Abstract
A novel ABC transporter gene, vga(C), was identified on the 14,365-bp multiresistance plasmid pKKS825 in a porcine methicillin (meticillin)-resistant Staphylococcus aureus isolate of sequence type 398. The vga(C) gene encodes a 523-amino-acid protein which confers resistance not only to streptogramin A antibiotics but also to lincosamides and pleuromutilins. Plasmid pKKS825 also carries the resistance genes aadD, tet(L), and dfrK, which may enable the coselection of vga(C) under selective pressure by kanamycin/neomycin, tetracyclines, and trimethoprim.
Methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) strains of sequence type 398 (ST398) are predominantly found in pigs, where they commonly occur as colonizers and are only rarely associated with infections (19-22). In the BfT-GermVet study 2004-2006, 248 coagulase-positive and -variable staphylococci associated with acute disease conditions in pigs and cats/dogs were investigated for their susceptibility to 24 antimicrobial agents (16). Only five MRSA strains had been identified, all of which were from pigs and were assigned to ST398 (16, 18). These five porcine MRSA ST398 strains were multiresistant and originated from infections of the skin or the urinary/genital tract, including metritis-mastitis-agalactia syndrome (16, 18). Further screening of these strains for transferable antimicrobial resistance led to the identification of the multiresistance plasmid pKKS825. The strain carrying this plasmid originated from a pig suffering from a skin infection and was isolated in September 2004 in Lower Saxony, Germany (18).
Southern blot hybridization experiments revealed that plasmid pKK825 carried at least the tetracycline resistance gene tet(L) and the recently discovered trimethoprim resistance gene dfrK. Plasmid pKKS825 was transferred by protoplast transformation into the recipient strain Staphylococcus aureus RN4220 and transformants were selected on regeneration plates supplemented with either 20 μg/ml tetracycline or 30 μg/ml trimethoprim (10). The plasmid was sequenced completely on both strands by primer walking, starting with primers from the dfrK gene. Susceptibility testing of the pKKS825 transformants was conducted by either broth microdilution or broth macrodilution according to CLSI recommendations (5). S. aureus ATCC 29213 served as the quality control strain.
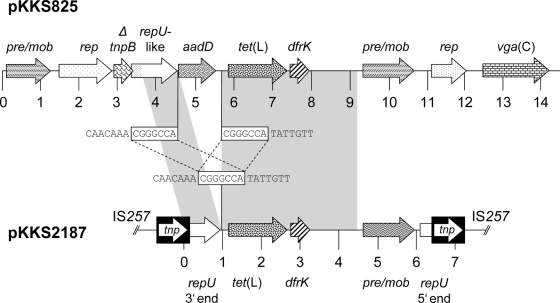
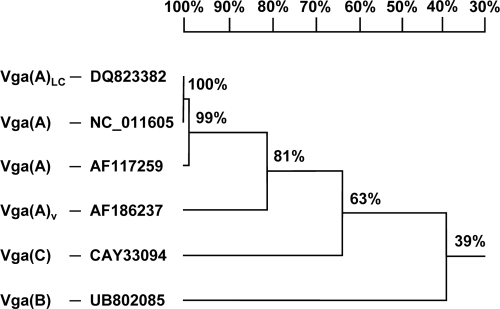
A sequence analysis revealed that plasmid pKKS825 consisted of 14,365 bp and exhibited 10 reading frames of >150 amino acids (aa). The schematic presentation of plasmid pKKS825 is shown in Fig. 1. The initial 3,215 bp corresponded closely (99.4% identity) to the sequence of plasmid pSCFS1 from Staphylococcus sciuri (9). This segment contained reading frames for a 408-aa protein with homology to putative recombinase/mobilization proteins, a 453-aa plasmid replication protein and a 167-aa protein whose N-terminal 73 aa corresponded exactly to the TnpB transposase protein of Tn554. Further downstream, a reading frame for a 393-aa protein was detected whose C-terminal 217 aa were indistinguishable from the RepU sequences of plasmids pUB110 (11), pBC16 (13, 14), and pKKS2187 (8). Three antimicrobial resistance genes were detected downstream of this repU-related reading frame: (i) the aadD gene coding for a 256-aa aminoglycoside adenyltransferase which confers