Abstract
In Arabidopsis thaliana, trichome cells are specialized unicellular structures with uncertain functions. Based on earlier observations that one of the genes involved in cysteine biosynthesis (Atcys-3A) is highly expressed in trichomes, we have extended our studies in trichome cells to determine their capacity for glutathione (GSH) biosynthesis. First, we have analyzed by in situ hybridization the tissue-specific expression of the genes Atcys-3A and sat5, which encode O-acetylserine(thio)lyase (OASTL) and serine acetyltransferase (SAT), respectively, as well as gsh1 and gsh2, which encode γ-glutamylcysteine synthetase and glutathione synthetase, respectively. The four genes are highly expressed in leaf trichomes of Arabidopsis, and their mRNA accumulate to high levels. Second, we have directly measured cytoplasmic GSH concentration in intact cells by laser-scanning microscopy after labeling with monochlorobimane as a GSH-specific probe. From these measurements, cytosolic GSH concentrations of 238 ± 25, 80 ± 2, and 144 ± 19 μM were estimated for trichome, basement, and epidermal cells, respectively. Taking into account the volume of the cells measured using stereological techniques, the trichomes have a total GSH content more than 300-fold higher than the basement and epidermal cells. Third, after NaCl treatment, GSH biosynthesis is markedly decreased in trichomes. Atcys-3A, sat5, gsh1, and gsh2 mRNA levels show a decrease in transcript abundance, and [GSH]cyt is reduced to 47 ± 5 μM. These results suggest the important physiological significance of trichome cells related to GSH biosynthesis and their possible role as a sink during detoxification processes.
Keywords: cysteine, glutathione, two-photon laser-scanning microscopy, O-acetylserine(thiol)lyase, sulfate assimilation
Trichomes are specialized unicellular or multicellular structures derived from the epidermal cell layer. Multicellular trichomes occur in many different species and often form glands that secrete various compounds, including organic acids, polysaccharides, terpenes, nectar, or salt (1). In Arabidopsis thaliana, trichomes form on the stem and leaf surface to an extent depending on the ecotype. Leaf trichomes originate from a single epidermal cell that differentiates to form a characteristic three-branched structure with unknown function (2, 3). Recently, it has been demonstrated that natural enemies impose selection for increased trichome density in Arabidopsis (4). However, it is not known whether these nonglandular trichomes produce toxic molecules against pest attack or act as a simple physical barrier to animals trying to cross the leaf surface.
There is also evidence that trichomes may play additional or alternative roles in both the detoxification of heavy metals and in responses to various other stress conditions. In Indian mustard and Alyssum lesbiacum, heavy metals such as cadmium and nickel are preferentially accumulated in leaf trichomes (5, 6), respectively, and metallothionein-like genes are also expressed predominantly in trichomes of Arabidopsis and Vicia faba (7, 8). Histochemical analysis of plants treated with salt, mannitol, or abscisic acid (ABA) shows that significant activity of a βglucuronidase (GUS) reporter gene can be driven in trichomes by promoters from stress-regulated genes, such as the osmotin osm, the cold-induced kin1, and cor6.6 and the desiccation-responsive rd29 (9–11).
Recently, we have shown that a gene involved in the synthesis of cysteine, Atcys-3A, is also highly expressed in Arabidopsis trichomes (12, 13). This gene encodes the cytosolic isoform of O-acetylserine(thiol)lyase, the enzyme that catalyzes the formation of cysteine. Synthesis of cysteine is the final step in the sulfate assimilation pathway that is typically associated with photosynthetic cells (14) and is one of the key precursor steps required for synthesis of glutathione (GSH) (15). The reduced form of glutathione is synthesized in two ATP-dependent steps: first, γ-glutamylcysteine (γ-EC) is synthesized from l-glutamate and cysteine, catalyzed by γ-glutamylcysteine synthetase (γ-ECS); second, glycine is added to the C-terminal end of γ-EC in a reaction catalyzed by glutathione synthetase (GS) (16).
