dendrogram to indicate divergence of further branches; these markers show a shared percentage of similarity of PFGE pattern for the isolates associated with each branch of the dendrogram (seen on the right).
Selection of representative isolates.
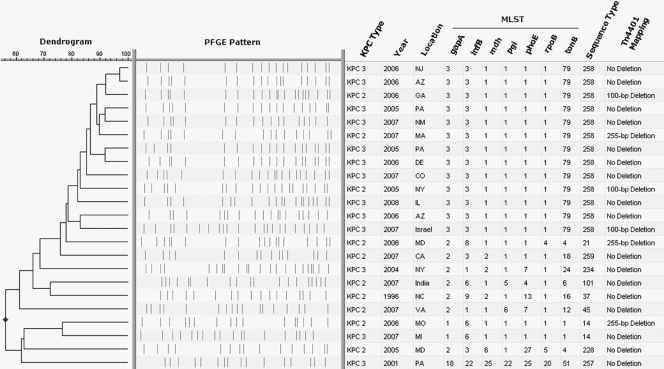
A dominant PFGE pattern with ≥80% similarity was observed in 172 isolates (69.3% of the database). To understand the significance of PFGE pattern similarities and differences, we selected 23 KPC-positive K. pneumoniae isolates for further analysis. Representative isolates were selected on the basis of variation in PFGE pattern and diversity of geographic location (Fig. 2). Of these 23 isolates, 13 were selected to represent the observed dominant PFGE pattern with ≥80% similarity and were isolated from Arizona, Colorado, Delaware, Georgia, Illinois, Massachusetts, New Jersey, New Mexico, New York, Pennsylvania, and Israel. The other 10 isolates displayed more variation in PFGE pattern, ranging from 56% to 76% similarity, and were originally isolated from California, Maryland, Michigan, Missouri, North Carolina, New York, Pennsylvania, Virginia, and India.
FIG. 2.
The dendrogram is based on similarity of PFGE patterns from 23 representative K. pneumoniae isolates. The table to the right illustrates results from blaKPC sequence analysis and MLST, along with PCR and sequence analysis of the nonconserved region of the Tn4401 element (16).
MLST.
MLST was performed on the 23 representative isolates according to the protocol described on the K. pneumoniae MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). These experiments were performed at a separate institution where researchers were blinded to previously established PFGE typing results on the 23 representative isolates. MLST results were compared to the international K. pneumoniae MLST database created in 2005 at the Pasteur Institute in Paris, France (10).
Sequencing of the blaKPC gene.
The KPC subtype of the 23 isolates was determined by amplification of a 1,011-bp PCR product and bi-directional DNA sequence analysis using previously described primers (21).
Surrounding genetic environment.
The Tn4401 regions that flank the blaKPC gene were initially analyzed by multiplex PCR using recently described primers (16). To focus on the only documented nonconserved region of the Tn4401 structure, located between the istB and blaKPC genes (16), we performed an additional PCR and subsequent bi-directional DNA sequence analysis using a previously described forward primer, 5′-TGA CCC TGA GCG GCG AAA GC-3′ (16), and reverse primer, 5′-CAC AGC GGC AGC AAG AAA GC-3′.
Plasmid analysis.
E. coli DH10B (Invitrogen, Carlsbad, CA) was transformed by electroporation (Gene Pulser Xcell; Bio-Rad, Hercules, CA) with plasmid DNA prepared from the K. pneumoniae isolates, using a Qiagen plasmid midi kit (Qiagen, Chatsworth, CA). Transformants, selected on LB agar plates containing 2 μg/ml ceftazidime and 1 μg/ml ertapenem, were screened by PCR for the presence of the blaKPC gene. Plasmid DNA from transformants were compared by restriction digestion with HindIII and gel electrophoresis (Fig. 3).
