Abstract
CMY-30, a Val211Gly mutant of CMY-2 cephalosporinase, was derived by mutagenesis. The hydrolytic efficiency of CMY-30 against expanded-spectrum cephalosporins was higher than that of CMY-2 due to increased kcat values. Findings indicate a role of the Ω loop residue 211 in determining the substrate specificities of CMYs also corroborated by modeling studies.
The molecular class C β-lactamases (AmpCs), also classified as group 1 in the Bush et al. scheme (3), share significant structural and functional similarities despite diversity in amino acid sequences. AmpCs are highly effective against early generation cephalosporins, while their catalytic efficiencies against newer β-lactams, such as expanded-spectrum cephalosporins (ESCs), cefepime, and aztreonam, are low (9). Yet, the emergence of natural AmpC variants with enhanced activity against the latter β-lactams is increasingly reported (9, 13). Most of these β-lactamases, designated ESACs (extended-spectrum AmpCs), have been derived from “wild-type” enterobacterial AmpCs through substitutions, insertions, and deletions in domains such as the R2 and Ω loops that affect enzyme-substrate interactions (11).
We have recently reported on ACC-4, a plasmid-encoded ESAC from Escherichia coli that differed from the Hafnia alvei-originated cephalosporinase ACC-1 by a Gly-for-Val substitution at the Ω loop position 211. This change caused a measurable increase in the catalytic efficiency against ESCs (17). Amino acid sequence alignments showed that position 211 is occupied by Val in the vast majority of AmpCs (17). Also, studies with AmpC from E. coli have indicated an interaction of the side chain of Val211 with the R1 group of ESCs (22). In this work, we studied the effect of the replacement of Val211 with a Gly on the substrate specificity of the Citrobacter freundii-originated CMY-2, which is the most widespread plasmid-mediated cephalosporinase (19). Of note, CMY-30, a CMY-2 variant with a Gly211, has recently been identified in a clinical E. coli strain isolated in New Zealand (20).
Construction of blaCMY-30.
The blaCMY-30 gene differs from the prototype blaCMY-2 by a single nucleotide, a T-to-G transversion in codon 211 (nucleotide [nt] 692 in blaCMY-2, nt 1924 to 3069 in GenBank Accession No. X91840) resulting in a Gly for Val substitution. Plasmid pB-cmy2, a previously described derivative of the high-copy-number vector pBCSK(+) (Stratagene, La Jolla, CA) with a 1,436-bp insert containing blaCMY-2 and the respective promoter provided by ISEcp1 (27), was used in mutagenesis experiments. A CMY-30-encoding plasmid (pB-cmy30) was obtained using a QuikChange mutagenesis kit (Stratagene) and two mutagenic primers, VG-F (5′-GAAGGGAAGCCCGTACACGGTTCTCCGGGACAACTTGAC-3′) and VG-R (5′-GTCAAGTTGTCCCGGAGAACCGTGTACGGGCTTCCCTTC-3′) (nt 2596 to 2634 in X91840; underlined nucleotides correspond to codon 211 in blaCMY-30). The CMY-encoding sequences were also cloned into the tetracycline-resistant vector pACYC184 (low copy number), yielding plasmids pAC-cmy30 and pAC-cmy2 encoding CMY-30 and CMY-2, respectively. The identity of each blaCMY-containing insert was verified by the sequencing of at least two PCR products. E. coli DH5α was used as a host in cloning experiments. E. coli MC4100, an ampC-deficient laboratory strain (8), was used to express CMY β-lactamases and in comparative determinations of β-lactam MICs conferred by CMY-30 and CMY-2. The production of CMY β-lactamases was confirmed by isoelectric focusing. Both enzymes exhibited apparent isoelectric points of approximately 9.0 (data not shown).
Effects of Val211Gly on interaction with β-lactams. (i) Resistance phenotypes.