In plants, glutathione is the predominant non-protein thiol and has physiologically important roles acting as a mobile pool of reduced sulfur (17), as an antioxidant in stress responses, and in the regulation of plant growth and development (15). Glutathione also plays an important physiological role in detoxification by targeting xenobiotics and toxic compounds (cytotoxins) into the vacuole. Cytotoxins are conjugated with GSH by the action of glutathione S-transferases and are transported from the cytosol to the vacuole by specific glutathione S-conjugate (GS-X) pumps. This mechanism functions in herbicide detoxification, cell pigmentation, storage of antimicrobial compounds, and regulation and transport of heavy metal chelates (18).
In this article, we have extended our studies on trichome cells to determine, first, whether high levels of all GSH biosynthetic enzymes are present in the trichomes; second, whether this biosynthetic capacity leads to constitutively higher levels of cytoplasmic GSH; and, third, whether levels of gene expression and GSH are affected by salt stress.
Experimental Procedures
Plant Material and Treatments.
Arabidopsis thaliana var. Columbia plants were grown on moist vermiculite supplemented with Hoagland medium or soil at 20°C in light and 18°C in dark, under a 16-h white light/8-h dark photoperiod. Thirty days after germination, plants were subjected to salt stress treatments by adding Hoagland medium supplemented with NaCl (0.17 M) for 1–4 days as indicated. For some GSH measurements, the seeds were also germinated on phytagel plates (Murashige and Skoog basal medium supplemented with 1% sucrose and 1% phytagel, Sigma) for 10–15 days with or without NaCl (0.17 M).
In Situ Hybridizations.
Probes for in situ hybridization were labeled with uridine 5′-[α-thio][35S]triphosphate. The gene-specific probes used were as follows: Atcys-3A cDNA fragment for the cytosolic isoform of O-acetylserine(thiol)lyase (X84097) (12); sat5 cDNA fragment for a putative cytosolic serine acetyltransferase (Z34888) (19); gsh1 cDNA fragment for the γ-glutamylcysteine synthetase (Z29490) (20), and gsh2 for the glutathione synthetase (X83411) (21). The cDNA fragments were amplified by PCR by using standard M13 reverse and forward primers and subcloned into pBluescriptII KS. About 1 μg of the PCR product was used as template to synthesize 35S-labeled RNA using T7 RNA polymerase or T3 RNA polymerase.
Mature Arabidopsis plants (30 days) grown under standard conditions or treated with 0.17 M NaCl for 24 h were used for in situ hybridization. The leaves were cut into small pieces, fixed in 4% (vol/vol) formaldehyde, and embedded in wax, and sections were prepared as described (12). To enhance penetration into the tissue sections, the probes were partially hydrolyzed to an average length of 150–200 nt by heating at 60°C in 0.1 M carbonate buffer (pH 10.2). The hybridization protocol was followed according to Cox and Golberg (22). For autoradiography, slides were coated with Amersham Hypercoat LM-1 nuclear emulsion and exposed 5–10 days at 4°C. The samples were then developed in Kodak D19 developer prechilled at 14°C, and fixed in 30% (wt/vol) sodium thiosulfate for 5 min. After developing, the tissue sections were stained with 0.05% (wt/vol) toluidine blue in water for 0.5 min, rinsed, dehydrated, and air dried.
For comparison, the tissue sections from control and stressed plants were fixed onto a single glass slide and hybridized with the same labeled probe.
Glutathione Determination by Laser Scanning Microscopy.
GSH levels were visualized in intact trichome cells of Arabidopsis after in situ conjugation with monochlorobimane (MCB) or monobromobimane (MBB) (Molecular Probes) to give a fluorescent GSH-bimane (GSB) adduct. Leaves were carefully cut into small pieces (4–9 mm2), and the pieces were placed into an Eppendorff tube containing 1 ml of 100 μM MCB, 50 μM propidium iodide (PI), and 5 mM sodium azide. PI was included to label the cell walls and nuclei in dead cells. Azide was used to deplete ATP levels and thereby prevent ATP-dependent vacuolar sequestration of GSB. The leaves were vacuum infiltrated with this solution for 5 min and incubated for further 20 min in the same solution after release of the vacuum. Leaf pieces were either washed briefly in distilled water before gently securing the abaxial surface of the leaf with double-sided adhesive tape to the base of a 60-mm Petri dish or mounted in the dye solution on a slide using a spacer between slide and coverslip to avoid squashing the trichomes. Samples were observed either with an Olympus universal plan fluorite (LUMPLFL) ×60 0.9 numerical aperture (NA) water immersion objective or an Olympus universal plan apochromat (UPLAPO) ×60 1.2 NA water immersion objective, on an Olympus BX50WI microscope attached to a Bio-Rad MRC-1024MP laser scanning microscope (Bio-Rad).
