Abstract
Although flaviviruses cause significant human diseases, no effective therapy is currently available. Host factors essential for viral replication are potential targets for antiviral development. Here we report that cyclophilins (CyPs), a family of cellular peptidyl-prolyl isomerases (PPIases), play a role in flavivirus replication. Huh-7.5 cells with knockdown of different isoforms of CyP were less efficient than parental cells in supporting flavivirus replication, including West Nile virus (WNV), dengue virus, and yellow fever virus. The low viral replication in CyP A (CyPA) knockdown cells could be rescued by trans supplying of a wild-type CyPA but not by trans supplying of a mutant CyPA (defective in the PPIase activity), indicating that the isomerase activity of CyPA is critical for viral replication. Immunoprecipitation and biochemical pulldown analyses showed that CyPA interacts with WNV genomic RNA and viral NS5 protein in the replication complex. Furthermore, antiviral experiments demonstrated that cyclosporine (Cs; an 11-amino-acid cyclic peptide known to block the PPIase activity of CyPA) inhibits flavivirus replication in cell culture at nontoxic concentrations. Time-of-addition and transient replicon results indicated that Cs inhibits flavivirus at the step of viral RNA synthesis. Biochemical analysis showed that Cs directly blocks the interaction between CyPA and WNV NS5 protein. Our results suggest that host CyPA is a component of flavivirus replication complex and could be targeted for potential antiviral development.
The family Flaviviridae includes three genera: Flavivirus, Pestivirus, and Hepacivirus. Many members from the genus Flavivirus are arthropod-borne and cause significant human morbidity and mortality, such as the four serotypes of dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus. The DENV alone poses a risk to 2.5 billion people worldwide and causes 50 to 100 million human cases each year. YFV and JEV infect 200,000 and 50,000 people every year, respectively (17). Since the outbreak of WNV in New York City in 1999, the virus has caused thousands of human infections in the United States, representing the largest meningoencephalitis outbreak in the Western Hemisphere and the biggest WNV outbreak ever reported (23). No effective antiviral therapy has been approved for clinical treatment of flavivirus infections. Human vaccines are currently available only for YFV, JEV, and TBEV (17). Understanding the molecular mechanism of viral replication is essential for the prevention and treatment of flavivirus infections.
Flaviviruses are spherical in shape, with a diameter of 50 nm (24). The viral genome is a single-stranded, plus-sense RNA, of about 11,000 nucleotides (nt) in length (27). The single open reading frame of the viral genome encodes a polyprotein, which is processed by viral and cellular proteases into three structural proteins (capsid [C], premembrane [prM] or membrane [M], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (27). The structural proteins are involved in viral particle formation. The NS proteins are responsible for viral RNA replication but have also been shown to function in viral assembly (25, 30, 39) and to evade host immune response (3, 18, 29, 37, 38). NS3 acts as a protease (with NS2B as a cofactor) (15, 26), a nucleotide triphosphatase (51, 54), an RNA triphosphatase (2), and a helicase (6). NS5 functions as a methyltransferase (13, 43, 57) and an RNA-dependent RNA polymerase (RdRp) (1, 19). Upon translation, viral NS proteins and host factors form the replication complex to synthesize minus-sense RNA, which, in turn, serves as a template for synthesis of more plus-sense RNA (27).
Cyclophilins (CyPs) are cellular peptidyl-prolyl isomerases (PPIases) which catalyze the isomerization of peptide bonds from trans to cis form at proline residues and facilitates protein folding. CyPs were shown to be critical for the replication of human immunodeficiency virus (HIV) and hepatitis C virus (HCV). Specifically, CyP A (CyPA) and CyPB were reported to bind specifically to the HIV-1 Gag polyprotein (33), as well as bind to the HCV NS5B (53, 56). These virus-host interactions have been targeted for antiviral development, as exemplified by cyclosporine (Cs) and its derivatives, which bind to CyPs with high affinity and block its PPIase activity (40). CsA is a 11-amino-acid cyclic peptide that has been used as an immunosuppressant drug in patients after organ transplants (47). The immunosuppression function of Cs is partially explained by the fact that the Cs/CyP complex binds to and inhibits calcineurin, a cellular phosphatase and a key mediator of T-cell activation (28). Cs was found to inhibit HIV-1 (4, 40) and HCV replication (52). Furthermore, analogues of Cs (NIM811 and DEBIO-025) that bind to CyPs with high affinity but lack calcineurin-mediated immunosuppressive activity are currently in clinical development for HCV treatment (16, 35). DEBIO-025 showed a potent anti-HCV effect in patients coinfected with hepatitis C and HIV (16).