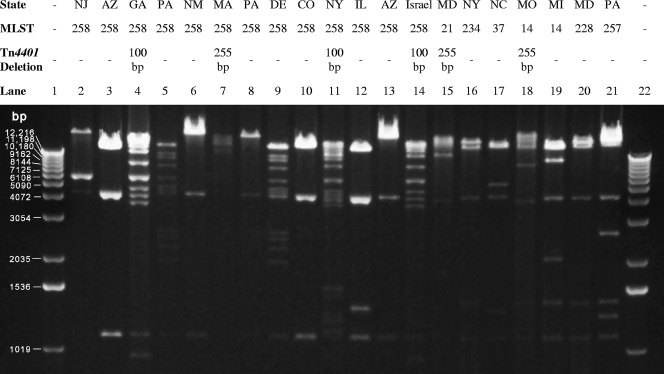
FIG. 3.
HindIII digests of blaKPC-containing plasmid DNA from E. coli transformants. State (or country) of origin, MLST type, and Tn4401 deletions are shown at the top of each lane. Lanes 1 and 22 contain a 1-kb ladder size standard.
RESULTS AND DISCUSSION
Comparison of PFGE and MLST results.
Of the isolates tested, the MLST results strongly agreed with the PFGE data and showed that the isolates with ≥80% similarity in PFGE patterns (n = 13) all shared the same MLST sequence type (ST) (Fig. 2). Furthermore, this MLST type did not match any of the previously recorded 256 STs in the international MLST database for K. pneumoniae isolates (10). This novel ST that represents the dominant PFGE pattern in the CDC's K. pneumoniae database was designated ST 258 and is a single-locus variant of ST 11; the latter has been described in Hungarian K. pneumoniae isolates that produce the CTX-M-15 extended-spectrum β-lactamase (8). All of the other detected STs that shared <80% similarity in PFGE patterns with those of ST 258 (n = 10) were neither single- nor double-locus variants of ST 258, and none of them have been associated with the dissemination of either extended-spectrum β-lactamases or carbapenemases. Two other novel STs were detected: (i) ST 257, later shown to be a double-locus variant of ST 334; and (ii) ST 259, a single-locus variant of ST 105. The discriminatory power of MLST being lower than that of PFGE represents an advantage in the identification of international clonal complexes, a point that has been previously made regarding P. aeruginosa (11, 12).
MLST and PFGE data were helpful in identifying possible dissemination of isolates in a limited geographic region. We found that isolates from Missouri (2006) and Michigan (2007) which share >76% similarity in PFGE patterns were of the same MLST type, ST 14 (Fig. 2). ST 14 is the primary founder of clonal complex 14, which comprises four other STs, including ST 15. Like ST 11, ST 15 has been associated with the spread of CTX-M-15-producing K. pneumoniae in Hungary (8), whereas ST 14 has been described in a blood culture isolate from Italy with high-level resistance to ceftazidime (10). When the entire KPC-producing K. pneumoniae PFGE database was examined for similar PFGE patterns, three isolates from Illinois (2008) with patterns with a range of >76% to >97% similarity to those of the Missouri and Michigan isolates were identified (Fig. 1). The finding of isolates that share common PFGE patterns and MLST types that are distant from the dominant PFGE pattern seen in the database appears to represent dissemination of a different strain in the midwestern United States (2).
Characterizing KPC subtype and the genetic environment that surrounds the blaKPC gene.
Currently, seven different variants of the KPC enzyme (KPC-2 through KPC-8) have been reported in GenBank. KPC-1 was recently shown to have a sequence identical to that of KPC-2 (33). Sequencing of the blaKPC gene showed that 10 of the 23 isolates produced KPC-2 and 13 of the 23 isolates produced KPC-3 (Fig. 2). Although these isolates differed in KPC subtype, the difference between blaKPC-2 and blaKPC-3 is a single base change. An interesting correlation between KPC subtype and MLST type was observed: the majority of the 13 isolates determined to be ST 258 (n = 10) produced KPC-3, and the majority of the 10 isolates that were not ST 258 (n = 7) produced KPC-2. This observation could suggest an association between isolates that both are ST 258 and possess a KPC-3; however, a larger number of isolates would need to be tested in order to make this conclusion.