resistance to kanamycin and neomycin, (ii) the tet(L) gene encoding a 459-aa protein of the major facilitator family and mediating resistance to tetracyclines, and (iii) the dfrK gene coding for a recently described dihydrofolate reductase of 163 aa which confers trimethoprim resistance. This arrangement ΔrepU-tet(L)-dfrK was almost identical (99.5% identity) to that of plasmid pKKS2187 (8), which also originated from a porcine MRSA ST398 strain. An analysis of the pKKS825 sequence, however, showed that a 1,036-bp segment encompassing the aadD gene was inserted between the repU and the tet(L) gene sequences. At the junctions of pKKS2187-homologous to -nonhomologous sequences, direct duplications of 7 bp were detected (Fig. 1). Since this 7-bp sequence, 5′-CGGGCCA-3′, was also present in pKKS2187, it might have served as the integration of the aadD region into a pKKS2187-like repU-tet(L) sequence. Further downstream of this resistance gene region, another two reading frames were detected: one coding for a 409-aa protein which was distantly related to the Mob proteins of pBC16 and pUB110 (37.8% identity and 60.0% similarity) (11, 13) and the other for a putative plasmid replication protein of 271 aa which was related to the Rep protein of Macrococcus caseolyticus (48.9% identity and 67.5% similarity) (3). The largest reading frame of plasmid pKKS825 coded for a protein of 523 aa. This protein revealed the typical features of a class 2 ABC transporter consisting of two ATP-binding cassettes, each with the Walker A and B motifs as well as the signature motif involved in ATP binding and hydrolysis (15). Structural comparisons identified closest similarity to the similar-sized Vga(A) variants of staphylococci (62.3% to 65.5% amino acid identity) (2, 4, 6, 7, 12) and only 39.2% identity to Vga(B) (1). Based on this comparatively low identity to previously known Vga proteins, the novel ABC transporter was designated Vga(C) by the MLS nomenclature center (http://faculty.washington.edu/marilynr/). The homology tree shown in Fig. 2 identified the Vga(C) protein on a separate branch between the Vga(A) variants and the Vga(B) protein. Since some of the Vga(A) variants had been reported to also mediate reduced susceptibility or resistance to lincosamides and/or pleuromutilins (4, 6, 12), S. aureus RN4220 and its pKKS825 transformant were comparatively investigated for their MICs to virginiamycin M1, lincomycin, pirlimycin, clindamycin, tiamulin, and valnemulin in addition to neomycin, kanamycin, trimethoprim, tetracycline, and, for control purposes, erythromycin (Table 1). Based on the MIC increases seen in the presence of plasmid pKKS825, Vga(C) mediated resistance not only to the streptogramin A antibiotic virginiamycin M1 but also to the lincosamides lincomycin, pirlimycin, and clindamycin as well as the pleuromutilins tiamulin and valnemulin. A comparative analysis of S. aureus RN4220 transformants harboring plasmid pKKS2187, which carries only tet(L) and dfrK (8), or plasmid pSTS7, which carries only tet(L) and aadD (17), confirmed the expected increases in the MICs of tetracycline and trimethoprim or tetracycline and kanamycin/neomycin, respectively, while no increases in the MICs of streptogramin A, lincosamides, or pleuromutilins were seen. This observation confirmed that Vga(C) mediates resistance to streptogramin A antibiotics, lincosamides, and pleuromutilins.
FIG. 1.