In the present study, we show that CyP functions in flavivirus replication. Knockdown of CyP expression in Huh-7.5 cells reduced flavivirus replication; the replication suppression could be rescued by trans supplying of a wild-type (WT) CyPA. An antibody against CyPA could immunoprecipitate WNV RNA from replication complexes, and recombinant CyPA could pull down viral NS5 protein. The functional role of CyPs in viral replication was further supported by the results that Cs inhibited flaviviruses in cell culture. Time-of-addition and transient replicon results indicate that Cs inhibits flavivirus at the step of RNA synthesis. Biochemical analysis showed that Cs could directly suppress the interaction between CyPA and flavivirus NS5 protein. The results suggest that Cs and its clinical candidates are potentially useful for the treatment of flavivirus infections.
MATERIALS AND METHODS
Viruses, compounds, and antibodies.
We used the following viruses: WNV (strain 3356), YFV (17D vaccine strain), DENV-2 (strain New Guinea C), Western equine encephalitis virus (WEEV; strain Cova 746), and vesicular stomatitis virus (VSV; New Jersey serotype). The sources of these viruses were reported previously (42). Cs was purchased from Alexis Corp. and was dissolved in 100% ethanol for antiviral experiments. Antibodies against Myc and CyPA were obtained from Santa Cruz Biotechnology and Biomol International LP, respectively. Monoclonal antibody against WNV NS5 was generated by immunizing mice in-house with recombinant NS5 protein.
CyP knockdown cell lines.
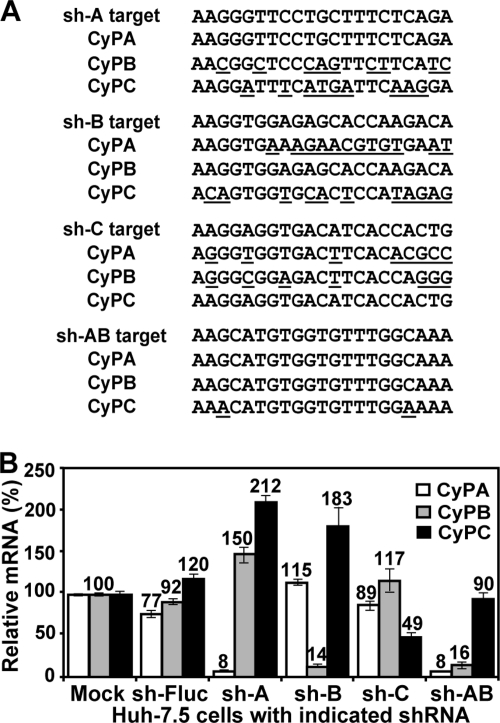
An HIV-based lentiviral vector was used to express all short hairpin RNAs (shRNAs) (50). Huh-7.5 cells were used to establish the following cell lines, each of which contained a stably expressed shRNA directed at a distinct CyP isoform: sh-A161 for CyPA, sh-B710 for CyPB, and sh-C454 for CyPC. A control cell line sh-Fluc, with a shRNA targeting firefly luciferase (Fluc), was also established. These cell lines were generated and reported previously (44, 56). A similar protocol was used in the present study to prepare a new cell line sh-AB, with both CyPA and CyPB knockdown. Figure 1A summarizes the target sequences of sh-A, sh-B, sh-C, and sh-AB and their alignments with the three isoforms of CyP. Stable cells expressing shRNAs were obtained by selection with 1 μg/ml of puromycin (MP Biomedicals) for 3 weeks. The knockdown cells were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum, 0.1 mM nonessential amino acids, and 1 μg/ml of puromycin. Huh-7.5 cells were maintained in a similar medium without puromycin.
FIG. 1.
Characterization of Huh-7.5 cells with knockdown of various CyP isoforms. (A) Sequence alignments of shRNA targets of different CyP isoforms. Mismatched nucleotide were underlined. The sequences of CyPA, CyPB, and CyPC are derived from GenBank accession numbers Y00052 (CyPA), NM_000942 (CyPB), and NM_000943 (CyPC). (B) Silencing of CyP mRNAs in stable Huh-7.5 cells transduced with shRNAs. Equal amounts of total cellular RNA (1 μg) were subjected to real-time RT-PCR using primer-probe sets specific for CyPA, CyPB, CyPC, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (56). The amounts of CyPs in Huh-7.5 cells (mock) were set as 100%. The percentages of CyPs in other cell lines were calculated and indicated after normalization to the GAPDH levels. The data represent means and standard deviations (n ≥ 3).