The Tn4401 regions that flank the blaKPC gene were initially analyzed by multiplex PCR and resulted in the observation of three distinct band patterns. We then performed an additional focused PCR and subsequent bi-directional DNA sequencing on the only documented nonconserved region of the Tn4401 structure, located between the istB and blaKPC genes (16). We found that 17 of the isolates produced a 703-bp PCR product (no deletion), three of the isolates produced a 604-bp PCR product (100-bp deletion), and three of the isolates produced a 448-bp PCR product (255-bp deletion) (Fig. 2). This observation may explain the three distinct band patterns that were observed in the preliminary multiplex PCR. Previous reports describe a 100-bp deletion in the nonconserved region of the Tn4401 element (GenBank accession no. EU176011) (16) that is identical to the 100-bp deletion seen in this study. Previous reports also describe a 215-bp deletion (GenBank accession no. DQ989640) that is encompassed by the 255-bp deletion that we observed.
The 13 isolates that were ST 258 demonstrated all three of the observed variable regions between the istB and blaKPC genes (Fig. 2). It is interesting to note that all ST 258 isolates from the United States that contained a KPC-3 (n = 9) had no deletion in the variable region of Tn4401 but the ST 258 isolate from Israel that also contained a KPC-3 exhibited a 100-bp deletion in this region. In addition, the three ST 258 isolates that contained a KPC-2 demonstrated both a 100-bp deletion (n = 2) and a 255-bp deletion (n = 1) in this variable region. This 255-bp deletion was also seen in K. pneumoniae isolates with unrelated MLST types from Massachusetts, Maryland, and Missouri, all of which contained a KPC-2 (Fig. 2).
Restriction analysis of blaKPC-containing plasmids.
To analyze plasmids carrying the blaKPC gene, plasmid DNA was prepared from the 23 isolates, and transfer to E. coli DH10B by electroporation was attempted. Only 20 transformations were successful, despite repeated attempts. The inability to transfer blaKPC by transformation with plasmid DNA from the California, Virginia, and India strains, which each displayed PFGE and MLST profiles different from those of other study isolates, could be due to a chromosomal location of the gene. Chromosomal insertion of blaKPC was reported in a P. aeruginosa isolate from Colombia (26).
Restriction profiles of the blaKPC-containing plasmids isolated from E. coli transformants resulted in several diverse band patterns (Fig. 3). In particular, the 13 isolates that were ST 258 (Fig. 3, lanes 2 to 14) exhibited at least five different plasmid restriction profiles. Similar plasmid restriction profiles were observed between isolates with different MLST types (e.g., Fig. 3, lanes 16 and 20). Plasmid restriction profiles did not correlate with KPC subtype data; however, it appears that isolates with similar deletions in the Tn4401 element may correlate with similar restriction profiles.
Pairs of ST 258 isolates with similar molecular characteristics (plasmid restriction profiles, KPC subtype, and Tn4401 profiles) were observed in New Mexico and Arizona (Fig. 3, lanes 6 and 13), Arizona and Colorado (lanes 3 and 10), and Pennsylvania and Delaware (lanes 5 and 9) isolates. These findings may represent regional dissemination of ST 258 isolates. Overall, ST 258 isolates demonstrated diversity in these molecular characteristics. This could mean that a single ST 258 isolate acquired a blaKPC-containing plasmid and then evolved with possible transposition of Tn4401 from one plasmid to another, or that multiple ST 258 isolates acquired plasmids carrying the blaKPC gene at different times.
Outbreaks of KPC-producing K. pneumoniae were first observed in health care settings in the northeastern regions of the United States a decade ago (5, 24, 31); these extensively drug-resistant organisms have now spread throughout the country. The CDC's collection of isolates contains strains of KPC-positive K. pneumoniae from throughout the continental United States as well as isolates from India and Israel. Our findings indicate that strains of a single lineage, ST 258, account for almost 70% of all isolates in the CDC's KPC-producing K. pneumoniae PFGE database (Fig. 1). In addition to these related organisms being isolated in 10 different states representing the Northeast, the South, the Midwest, and the western regions of the United States (Fig. 4), a representative KPC-3-producing K. pneumoniae isolate from an outbreak in Israel (19) was also determined to be ST 258. This Israeli isolate had plasmid restriction and Tn4401 profiles that were similar to those of ST 258 isolates from the United States (e.g., ST 258 from Georgia and New York [Fig. 3, lanes 4 and 11, respectively]). KPC-producing ST 258 was also recently identified in isolates from Norway and Sweden obtained from patients with prior hospitalization in Greece and Israel (23). These findings suggest possible international dissemination of KPC-producing ST 258.