β-Lactam MICs for E. coli MC4100 strains producing CMY-30 and CMY-2 under isogenic conditions were determined in parallel by a microdilution technique using Mueller-Hinton (MH) broth and an inoculum of 5 × 105 CFU/ml. CMY-producing clones both exhibited high-level resistance to penicillins (amoxicillin [amoxicilline], ticarcillin, and piperacillin), early generation cephalosporins (cephalothin [cefalotin], cefaclor, and cefuroxime), and cefoxitin. The MICs of the latter drugs, however, exceeded the dilutions tested, not allowing for an accurate phenotypic comparison. Combining clavulanic acid with amoxicillin and ticarcillin did not significantly affect susceptibility, while tazobactam reduced piperacillin MICs at levels below the resistance breakpoint. Also, both CMY-producing E. coli strains were highly susceptible to cefepime and imipenem, and the respective MIC pairs were similar. On the other hand, CMY-30 conferred significantly higher levels of resistance to ESCs and aztreonam than CMY-2. The most pronounced MIC increase was observed with cefotaxime (four doubling dilutions), followed by ceftazidime and aztreonam (at least three and three doubling dilutions, respectively) (Table 1). Therefore, a comparison of resistance phenotypes suggested a higher catalytic efficiency of CMY-30 over CMY-2 against the tested oxyimino-β-lactams, except cefepime.
TABLE 1.
β-Lactam resistance levels conferred by CMY-30 and CMY-2 in an isogenic background
| β-Lactam | MIC (μg/ml) for blaCMY-carrying E. coli MC4100 clone:
|
||
|---|---|---|---|
| pAC-cmy30 (CMY-30) | pAC-cmy2 (CMY-2) | pACYC184 (−) | |
| Amoxicillin | >512 | >512 | 4 |
| Amoxicillin + clavulanic acida | >512 | >512 | 4 |
| Ticarcillin | >512 | >512 | 2 |
| Ticarcillin + clavulanic acida | >512 | >512 | 1 |
| Piperacillin | 64 | 128 | 0.5 |
| Piperacillin + tazobactamb | 16 | 16 | 0.5 |
| Cephalothin | >512 | >512 | 2 |
| Cefaclor | >512 | >512 | 2 |
| Cefoxitin | >512 | >512 | 2 |
| Cefuroxime | >512 | >512 | 1 |
| Cefotaxime | 256 | 16 | 0.12 |
| Ceftazidime | >512 | 128 | 0.25 |
| Aztreonam | 128 | 16 | 0.12 |
| Cefepime | 0.25 | 0.25 | 0.06 |
| Imipenem | 0.25 | 0.25 | 0.12 |
Clavulanic acid at a fixed concentration of 2 μg/ml.
Tazobactam at a fixed concentration of 4 μg/ml.
(ii) Kinetic and inhibition studies.
E. coli MC4100 strains harboring the high-copy-number plasmids pB-cmy30 and pB-cmy2 were chosen for enzyme purification due to higher cephalosporinase production (7- to 10-fold-higher specific activity against cephalothin) compared to that of their counterparts carrying pACYC184 derivatives. β-Lactamases were released from bacterial cells suspended in Tris buffer (20 mM; pH 7.4) by sonication. The purification of cephalosporinases was carried out by two ion-exchange chromatography steps using Q- and S-Sepharose columns (17). The purity of the final preparations was >95% as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The hydrolysis of penicillin G, cephalothin, nitrocefin, cefoxitin, cefotaxime, and ceftazidime was studied by spectrophotometry. The respective extinction coefficients and wavelengths have been reported elsewhere (21). Kinetic parameters were calculated as described previously (2, 6). The results are presented in Table 2. The replacement of Val211 by Gly did not significantly affect the kinetic parameters for penicillin G, cephalothin, and cefoxitin. A reduction in the catalytic efficiency (kcat/Km) of CMY-30 compared to CMY-2 against nitrocefin was noticed due to an approximately threefold decrease in the kcat value, while the Km values remained similar. The change of Val211 to Gly, however, induced notable changes in the kinetic parameters for cefotaxime and ceftazidime that were in line with the respective MIC differences (Table 1). CMY-30 exhibited higher hydrolytic efficiencies than CMY-2 against both these ESCs (approximately four- and sixfold, respectively). This was due to significant increases in the kcat values that were partly compensated by the raising of the respective Km values.
TABLE 2.