WNV and DENV-1 replicon-containing cells.
The replicon-harboring Vero cells for WNV and DENV-1 were established previously (32, 42). Both replicons (WNV-Rluc-Neo-Rep and DENV-1-Rluc-Neo-Rep) contained dual reporters, a Renilla luciferase (Rluc) and a neomycin phosphotransferase (Neo). Both replicon cell lines were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum plus 1 mg/ml of G418. All cells in the present study were maintained in 5% CO2 at 37°C.
Packaging of VLPs.
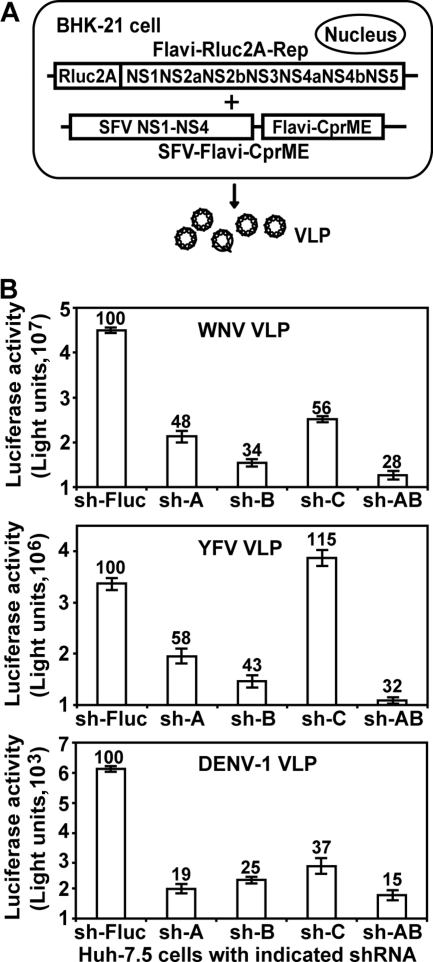
Flavivirus viruslike particles (VLPs) were prepared by trans supplying of viral structural proteins to Renilla luciferase-reporting replicon (Rluc2A-Rep) (46). Briefly, 10 μg of replicon RNAs of WNV, DENV-1, or YFV were electroporated into 8 × 106 BHK-21 cells. Each of the WNV, DENV-1, and YFV replicons has a Renilla luciferase and the foot-and-mouth disease virus 2A sequence substituting for the viral structural genes. The WNV and DENV-1 replicons were described before (31, 41); the YFV replicon was a generous gift from Richard Kuhn (Purdue University) (22). We used an alphavirus Semliki Forest virus (SFV) replicon to express flavivirus structural genes (SFV-CprME-Rep) (22, 41). At 24 h posttransfection (p.t.) of replicon RNAs, the cells were electroporated again with 10 μg of SFV-CprME-Rep RNAs (expressing homologous viral structural proteins). At 24 h after the second transfection, culture fluids were harvested, divided into aliquots, and stored at −80°C. Both flavivirus Rluc2A-Rep RNA and SFV-CprME-Rep RNA were in vitro transcribed from linearized DNAs, using a T7 and a SP6 mMESSAGE mMACHINE kit (Ambion), respectively.
We used indirect immunofluorescence assay (IFA) to estimate the VLP titers. Vero cells in a four-chamber slide (Nalge Nunc International) were infected with serial dilutions of VLP samples, IFA was performed on the infected cells at 24 h postinfection (p.i.), and IFA-positive cell foci were counted. The VLP titers were estimated by the number of positive IFA-positive foci and expressed in focus-forming units (FFU)/ml. Immune mouse ascetic fluid of WNV, DENV-1, and YFV (American Type Culture Collection) and goat anti-mouse immunoglobulin G (IgG) conjugated with Texas Red were used as primary and secondary antibodies, respectively.
Co-IP and RT-PCR.