The fluorescent GSB conjugate was visualized in either single optical sections or serial optical sections of leaves. For two-photon laser scanning microscopy, GSB was excited at 770 nm, with pulse widths of approximately 100 femtoseconds by using a Tsunami mode-locked Ti:Sapphire laser (Spectra-Physics) with a 5-W pump laser (Millenia V, Spectra-Physics). Emitted light was collected on internal detectors with open pinholes by using Bio-Rad filter blocks TS1 (650DCSPXR Special) and T2A (560DRLP) and emission filters 585LP (PI, red) and 522DF35 (GSB, green). For confocal laser-scanning microscopy, GSB was excited with a HeCd laser (Liconix, Sunnyvale, CA) at 442 nm and detected with the same combination of emission filters. Typically, the pixel spacing was 0.2–0.3 μm, and each section was Kalman filtered over 3–4 frames. Average fluorescence levels were measured from user-defined regions of interest (ROI) in each optical sections by using LaserSharp software (Bio-Rad). After subtraction of the average background signal, fluorescence intensities were calibrated against known concentrations of GSB. A 10-mM stock solution of GSB was prepared by nonenzymatic conjugation of excess GSH to 10 mM monobromobimane and imaged with identical instrument settings. Measurements of total cell volume were performed by using Digital Stereology (Kinetic Imaging, Liverpool, U.K.). The Cavalieri estimator of volume was applied to stacks of optical (x, y) sections collected with a z-distance of 1 μm. For the analysis, every third section was taken, starting from a randomized position. Image montages were assembled with Photoshop (Adobe Systems, Mountain View, CA).
Results
Trichome-Specific Expression of Cysteine and Glutathione Biosynthesis Genes.
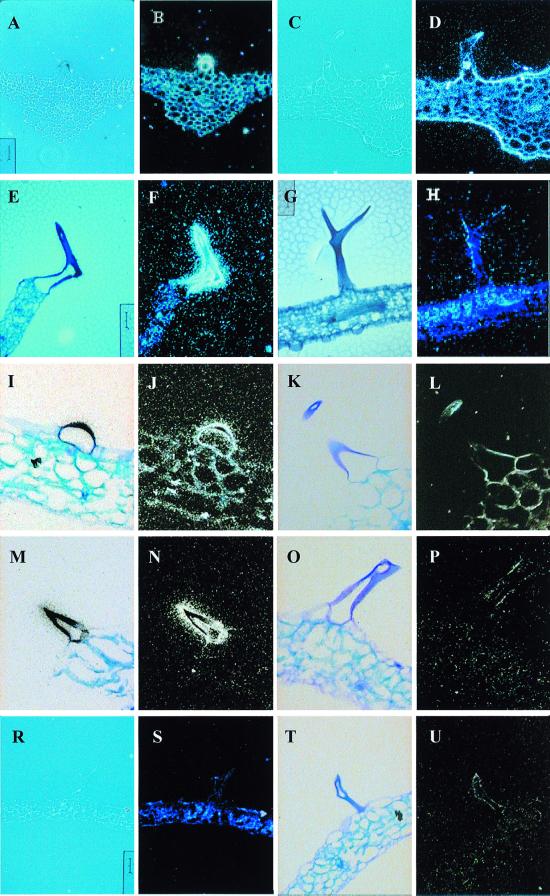
Analysis by in situ hybridization of the tissue-specific expression of the Atcys-3A gene from Arabidopsis thaliana showed a high level of expression in stem and leaf trichomes (12). Further analysis of sat5, which encodes for the enzyme that provide the carbon skeleton for cysteine biosynthesis, showed the same pattern of expression as Atcys-3A, with both transcripts being highly abundant in trichome cells (Fig. 1 A and B and E and F). The positive hybridization signal was detected as black spots under bright field and as white bright spots under dark field. We observed a high level of transcript throughout the different stages of trichome development, although only one representative trichome image is shown.