Kinetic parameters of CMY-30 and CMY-2 cephalosporinases for various β-lactam substratesa
| β-Lactam | Kinetic parameters for:
|
|||||
|---|---|---|---|---|---|---|
| CMY-30 (Gly211)
|
CMY-2 (Val211)
|
|||||
| kcat (s−1) | Km (μM) | kcat/Km (μM−1· s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) | |
| Penicillin Gb | 23 ± 3 | 0.6 ± 0.1 | 38 | 24 ± 3 | 0.6 ± 0.08 | 40 |
| Cephalothin | 256 ± 14 | 26 ± 3.0 | 10 | 267 ± 10 | 17 ± 1.0 | 16 |
| Nitrocefin | 333 ± 30 | 20 ± 1.8 | 17 | 867 ± 40 | 21 ± 1.5 | 41 |
| Cefoxitinb | 0.28 ± 0.02 | 0.17 ± 0.02 | 1.6 | 0.35 ± 0.04 | 0.15 ± 0.02 | 2.3 |
| Cefotaximeb | 1.7 ± 0.1 | 0.3 ± 0.02 | 5.7 | <0.01 ± 0.002 | 0.005 ± 0.001 | <2 |
| Ceftazidimeb | 0.4 ± 0.05 | 0.14 ± 0.02 | 2.9 | 0.01 ± 0.002 | 0.02 ± 0.003 | 0.5 |
Values are the means of four independent measurements.
For these β-lactams, the Km value was determined as a Ki value by using cephalothin as the reporter substrate.
Inhibitory activities of cloxacillin, aztreonam, Ro 48-1220, and tazobactam were assessed using cephalothin as a reporter substrate (17) and expressed as Ki values. The most potent inhibitor for both cephalosporinases was cloxacillin, followed, in descending order, by aztreonam and Ro 48-1220, as is typical for this enzyme group. CMY-30, however, was less susceptible to inhibition by cloxacillin and aztreonam than CMY-2. Tazobactam was the weakest inhibitor for both enzymes (Table 3).
TABLE 3.
Inhibition profile of CMY-30 cephalosporinase and comparison with CMY-2
| Inhibitor |
Ki (μM) for:
|
|
|---|---|---|
| CMY-30 | CMY-2 | |
| Cloxacillin | 2.3 × 10−3 | 0.2 × 10−3 |
| Aztreonam | 17 × 10−3 | 2.1 × 10−3 |
| Ro 48-1220 | 200 × 10−3 | 100 × 10−3 |
| Tazobactam | >1,000 × 10−3 | 800 × 10−3 |
(iii) Bioassay for aztreonam hydrolysis.
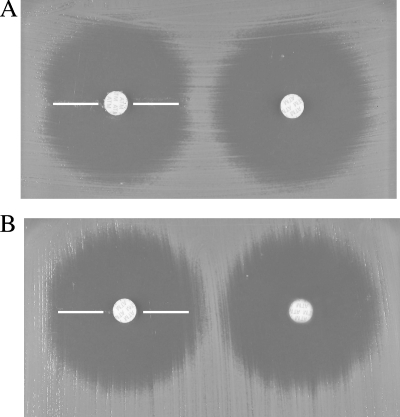
Although inhibition data probably indicated diminished affinity of aztreonam for CMY-30 compared to CMY-2, isogenic MICs showed that the Gly-for-Val211 substitution increased catalytic efficiency against this β-lactam. To indirectly compare aztreonam hydrolysis by CMY-30 and CMY-2, we employed a bioassay using E. coli DH5a as a susceptible indicator strain, aztreonam disks (30 μg), and solutions containing low concentrations of purified cephalosporinases (additional details are included in the legend to Fig. 1). Distortions of the inhibition zones caused by the enzymes indicated a higher efficiency of CMY-30 against aztreonam (Fig. 1).
FIG. 1.
Biological assay comparing the efficacy of CMY-30 (A) with CMY-2 (B) against aztreonam. Suspensions of E. coli DH5α (105 CFU per ml) and disks of aztreonam were applied in MH agar. Solution containing 700 ng of purified cephalosporinase in 10 μl of phosphate buffer was then placed on the culture as shown by the thin white lines. Plates were read after incubation for 18 h at 35°C. A distortion of the inhibition zone was seen with CMY-30. Control aztreonam disks are on the right in each panel.