WNV-Rluc-Neo-Rep Vero cells (5 × 106) were seeded into a T-75 flask 1 day before the coimmunoprecipitation (Co-IP) experiment. After 24 h, the cells were lysed in 1 ml of immunoprecipitation buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 0.5% NP-40). Two hundred units of RNaseOUT (Invitrogen) were added to the supernatant after centrifugation at 12,000 × g for 15 min. The supernatant was then added to 50 μl of 75% protein G slurry containing either anti-CyPA or rabbit IgG. The binding was allowed to proceed at 4°C overnight, after which the protein G-beads were washed with immunoprecipitation buffer four times. RNA was extracted from the beads with an RNeasy kit (Qiagen). Reverse transcription-PCR (RT-PCR) was then performed to detect the RdRp region of WNV NS5, using the primers 8706-forward (5′-CATGGCCATGACTGACACTACTC-3′) and 9093-reverse (5′-CTTGGCCTTTCCGAACTCTCCG-3′) (45).
Viral growth kinetics.
The knockdown cell lines, sh-AB and sh-Fluc, were seeded in a 12-well plate (4 × 105 cells/well). At 24 h after seeding, the cells were infected with WNV, DENV-2, YFV, WEEV, or VSV at an multiplicity of infection (MOI) of 0.1. For VSV, samples of culture medium were collected at 12, 16, and 24 h p.i. For the other four viruses, samples were collected on 16 h, 24 h, 48 h p.i. All virus titers were determined by a double-layer plaque assay on Vero cells, as described previously (42).
Transient replicon assay.
Portions (10 μg) of luciferase replicon (Rluc2A-Rep) RNA of WNV, YFV, or DENV-1 were electroporated into Huh-7.5 cells (46). The cells were seeded in a 12-well plate (4 × 105 cells/well) and immediately treated with 8 μM Cs or treated with 1% ethanol as a control. At various time points, the cells were washed once with cold phosphate-buffered saline (PBS) and added with 250 μ1 of 1× lysis buffer (Promega). The plates containing the lysis buffer were sealed with Parafilm and stored at −80°C. Once the samples for all time points had been collected, 20 μl of cell lysis was transferred to a 96-well plate and assayed for luciferase signals in a Turner BioSystems luminometer (Promega).
Recombinant CyPA production and GST binding assay.
The recombinant protein of CyPA, with a N-terminal glutathione S-transferase (GST) tag, was prepared as described previously (56). For the GST pulldown assay, 20 μg of GST or GST-CyPA was brought to a final volume of 200 μl with binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM dithiothreitol, 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 5% glycerol). WNV-Rluc-Neo-Rep Vero cells (8 × 105) were lysed in 400 μl of immunoprecipitation buffer supplemented with 1 mM dithiothreitol and 1 mM EDTA. Then, 50 μl portions of the cell lysate were added to the tubes containing GST or GST-CyPA. The samples were mixed by rotating at 4°C for 1 h. Glutathione-Sepharose 4B beads (25 μl of a 50% slurry; GE Healthcare) were then added to each sample. After the tubes were rotated at 4°C for 30 min, the beads were washed three times with binding buffer and then pelleted at 500 × g for 5 min. Proteins bound to the beads were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting. To examine the effects of Cs on interaction between CyPA and WNV NS5, we incubated Cs with the recombinant GST-CyPA and replicon cell lysates; the GST pulldown experiment was performed exactly as described above.
Coelectroporation of WNV replicon RNA and plasmid DNA expressing CyPA.
A 5 μg portion of WNV Rluc2A-Rep RNA was mixed with 9 μg of an empty plasmid pcDNA3.1, a plasmid expressing WT CyPA, or a plasmid expressing mutant (MT) CyPA. The MT CyPA contained an Ala substitution for Arg at amino acid position 55 of CyPA, resulting in an inactivation of PPIase (Arg55Ala; AGA→GCA) (58). The N termini of both WT and MT CyPAs were fused with three copies of Myc tag (3Myc) for antibody detection. The RNA and DNA mixtures were electroporated into 7.5 × 106 of sh-A cells, which were then plated onto a 12-well plate (4 × 105 cells per well). The cells were assayed for luciferase activities at various time points p.t. (49).
VLP and virus titer reduction assays.
For the virus titer reduction assay, Huh-7.5 cells were seeded in a 12-well plate (4 × 105 cells per well). At 24 h after seeding, the cells were infected with WNV, DENV-2, YFV, WEEV, or VSV at an MOI of 0.1. The infected cells were immediately treated with Cs at 8 or 20 μM. Virus titers in culture medium were quantified by plaque assays at the indicated time points (42).
MTT assay.