Figure 1.
In situ transcript localization in leaf trichomes. Transverse sections (10 μm) of Arabidopsis thaliana leaf were hybridized with 35S-labeled antisense (A–P) or sense (R–U) RNA probes. Positive signal is detected as black spots under bright field and as white spots under dark field. The effect of NaCl on transcript abundance was studied in leaves from plants that were treated with 170 mM NaCl for 24 h before fixation. (A) Bright- and (B) dark-field micrograph of untreated leaf, hybridized with antisense Atcys-3A 35S-RNA. (C) Bright- and (D) dark-field micrograph of salt-treated leaf, hybridized with antisense Atcys-3A 35S-RNA. (E) Bright- and (F) dark-field micrograph of untreated leaf, hybridized with antisense sat5 35S-RNA. (G) Bright- and (H) dark-field micrograph of salt-treated leaf, hybridized with antisense sat5 35S-RNA. (I) Bright- and (J) dark-field micrograph of untreated leaf, hybridized with antisense gsh1 35S-RNA. (K) Bright- and (L) dark-field micrograph of salt treated leaf, hybridized with antisense gsh1 35S-RNA. (M) Bright- and (N) dark-field micrograph of untreated leaf, hybridized with antisense gsh2 35S-RNA. (O) Bright- and (P) dark-field micrograph of salt-treated leaf, hybridized with antisense gsh2 35S-RNA. (R) Bright- and (S) dark-field micrograph of untreated leaf, hybridized with sense Atcys-3A 35S-RNA. (T) Bright- and (U) dark-field micrograph of untreated leaf, hybridized with sense gsh2 35S-RNA.
Cysteine, the final metabolite of the sulfur assimilation pathway, is rapidly metabolized into glutathione, the major non-protein thiol within the cells. To analyze the glutathione biosynthesis pathway in trichome cells, in situ hybridization with gene-specific probes against gsh1 and gsh2 was also performed. Both of these genes were also expressed to a high level in leaf trichomes of mature Arabidopsis plants (Fig. 1 I and J and M and N, respectively). Thus, the mRNA encoding the four genes responsible for cysteine and glutathione biosynthesis in Arabidopsis are all highly expressed in leaf trichomes. Cross sections of leaves hybridized with sense probes for Atcys-3A (Fig. 1 R and S) and gsh2 (Fig. 1 T and U) are also included in the figure to show the background signal that was routinely obtained.
Glutathione Determination in Trichomes Cells.
The high level of transcript expression detected in trichome cells for the genes involved in cysteine and GSH biosynthesis suggests that these thiols could be produced to a high level in this particular plant cell type. To address this point, we measured the level of GSH in the cytoplasm by two-photon laser-scanning microscopy (23) or confocal laser scanning microscopy, after conjugation with MCB to give a fluorescent GSB conjugate (24). This technique allows quantitative measurements of GSH in intact tissues with subcellular resolution (25–27). Monochlorobimane is cell permeant and nonfluorescent until conjugated to thiol groups, when it becomes fluorescent with an excitation peak at 395 nm and membrane impermeant. In plant tissues, rates of conjugation are negligible unless catalyzed by a glutathione S-transferase, which confers specificity for GSH (24). To prevent subsequent transfer of GSB to the vacuole, sodium azide (NaN3) was included in the labeling protocol to prevent ATP synthesis and thereby inhibit the ATP-dependent vacuolar GS-X conjugate pump (27). In leaf trichomes, GSB fluorescence increased in the cytoplasm to a plateau over about 20 min after the start of labeling and was visualized by using two-photon laser-scanning microscopy (pseudocolored green in Fig. 2) together with PI (pseudocolored red in Fig. 2). PI labeled the cell walls and also served as a marker for dead or dying cells when the dye gained access to the nucleus and labeled the DNA (26, 27). The absence of nuclear PI signal indicated that the cells remained viable during the loading and imaging period.