Modeling studies.
Modeling was based on the crystal structure of AmpC from E. coli (78% identity with CMY-2) bound to ceftazidime (β subunit of Protein Data Bank [PDB] entry 1IEL) (22). CMY models were built by comparative protein modeling by the satisfaction of partial restraints and refined using simulated annealing with the MODELLER software package (5). The stereochemistries of the models were evaluated by the program PROCHECK (10). The root mean square deviation of Cα positions between the CMY-2 model and the crystal structure of CMY-2 bound to citrate (C. Bauvois, L. Jacquamet, S. Fieulaine, J.-M. Frère, M. Galleni, and J.-L. Ferrer, PDB entry 1ZC2) was 0.6 Å. The respective value between the CMY-2 and CMY-30 models was 0.16 Å.
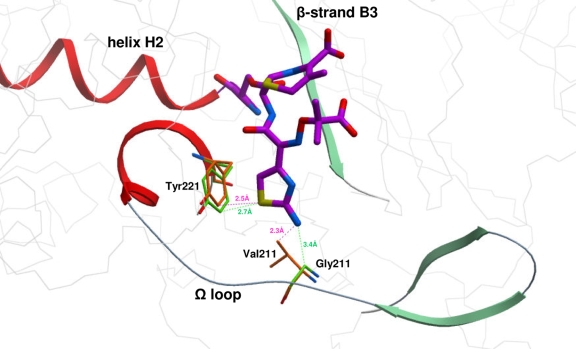
The superpositions of the CMY-2 and CMY-30 models to the AmpC-ceftazidime complex (residues 10 to 350) indicated a similar position of the catalytic Ser64 and only minor differences in the positioning of important residues, including Lys315 and Ser318 located in the β3 strand. Ceftazidime, at the superposed structures, was then modeled to adopt the catalytically competent conformation of loracarbef, a preferable substrate for AmpC (PDB entry 1FCN) (18). Atoms of the open lactam ring of loracarbef as those of the carbacephem ring were used as a template during the positioning of the atoms of the open lactam and dihydrothiazine rings of ceftazidime. A conformational search for the unbound state of ceftazidime was carried out in order to find possible conformations for the oxyimino moiety while keeping the remaining part of the molecule restrained at the positions described above. Of a total of 100 generated structures, those that exhibited acceptable contacts with the residues of the binding pocket and good distances between N10-Ala/Ser318O and O12-Asn152Nδ2 were selected for analysis. The selected solutions that satisfied the criteria described above shared similar conformation. A representative complex structure is shown in Fig. 2. At this potential conformation, N19 of the aminothiazole ring of ceftazidime appeared too close (2.3 Å) to the Cγ2 of Val211 in both CMY-2 and AmpC. Furthermore, the Cδ1 of Tyr221 in CMY-2 and AmpC appeared at short distances from aminothiazole's S16 (2.5 and 2.4 Å, respectively). In CMY-30, however, the R1 side chain of ceftazidime appeared to be more remotely positioned from either Gly211 or Tyr221 (the calculated distance between N19 and Gly211 Cα was 3.4 Å, and that of S16 and Tyr221 Cδ1 was 2.7 Å), therefore reducing the possibility for steric clashes (Fig. 2).
FIG. 2.
Ceftazidime covalently bound to Ser-64 of the superposed structures of CMY-2 and CMY-30. Ceftazidime has been modeled in a catalytically competent conformation similar to that adopted by loracarbef (see text). The view is focused on the possible steric constraints between the aminothiazole substituent of ceftazidime and the Ω loop residues 211 and 221. Calculated distances between N19-CMY2-Val211 Cγ2 (2.3 Å) and S16-CMY2-Tyr221 Cδ1 (2.5 Å) are shown by purple dashed lines. Green dashed lines indicate distances between N19-CMY30-Gly211 Cα (3.4 Å) and S16-CMY30-Tyr221 Cδ1 (2.7 Å). Carbon atoms of CMY-2 residues are colored orange, and those of CMY-30 are colored green. Ceftazidime and Ser64 carbon atoms are colored magenta.