Comparison of the plasmid pKKS825 (EMBL accession no. FN377602) identified in the present study with the tet(L)-dfrK segment of plasmid pKKS2187 (accession no. FM207105). The arrows indicate the extents and directions of the transcription of the genes rep, repU (plasmid replication), tet(L) (tetracycline resistance), dfrK (trimethoprim resistance), pre/mob (plasmid recombination/mobilization), and ΔtnpB (truncated transposase B). The different rep and pre/mob genes are displayed in different shadings to underline their structural differences. The IS257 elements in the map of pKKS2187 are shown as black boxes with the white arrows indicating the transposase tnp genes. The regions of >99% homology between pKKS825 and pKKS2187 are marked by gray shading. The sequences at the junctions of pKKS2187-homologous and -nonhomologous parts in pKKS825 are shown in comparison to the corresponding pKKS2187 sequence between the two maps; the 7-bp direct repeats are shown in boxes.
FIG. 2.
Homology tree of the staphylococcal Vga(A) (2, 4, 6, 7, 12), Vga(B) (1), and Vga(C) (this study) proteins. Database accession numbers of the various Vga proteins are indicated. The branching order follows the amino acid exchanges observed in a multisequence alignment. The percentages of identity are rounded up or down to the nearest integral percent values.
TABLE 1.
Comparative analysis of the MICs of S. aureus RN4220 and the S. aureus RN4220 transformants carrying the plasmids pKKS825, pKKS2187, and pSTS7
| Bacterial strain | Resistance genes | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIR M1 | TIA | VAL | CLI | PIR | LIN | ERY | TET | KAN | NEO | TMP | ||
| S. aureus RN4220 | 4 | 0.06 | 0.03 | ≤0.12 | 0.25 | 1 | 0.25 | 0.06 | 4 | ≤1 | 0.5 | |
| S. aureus RN4220(pKKS825) | aadD, tet(L), dfrK, vga(C) | 32 | ≥128 | ≥128 | 2 | 4 | 64 | 0.25 | 64 | 32 | 64 | ≥512 |
| S. aureus RN4220(pKKS2187) | tet(L), dfrK | 4 | 0.06 | 0.03 | ≤0.12 | 0.25 | 1 | 0.25 | 64 | 4 | ≤1 | ≥512 |
| S. aureus RN4220(pSTS7) | aadD, tet(L) | 4 | 0.06 | 0.03 | ≤0.12 | 0.25 | 1 | 0.25 | 64 | 32 | 64 | 0.5 |
Antimicrobial agents are abbreviated as follows: VIR M1, virginiamycin M1; TIA, tiamulin; VAL, valnemulin; CLI, clindamycin; PIR, pirlimycin; LIN, lincomycin; ERY, erythromycin; TET, tetracycline; KAN, kanamycin; NEO, neomycin; and TMP, trimethoprim.
The structural analysis of plasmid pKKS825 identified four different resistance genes which confer resistance to members of six different classes of antimicrobial agents. Given the fact that this plasmid was identified in an MRSA strain that additionally carried a mecA gene for β-lactam resistance, a tet(M) gene for resistance to tetracycline/minocycline, an erm(A) gene for combined resistance to macrolides, lincosamides, and streptogramin B antibiotics, and an aacA-aphD gene for resistance to gentamicin, kanamycin, and tobramycin (18), therapeutic options to control infections due to such an MRSA strain are limited. Thus, the monitoring of multiresistance plasmids, such as pKKS825, among MRSA strains is required to determine their prevalence and dissemination.
Nucleotide sequence accession number.
The sequence of the 14,365-bp plasmid pKKS825 has been deposited in the EMBL database under accession number FN377602.
Acknowledgments
We thank Kerstin Meyer for excellent technical assistance.
This study was financially supported by internal funding from the Friedrich-Loeffler-Institute.