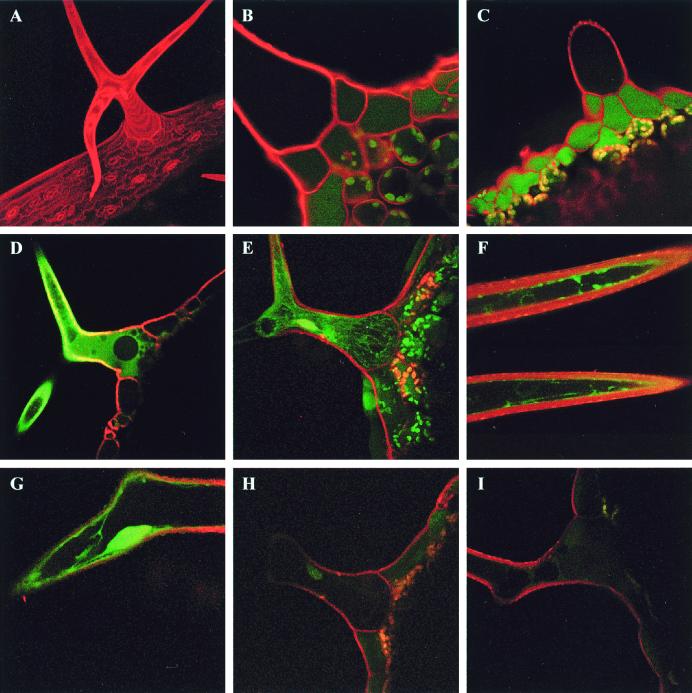
Figure 2.
Measurement of cytoplasmic GSH levels by two-photon laser scanning microscopy. GSH was visualized in intact trichome cells of Arabidopsis after in situ conjugation with MCB to give a fluorescent GSH-bimane conjugate. (A) Maximum projection of 21 optical sections of a leaf trichome stained with 50 μM PI. (B) Single optical section of a leaf trichome labeled with PI showing the low levels of autofluorescence in the trichome itself. (C) Leaf trichome labeled with MCB and PI in the absence of azide. Relatively little labeling is apparent in the trichome because the GSB has been efficiently transported from the cytoplasm into the vacuole, where the massive dilution effectively reduces the concentration to near background levels. (D–G) Representative single optical sections (D, F, and G) or a maximum projection of 20 optical sections (E) of leaf trichomes labeled with MCB and PI in the presence of 5 mM sodium azide to inhibit vacuolar sequestration of the GSB. Fluorescence is visible in the thin peripheral layer of cytoplasm adjacent to the cell wall and in cytoplasmic strands crossing the vacuole. (H and I) Representative images of leaf trichomes from salt-treated plants, labeled with MCB and PI in the presence of 5 mM sodium azide
GSB signal was localized in a thin layer of cytoplasm underneath the PI-stained cell wall, in cytoplasmic strands crossing the vacuole, and in the nucleus (Fig. 2 D–G). No detectable signal above background was found in the vacuole in the presence of azide. Imaging of nonstained leaf trichomes under the same laser-power and instrument settings did not show measurable autofluorescence in the cytoplasm or vacuoles of the trichome; however, significant blue-green autofluorescence was observed in the vacuole of the specialized basement cells surrounding the trichome (Fig. 2B) as well as other epidermal cells when excited with two-photon excitation at 770 nm (data not shown).
Calibration of the GSB fluorescence intensity against standard GSB concentrations imaged under identical conditions gave estimates of [GSH]cyt from 100 to 800 μM (238 ± 25 μM, n = 43 from 11 independent plants). For comparison, the cytoplasmic GSH concentration was also measured in trichome, basement, and epidermal cells by using confocal laser-scanning microscopy with excitation at 442 nm to avoid the autofluorescence produced by the two-photon laser in basement and epidermal cells. The results in Table 1 indicate that trichome cells showed 1.6- and 3.0-fold higher cytoplasmic GSH concentrations than epidermal and basement cells, respectively. The volume of trichome cells was approximately 200-fold higher than the other epidermal cells. Assuming the cytoplasm-to-vacuole ratio is similar in these cell types (1:9), the total GSH content of trichomes is approximately 300- and 600-fold higher than in epidermal or basement cells.