Conclusions.
ESCs and aztreonam, while not preferable substrates for class C β-lactamases, are hydrolyzed at meaningful rates in the periplasm of gram-negative microorganisms (24). Yet, high-level resistance usually requires increased enzyme quantities attained by the upregulation of chromosomal AmpCs or the multiple copies of plasmidic ampC genes (9, 19). The expanded activity of ESACs, apart from a direct effect on resistance to ESCs, may also have a positive impact on fitness under selective pressure by reducing the need for high-level AmpC production. ESACs occur thus far in a sporadic fashion and, apart from plasmid-mediated ACC-4 (17), CMY-19 (25), and CMY-30 (20), are chromosomal variants. This study shows that a single point mutation provides extended-spectrum properties in CMY-2, a cephalosporinase that has achieved global spread through epidemic strains and self-transferable plasmids (19). Moreover, the potential of CMY-2 to broaden its hydrolytic spectrum through changes at diverse positions other than 211 has been documented in a previous study (1). It is therefore plausible to hypothesize that the clinical significance of CMY-type ESACs will probably increase.
In keeping with the Ω loop ESACs from Enterobacter cloacae (4, 14, 16, 26), C. freundii (15, 23), and Serratia marcescens (7, 12), CMY-30 displays reduced affinity for ESCs, while kcat values, roughly reflecting deacylation rates, are high. It is considered that alterations at this domain increase the accessibility of β-lactams with large R1 groups (4, 9, 11, 26). As it was indicated by modeling, the lack of a side chain at position 211 in CMY-30 may abolish steric clashes with the aminothiazole ring of ceftazidime. This, in turn, may enable the antibiotic to adopt conformations that facilitate the deacylation process. Data thus far strongly support the hypothesis for a contribution of the conserved Val211 in determining the substrate specificity of class C β-lactamases (17, 22; this study). Nevertheless, further modeling, mutagenesis, and inhibition studies are required to better clarify the impact of mutations at this residue.
Acknowledgments
We thank Irene Siatravani for technical assistance.
This work was supported by the Hellenic Pasteur Institute.
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Barlow, M., and B. G. Hall. 2003. Experimental prediction of the evolution of cefepime resistance from the CMY-2 AmpC β-lactamase. Genetics 164:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauvois, C., A. Shimizu Ibuka, A. Celso, J. Alba, Y. Ishii, J.-M. Frère, and M. Galleni. 2005. Kinetic properties of four plasmid-mediated AmpC β-lactamases. Antimicrob. Agents Chemother. 49:4240-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crichlow, G. V., A. P. Kuzin, M. Nukaga, K. Mayama, T. Sawai, and J. R. Knox. 1999. Structure of the extended-spectrum class C β-lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry 38:10256-10261. [DOI] [PubMed] [Google Scholar]
- 5.Eswar, N., B. Webb, M. A. Marti-Renom, M. S. Madhusudhan, D. Eramian, M. Y. Shen, U. Pieper, and A. Sali. 2007. Comparative protein structure modeling using MODELLER, p. 2.9.1-2.9.31. In J. E. Coligan, B. M. Dunn, D. W. Speicher, P. T. Wingfield, and H. L. Ploegh (ed.), Current protocols in protein science. John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed]
- 6.Galleni, M., G. Amicosante, and J.-M. Frère. 1988. A survey of the kinetic parameters of class C β-lactamases. Cephalosporins and other β-lactam compounds. Biochem. J. 255:123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hidri, N., G. Barnaud, D. Decre, C. Cerceau, V. Lalande, J. C. Petit, R. Labia, and G. Arlet. 2005. Resistance to ceftazidime is associated with a S220Y substitution in the omega loop of the AmpC β-lactamase of a Serratia marcescens clinical isolate. J. Antimicrob. Chemother. 55:496-499. [DOI] [PubMed] [Google Scholar]