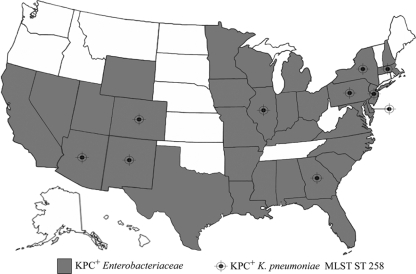
FIG. 4.
Map of the United States identifying states that currently have known KPC-producing Enterobacteriaceae (shaded in gray) (n = 33). States that have KPC-producing K. pneumoniae isolates identified by this study as MLST ST 258 are marked with a target symbol. We also identified ST 258 in an isolate from Israel. These data are based on a passive reporting system that relies on isolates voluntarily sent to the CDC for KPC testing. Thus, states on this map that are not marked to indicate known KPC-producing Enterobacteriaceae (white) have neither reported nor voluntarily sent isolates to the CDC.
Our study has identified another strain of KPC-producing K. pneumoniae, ST 14, which was isolated in various facilities in the midwestern United States. Although a variety of K. pneumoniae strains may harbor plasmids carrying the blaKPC gene, our data suggest that certain strains may be more widely distributed (both regionally and nationally) in the United States. These findings are reminiscent of the widespread dissemination of methicillin-resistant Staphylococcus aureus strain USA300, and the Clostridium difficile strain NAP1 (13, 28). Identification of related KPC-producing K. pneumoniae strains could represent either a strain that has successfully disseminated over a wide geographic region or a strain that more readily acquires or maintains this resistance mechanism. Nevertheless, the recognition of a common strain type of KPC-producing K. pneumoniae is an important step toward developing additional targeted strategies to prevent the spread of these pathogens (30).
Acknowledgments
We acknowledge Karen F. Anderson and Linda K. McDougal for their work on reference susceptibility testing of KPC-producing isolates and help in creating the K. pneumoniae PFGE database, respectively.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
We do not have a commercial or other kind of association that might be considered a conflict of interest regarding the information that is within this document.
Footnotes
Published ahead of print on 8 June 2009.
REFERENCES
- 1.Anderson, K. F., D. R. Lonsway, J. K. Rasheed, J. Biddle, B. Jensen, L. K. McDougal, R. B. Carey, A. Thompson, S. Stocker, B. Limbago, and J. B. Patel. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 45:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, T. J., P. Gerner-Smidt, and B. Swaminathan. 2006. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog. Dis. 3:20-31. [DOI] [PubMed] [Google Scholar]
- 3.Bratu, S., S. Brooks, S. Burney, S. Kochar, J. Gupta, D. Landman, and J. Quale. 2007. Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase KPC-2 in Brooklyn, New York. Clin. Infect. Dis. 44:972-975. [DOI] [PubMed] [Google Scholar]
- 4.Bratu, S., D. Landman, M. Alam, E. Tolentino, and J. Quale. 2005. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrob. Agents Chemother. 49:776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratu, S., M. Mooty, S. Nichani, D. Landman, C. Gullans, B. Pettinato, U. Karumudi, P. Tolaney, and J. Quale. 2005. Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob. Agents Chemother. 49:3018-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, J. C., H. W. Zhou, R. Zhang, and G. X. Chen. 2008. Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuzon, G., T. Naas, M. C. Demachy, and P. Nordmann. 2008. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in Klebsiella pneumoniae isolate from Greece. Antimicrob. Agents Chemother. 52:796-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damjanova, I., A. Toth, J. Paszti, G. Hajbel-Vekony, M. Jakab, J. Berta, H. Milch, and M. Fuzi. 2008. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type β-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005-the new “MRSAs”? J. Antimicrob. Chemother. 62:978-985. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande, L. M., P. R. Rhomberg, H. S. Sader, and R. N. Jones. 2006. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States medical centers: report from the MYSTIC program (1999-2005). Diagn. Microbiol. Infect. Dis. 56:367-372. [DOI] [PubMed] [Google Scholar]