Table 1.
Volume and cytoplasmic glutathione determination in trichome, epidermis, and trichome basement Arabidopsis leaf cells.
| Cells | Volume, mm3 | Control plants GSH, μM | Salt-treated plants GSH, μM |
|---|---|---|---|
| Trichome | 3.8 ± 0.8 × 10−3 | 238 ± 25 | 47 ± 5 |
| Epidermis | 2 ± 0.3 × 10−5 | 144 ± 19 | 266 ± 22 |
| Basement | 2 ± 0.3 × 10−5 | 80 ± 2 | 142 ± 14 |
Values are presented as mean ± SEM for 30–43 measurements (GSH) or means ± SD for 9 measurements (volume).
Affects of Salt Stress on Transcript Abundance and GSH Levels.
We have observed previously that the expression of the Atcys-3A gene is induced in bulk leaf tissues under salt stress and that this induction is under the control of the hormone ABA (13). In contrast, the normally high levels of Atcys-3A expression in trichomes was decreased under salt stress (ref. 13 and Fig. 1 C and D). In situ hybridization of the sat5, gsh1, and gsh2 mRNA in trichomes also showed a decrease in transcript abundance in plants treated with 170 mM NaCl for 24 h (Fig. 1 G and H, K and L, and O and P, respectively). Furthermore, treatment of Arabidopsis plants with NaCl substantially reduced cytoplasmic GSH concentrations in trichomes to the range of 10–100 μM (47 ± 5 μM, n = 30 from seven independent plants) (Fig. 2 H and I). Taken together, these data suggest that, under salt stress, control GSH biosynthesis is markedly different between adjacent cells in the leaf. The ratio of GSH content in trichome/epidermal and trichome/basement cells increases by 30- and 60-fold, respectively (Table 1).
To eliminate the possibility that changes in fluorescence in different cell types and in response to salt treatment were due to differences in glutathione S-transferase activity, required for conjugation of MCB, leaves were also labeled with monobromobimane, which does not require glutathione S-transferase activity for labeling. In this case, the labeling pattern and changes in response to salt were similar to those observed with MCB (data not shown).
Discussion
The cell-specific studies performed by in situ hybridization and laser-scanning microscopy have demonstrated a high level of GSH in Arabidopsis leaf trichomes in comparison with other mature leaf cells. These results may have two important physiological significances. First, it suggests that the glutathione biosynthesis pathway is highly active in this cell type. In situ hybridization experiments indicate that transcripts of all four genes required for cysteine and glutathione biosynthesis are highly abundant in trichomes. Although the reduction and assimilation of sulfate into cysteine is typically assigned to photosynthetically active plastids, in recent years it has become evident that nonphotosynthetic tissues contribute significantly to the sulfur assimilatory pathway (28, 29). Because trichomes do not contain green plastids, sulfate may be reduced to sulfide in non-green plastids, with the reducing power supplied from carbohydrate oxidation via the pentose phosphate pathway (29). The produced sulfide can then be incorporated by the O-acetylserine(thiol)lyase and SAT enzymes into cysteine, which, in turn, is used for GSH biosynthesis. We cannot exclude the possibility that sulfide or other intermediates in the pathway can be provided by the basement cells; nevertheless, irrespective of the source, glutathione is synthesized in trichomes.
The second important point of physiological significance arising from our observations is related to trichome function. The unicellular trichomes in Arabidopsis seem to be dispensable because mutants lacking trichomes are able to grow normally (gl1 mutants) (30). The total amount of GSH observed in trichomes, over 300-fold greater than in other epidermal cells, could be related to a cell-specific function. Although the concentration of GSH in chloroplasts has been measured as 1–5 mM (31), there have been relatively few previous estimates of GSH levels in the cytosol of aerial tissues. A value of 60 μM GSH was calculated for the cytosol of tobacco mesophyll cells (31) and 144 and 80 μM for the Arabidopsis epidermal and basement cells surrounding the trichome (this work), respectively, which is significantly lower than the average concentration (238 μM) determined for the trichome cytoplasm. Because trichomes cells are considerable larger than epidermal and basement cells, the differences in total GSH content are even greater. Even with the subcellular resolution achieved by using two-photon optical sectioning, the values measured in this study may still underestimate the true cytosolic GSH concentration if there are subresolution negatively stained organelles contained within the volume sampled by the two-photon or confocal optical probe. It is difficult to determine the precise size of this probe in an intact biological specimen (32); however, we would expect it to be roughly ovoid, with dimensions of 400 nm in the (x, y)-plane and 1200 nm along the z-axis.