- 8.Honoré, N., M. H. Nicolas, and S. T. Cole. 1989. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol. Microbiol. 3:1121-1130. [DOI] [PubMed] [Google Scholar]
- 9.Jacoby, G. A. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 11.Mammeri, H., and P. Nordmann. 2007. Extended-spectrum cephalosporinases in Enterobacteriaceae. Anti-Infect. Agents Med. Chem. 6:71-82. [Google Scholar]
- 12.Matsumura, N., S. Minami, and S. Mitsuhashi. 1998. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob. Agents Chemother. 42:176-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nordmann, P., and H. Mammeri. 2007. Extended-spectrum cephalosporinases: structure, detection, and epidemiology. Future Microbiol. 2:297-307. [DOI] [PubMed] [Google Scholar]
- 14.Nukaga, M., S. Haruta, K. Tanimoto, K. Kogure, K. Taniguchi, M. Tamaki, and T. Sawai. 1995. Molecular evolution of a class C β-lactamase extending its substrate specificity. J. Biol. Chem. 270:5729-5735. [DOI] [PubMed] [Google Scholar]
- 15.Nukaga, M., S. Kumar, K. Nukaga, R. F. Pratt, and J. R. Knox. 2004. Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotaxime in wild-type and extended spectrum enzymes. J. Biol. Chem. 279:9344-9352. [DOI] [PubMed] [Google Scholar]
- 16.Nukaga, M., K. Taniguchi, Y. Washio, and T. Sawai. 1998. Effect of an amino acid insertion into the omega loop region of a class C β-lactamase on its substrate specificity. Biochemistry 37:10461-10468. [DOI] [PubMed] [Google Scholar]
- 17.Papagiannitsis, C. C., L. S. Tzouvelekis, E. Tzelepi, and V. Miriagou. 2007. Plasmid-encoded ACC-4, an extended-spectrum cephalosporinase variant from Escherichia coli. Antimicrob. Agents Chemother. 51:3763-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patera, A., L. C. Blaszczak, and B. K. Shoichet. 2000. Crystal structure of substrate and inhibitor complexes with AmpC β-lactamase: possible implications for substrate-assisted catalysis. J. Am. Chem. Soc. 122:10504-10512. [Google Scholar]
- 19.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pope, C. E., P. E. Carter, and H. Heffernan. 4 May 2009. CMY-29 and CMY-30: two novel plasmid-mediated AmpC β-lactamases. Antimicrob. Agents Chemother. doi:10.1128/AAC.01586-08. [DOI] [PMC free article] [PubMed]
- 21.Power, P., M. Galleni, J. A. Ayala, and G. Gutkind. 2006. Biochemical and molecular characterization of three new variants of AmpC β-lactamases from Morganella morganii. Antimicrob. Agents Chemother. 50:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers, R. A., E. Caselli, P. J. Focia, F. Prati, and B. K. Shoichet. 2001. Structures of ceftazidime and its transition-state analogue in complex with AmpC β-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207-9214. [DOI] [PubMed] [Google Scholar]
- 23.Tsukamoto, K., R. Ohno, M. Nukaga, and T. Sawai. 1992. The effect of amino acid substitution at position 219 of Citrobacter freundii cephalosporinase on extension of its substrate spectrum. Eur. J. Biochem. 207:1123-1127. [DOI] [PubMed] [Google Scholar]
- 24.Vu, H., and H. Nikaido. 1985. Role of β-lactam hydrolysis in the mechanism of resistance of a β-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum β-lactams. Antimicrob. Agents Chemother. 27:393-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wachino, J., H. Kurokawa, S. Suzuki, K. Yamane, N. Shibata, K. Kimura, Y. Ike, and Y. Arakawa. 2006. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob. Agents Chemother. 50:534-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Z., Y. Yu, J. M. Musser, and T. Palzkill. 2001. Amino acid sequence determinants of extended-spectrum cephalosporin hydrolysis by the class C P99 β-lactamase. J. Biol. Chem. 276:46568-46574. [DOI] [PubMed] [Google Scholar]
- 27.Zioga, A., J. M. Whichard, S. D. Kotsakis, L. S. Tzouvelekis, E. Tzelepi, and V. Miriagou. 2009. CMY-31 and CMY-36 cephalosporinases encoded by ColE1 plasmids. Antimicrob. Agents Chemother. 53:1256-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]