- 10.Diancourt, L., V. Passet, J. Verhoef, P. A. Grimont, and S. Brisse. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giske, C. G., B. Libisch, C. Colinon, E. Scoulica, L. Pagani, M. Fuzi, G. Kronvall, and G. M. Rossolini. 2006. Establishing clonal relationships between VIM-1-like metallo-β-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J. Clin. Microbiol. 44:4309-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, J. K., S. M. Arduino, O. C. Stine, J. A. Johnson, and A. D. Harris. 2007. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 45:3707-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazakova, S. V., J. C. Hageman, M. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. K. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468-475. [DOI] [PubMed] [Google Scholar]
- 14.Miriagou, V., L. S. Tzouvelekis, S. Rossiter, E. Tzelepi, F. J. Angulo, and J. M. Whichard. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47:1297-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteiro, J., A. P. C. Henriques, A. F. Santos, D. G. C. Matos, G. Perano, M. D. Asensi, and A. C. Gales. 2007. Carbapenem-resistant Klebsiella pneumoniae outbreak: emergence of KPC-2 producing strains in Brazil, abstr. C2-1929, p. 141. Abstr. 47th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 16.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naas, T., P. Nordmann, G. Vedel, and C. Poyart. 2005. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49:4423-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, M. J. Schwaber, D. Schwartz, and Y. Carmeli. 2006. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob. Agents Chemother. 50:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navon-Venezia, S., A. Leavitt, M. J. Schwaber, J. K. Rasheed, A. Srinivasan, J. B. Patel, and Y. Carmeli. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasheed, J. K., J. W. Biddle, K. F. Anderson, L. Washer, C. Chenoweth, J. Perrin, D. W. Newton, and J. B. Patel. 2008. Detection of the KPC-2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and Klebsiella oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samra, Z., O. Ofir, Y. Lishtzinsky, L. Madar-Shapiro, and J. Bishara. 2007. Outbreak of carbapenem-resistant Klebsiella pneumoniae producing KPC-3 in a tertiary medical centre in Israel. Int. J. Antimicrob. Agents 30:525-529. [DOI] [PubMed] [Google Scholar]
- 23.Samuelsen, O., U. Naseer, S. Tofteland, D. H. Skutlaberg, A. Onken, R. Hjetland, A. Sundsfjord, and C. G. Giske. 2009. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63:654-658. [DOI] [PubMed] [Google Scholar]
- 24.Smith Moland, E., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolysing β-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51:711-714. [DOI] [PubMed] [Google Scholar]
- 25.Villegas, M. V., K. Lolans, A. Correa, C. J. Suarez, J. A. Lopez, M. Vallejo, J. P. Quinn, et al. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 50:2880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villegas, M. V., K. Lolans, A. Corrrea, J. N. Kattan, J. A. Lopez, J. P. Quinn, and the Colombian Nosocomial Resistance Study Group. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 51:1553-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walther-Rasmussen, J., and N. Høiby. 2007. Class A carbapenemases. J. Antimicrob. Chemother. 60:470-482. [DOI] [PubMed] [Google Scholar]
- 28.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
- 29.Wei, Z.-Q., X.-X. Du, Y.-S. Yu, P. Shen, Y.-G. Chen, and L.-J. Li. 2007. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob. Agents Chemother. 51:763-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodford, N. 2008. Successful, multiresistant bacterial clones. J. Antimicrob. Chemother. 61:233-234. [DOI] [PubMed] [Google Scholar]
- 31.Woodford, N., P. M. Tierno, Jr., K. Young, L. Tysall, M.-F. I. Palepou, E. Ward, R. E. Painter, D. F. Suber, D. Shungu, L. L. Silver, K. Inglima, J. Kornblum, and D. M. Livermore. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob. Agents Chemother. 48:4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenench-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2008. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 52:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yigit, H., A. M. Queenan, J. K. Rasheed, J. W. Biddle, A. Domenech-Sanchez, S. Alberti, K. Bush, and F. C. Tenover. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob. Agents Chemother. 47:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]