Considering the size of the trichome cells, with an estimated volume of 3.8 × 10−3 mm3, and that the vacuole may account for 90–95% of the total volume, each trichome has a substantial storage capacity to act as a sink during detoxification of xenobiotics and/or heavy metals. From our data, it seems likely that the trichome functions efficiently as a detoxification system under normal growth conditions, removing cytotoxins and xenobiotic compounds (including MCB) via GSH conjugation and sequestration in the vacuole. This mechanism was observed directly in this study when the GSB conjugate was transferred completely into the vacuole in the absence of azide in the labeling medium.
What is far less clear is why this system should be coordinately down-regulated under salt-stress conditions, particularly when the overall level of Atcys-3A and sat5 transcript abundance increase by 3-fold and cys/GSH levels by 2- and 2.5-fold, respectively, in other cell types in the leaf lamina of salt stressed plants (13). In addition, GSH content in basement and epidermal cells also increased on salt treatment (Table 1). One hypothesis could be that sodium ions are accumulated in the trichome cells to toxic levels during salt stress and progressively inhibit the transcriptional and biosynthetic machinery of the cell. We suggest that plant response to NaCl may trigger a specific inhibitory response in trichomes to limit the high expression of these genes to a basal level for metabolic maintenance, whereas increased transcription is simultaneously induced in other cell types. Although we do not yet appreciate the full functional significance of these shifts in metabolic poise, the combination of molecular biology and in situ imaging techniques are providing powerful new tools to analyze complex regulatory pathways at a single cell level.
Acknowledgments
We thank Dr. John L. Wray (Univerity of St. Andrews) for sat5 probe, Drs. Pascaline Ullmann and Laurence Gondet (Centre National de la Recherche Scientifique, Strasbourg) for the gsh1 and gsh2 probes, and Mr. Nick White and the Bio-Rad Biological Microscopy Unit for help with the two-photon laser scanning microscopy. This work was funded by Dirección General de Investigación Cientifica y Ténica Grant PB96-1367 (Spain), and Junta de Andalucia (Spain) Grant CVI0118. G.G.-A. and L.C.R. are indebted to the COST-829 Action and the European Molecular Biology Organization (EMBO) for short-term fellowships.
Abbreviations
- GSH
glutathione
- MCB
monochlorobimane
- GSB
glutathione-bimane
- PI
propidium iodide
- SAT
serine acetyltransferase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190334497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190334497
References
- 1.Paniagua R, Nistal M, Sesma P, Alvarez-Uria M, Fraile B. Citologia e Histologia Vegetal y Animal. Interamericana, Madrid: McGraw-Hill; 1993. pp. 619–777. [Google Scholar]
- 2.Hülskamp M, Miséra S, Jürgens G. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 3.Larkin J C, Young N, Prigge M, Marks M D. Development. 1996;122:997–1005. doi: 10.1242/dev.122.3.997. [DOI] [PubMed] [Google Scholar]
- 4.Mauricio R, Rausher M D. Evolution. 1997;51:1435–1444. doi: 10.1111/j.1558-5646.1997.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 5.Salt D E, Prince R C, Pickering I J, Raskin I. Plant Physiol. 1995;109:1427–1433. doi: 10.1104/pp.109.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kramer U, Grime G W, Smith J A C, Hawes C R, Baker A J M. Nucl Instr Methods Phys Res. 1997;130:345–350. [Google Scholar]