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Allignet, J., and N. El Solh. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202:133-138. [DOI] [PubMed] [Google Scholar]
- 2.Allignet, J., and N. El Solh. 1999. Comparative analysis of staphylococcal plasmids carrying three streptogramin-resistance genes: vat-vgb-vga. Plasmid 42:134-138. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., K. Kuwahara-Arai, I. Uchiyama, F. Takeuchi, T. Ito, and K. Hiramatsu. 2009. Complete genome sequence determination of a Macrococcus caseolyticus strain JSCS5402 reflecting the ancestral genome of the human pathogenic staphylococci. J. Bacteriol. 191:1180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesneau, O., H. Ligeret, N. Hosan-Aghaie, A. Morvan, and E. Dassa. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob. Agents Chemother. 49:973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CLSI. 2008. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals. Approved standard, 3rd ed. CLSI document M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Gentry, D. R., L. McCloskey, M. N. Gwynn, S. F. Rittenhouse, N. Scangarella, R. Shawar, and D. J. Holmes. 2008. Genetic characterization of Vga ABC proteins conferring reduced susceptibility of Staphylococcus aureus to pleuromutilins. Antimicrob. Agents Chemother. 52:4507-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haroche, J., J. Allignet, C. Buchrieser, and N. El Solh. 2000. Characterization of a variant of vga(A) conferring resistance to streptogramin A and related compounds. Antimicrob. Agents Chemother. 44:2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadlec, K., and S. Schwarz. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L). Antimicrob. Agents Chemother. 53:776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kehrenberg, C., K. K. Ojo, and S. Schwarz. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 54:936-939. [DOI] [PubMed] [Google Scholar]
- 10.Kehrenberg, C., and S. Schwarz. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenzie, T., T. Hoshino, T. Tanaka, and N. Sueoka. 1987. Correction. A revision of the nucleotide sequence and functional map of pUB110. Plasmid 17:83-85. [DOI] [PubMed] [Google Scholar]
- 12.Novotna, G., and J. Janata. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob. Agents Chemother. 50:4070-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oskam, L., D. J. Hillenga, G. Venema, and S. Bron. 1991. The large Bacillus plasmid pTB19 contains two integrated rolling-circle plasmids carrying mobilization functions. Plasmid 26:30-39. [DOI] [PubMed] [Google Scholar]
- 14.Palva, A., G. Vigren, M. Simonen, H. Rintala, and P. Laamanen. 1990. Nucleotide sequence of the tetracycline resistance gene of pBC16 from Bacillus cereus. Nucleic Acids Res. 18:1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider, E., and S. Hunke. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1-20. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz, S., E. Alešík, C. Werckenthin, M. Grobbel, A. Lübke-Becker, L. H. Wieler, and J. Wallmann. 2007. Antimicrobial susceptibility of coagulase-positive and coagulase-variable staphylococci from various indications of swine, dogs and cats as determined in the BfT-GermVet monitoring program 2004-2006. Berl. Münch. Tierärztl. Wochenschr. 120:372-379. [PubMed] [Google Scholar]
- 17.Schwarz, S., P. D. Gregory, C. Werckenthin, S. Curnock, and K. G. H. Dyke. 1996. A novel plasmid from Staphylococcus epidermidis specifying resistance to kanamycin, neomycin and tetracycline. J. Med. Microbiol. 45:57-63. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz, S., K. Kadlec, and B. Strommenger. 2008. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius detected in the BfT-GermVet monitoring programme 2004-2006 in Germany. J. Antimicrob. Chemother. 61:282-285. [DOI] [PubMed] [Google Scholar]
- 19.van Belkum, A., D. C. Melles, J. K. Peeters, W. B. van Leeuwen, E. van Duijkeren, X. W. Huijsdens, E. Spalburg, A. J. de Neeling, H. A. Verbrugh, and the Dutch Working Party on Surveillance and Research of MRSA-SOM. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Duijkeren, E., R. Ikawaty, M. J. Broekhuizen-Stins, M. D. Jansen, E. C. Spalburg, A. J. de Neeling, J. G. Allaart, A. van Nes, J. A. Wagenaar, and A. C. Fluit. 2008. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 126:383-389. [DOI] [PubMed] [Google Scholar]
- 21.van Duijkeren, E., M. D. Jansen, S. C. Flemming, H. de Neeling, J. A. Wagenaar, A. H. W. Schoormans, A. van Nes, and A. C. Fluit. 2007. Methicillin-resistant Staphylococcus aureus in pigs with exudative epidermitis. Emerg. Infect. Dis. 13:1408-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voss, A., F. Loeffen, J. Bakker, C. Klaassen, and M. Wulf. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]