- 7.García-Hernández M, Murphy A, Taiz L. Plant Physiol. 1998;118:387–397. doi: 10.1104/pp.118.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foley R C, Singh K B. Plant Mol Biol. 1994;26:435–444. doi: 10.1007/BF00039552. [DOI] [PubMed] [Google Scholar]
- 9.Kononowicz A K, Nelson E, Singh N, Hasegawa P M, Bressan R A. Plant Cell. 1992;4:513–524. doi: 10.1105/tpc.4.5.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Cutler A J. Plant Mol Biol. 1995;28:619–634. doi: 10.1007/BF00021188. [DOI] [PubMed] [Google Scholar]
- 11.Yamagushi-Shinozaki K, Shinozaki K. Mol Gen Genet. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- 12.Gotor C, Cejudo F J, Barroso C, Vega J M. Plant J. 1997;11:347–352. doi: 10.1046/j.1365-313x.1997.11020347.x. [DOI] [PubMed] [Google Scholar]
- 13.Barroso C, Romero L C, Cejudo F J, Vega J M, Gotor C. Plant Mol Biol. 1999;40:729–736. doi: 10.1023/a:1006285016296. [DOI] [PubMed] [Google Scholar]
- 14.Barroso C, Romero L C, Vega J M, Gotor C. Curr Top Phytochem. 1997;1:19–29. [Google Scholar]
- 15.May M J, Vernoux T, Leaver C, Van Montagu M, Inzé D. J Exp Bot. 1998;49:649–667. [Google Scholar]
- 16.Noctor G, Arisi A C M, Jouani L, Kunert K J, Rennenberg H, Foyer C H. J Exp Bot. 1998;49:623–647. [Google Scholar]
- 17.Herschbach C, Jouanin L, Rennenberg H. Plant Cell Physiol. 1998;39:447–451. [Google Scholar]
- 18.Rea P A, Li Z-S, Lu Y-P, Drozdowicz Y M, Martinoia E. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:727–760. doi: 10.1146/annurev.arplant.49.1.727. [DOI] [PubMed] [Google Scholar]
- 19.Ruffet M-L, Lebrun M, Droux M, Douce R. Eur J Biochem. 1995;227:500–509. doi: 10.1111/j.1432-1033.1995.tb20416.x. [DOI] [PubMed] [Google Scholar]
- 20.May M J, Leaver C J. Proc Natl Acad Sci USA. 1994;91:10059–10063. doi: 10.1073/pnas.91.21.10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullmann P, Gondet L, Potier S, Bach T J. Eur J Biochem. 1996;236:662–669. doi: 10.1111/j.1432-1033.1996.00662.x. [DOI] [PubMed] [Google Scholar]
- 22.Cox K H, Golberg R B. In: Plant Molecular Biology: A Practical Approach. Shaw C H, editor. Oxford: IRL; 1988. pp. 1–34. [Google Scholar]
- 23.Denk W, Strickler J, Webb W W. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 24.Coleman J O D, Randall R, Blake-Kalff M M A. Plant Cell Environ. 1997;20:449–460. [Google Scholar]
- 25.Sánchez-Fernández R, Fricker M, Corben L B, White N S, Sheard N, Leaver C J, Van Montagu M, Inzé D, May M J. Proc Natl Acad Sci USA. 1997;94:2745–2750. doi: 10.1073/pnas.94.6.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer A J, Fricker M D. J Microsc. 2000;198:174–181. doi: 10.1046/j.1365-2818.2000.00697.x. [DOI] [PubMed] [Google Scholar]
- 27.Fricker M D, May M, Meyer A J, Sheard N, White N S. J Microsc. 2000;198:162–173. doi: 10.1046/j.1365-2818.2000.00696.x. [DOI] [PubMed] [Google Scholar]
- 28.Barroso C, Vega J M, Gotor C. J Physiol Biochem. 1998;54:189–194. [PubMed] [Google Scholar]
- 29.Hell R. Planta. 1997;202:138–148. doi: 10.1007/s004250050112. [DOI] [PubMed] [Google Scholar]
- 30.Marks M D. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:137–163. doi: 10.1146/annurev.arplant.48.1.137. [DOI] [PubMed] [Google Scholar]
- 31.Rennenberg H. Phytochemistry. 1982;21:2771–2781. [Google Scholar]
- 32.White N S, Errington R J, Fricker M D, Wood J L. J Microsc. 1996;181:99–116. [Google Scholar]