A cell proliferation-based MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay (American Type Culture Collection) was used to examine the compound cytotoxicity. Approximately 2 × 104 Vero cells or Huh-7.5 cells in 100 μl were seeded in a 96 well-plate. After 16 h of incubation, the cells were treated with 0, 2.5, 5, 8, 10, 20, and 40 μM Cs. Cs was dissolved in 100% ethanol and was tested at a 1% final ethanol concentration. After 48 h of incubation, 10 μl of MTT reagent was added to each well. The cells were incubated for another 4 h, after which 100 μl of detergent reagent was added to each well. The plates were swirled gently and left in the dark at room temperature overnight. The absorbance was then recorded in a microliter plate reader (Molecular Devices Corp.) with a 550-nm filter.
Time-of-addition analysis.
Approximately 4 × 105 Huh-7.5 cells were seeded in a 12-well plate per well, followed by incubation for 24 h for cell attachment. The cells were infected with WNV or DENV-2 at an MOI of 3 for 1 h at 4°C, followed by three rounds of cold PBS washes to remove the unabsorbed virus. At different time points p.i., Cs (20 μM for WNV and 8 μM for DENV-2) was added to the infected cells without a medium change. Culture medium were collected at 24 h p.i. and assayed for virus titers by using plaque assays (42). As negative controls, ethanol was added to the infected cells at a final concentration of 1% at 0, 10, and 20 h p.i. to estimate its effect on viral production.
RESULTS
Knockdown of CyP isoforms reduces the replication of flavivirus VLPs.
To examine the role of CyP on flavivirus replication, we prepared a panel of CyP knockdown cell lines using shRNA (Fig. 1). Huh-7.5 cells with individual CyPA, CyPB, or CyPC knockdown were generated by using isoform-specific sh-A, sh-B, and sh-C, respectively; a double-knockdown (CyPA plus CyPB) cell line was established by using sh-AB (Fig. 1A); a control cell line was prepared by using shRNA targeting firefly luciferase (sh-Fluc). Figure 1A summarizes the target sequences of sh-A, sh-B, sh-C, and sh-AB and their alignments with the three isoforms of CyP. Real-time RT-PCR analysis showed a significant difference in mRNA expression of the CyP isoforms in the knockdown cells (Fig. 1B). The mRNA levels for all three CyPs in parental Huh-7.5 (set as 100% for each isoform) were similar to those in sh-Fluc cells. In sh-A cells, the mRNA level of CyPA was reduced by 92%, but the CyPB and CyPC mRNAs were increased by 50 and 112%, respectively. In sh-B cells, CyPB mRNA was decreased by 86%, whereas the CyPC mRNA was increased by 83%. In sh-C cells, the CyPC level dropped only by 51%, and the CyPA and CyPB expression did not change substantially (<20%). In the double-knockdown sh-AB cells, CyPA and CyPB levels were, respectively, decreased by 92 and 84%, and the CyPC level was not dramatically changed (10%). These results indicated that specific isoforms of CyP were knocked down in the cell lines; however, the expression of untargeted isoforms may increase in some of the knockdown cells (e.g., sh-A and sh-B cells), possibly to compensate for the decrease of the sh-RNA-targeted isoform.
We used VLPs containing a luciferase reporter to quantify the effect of CyP knockdown on flavivirus replication. VLPs of WNV, YFV, and DENV-1 were prepared by trans supplying luciferase replicons (Flavi-Rluc2A-Rep) with viral structure proteins (Fig. 2A). The structural proteins were expressed through an alphavirus SFV replicon (SFV-Flavi-CprME). The titers of VLPs were approximately 2.5 × 106, 2 × 105, and 2.4 × 103 FFU/ml for WNV, YFV, and DENV-1, respectively. Because of the dramatic difference in VLP titers, we infected the knockdown cells with WNV, YFV, and DENV-1 VLPs at 1, 0.1, and 0.01 FFU/cell, respectively. Luciferase signals (measured at 24 h p.i.) showed that VLP replication was reduced in almost all knockdown cells (Fig. 2B); the reduction of luciferase signal is statistically significant (Student t test P < 0.01). One exception was observed, in which the replication of YFV VLP in sh-C cells increased by 15% (Fig. 2B). Of the five cell lines, the sh-AB double-knockdown cells yielded the lowest luciferase signals (decreased by 68 to 85%) for all three flavivirus VLPs (Fig. 2B). Changing the FFU/cell of VLP of the above experiments did not substantially affect the relative luciferase activities among different CyP knockdown cells (data not shown). Taken together, the results indicate that CyPs, especially CyPA and CyPB, play a role in flavivirus replication.
FIG. 2.
Effects of CyP knockdown on flavivirus VLP infection. (A) Production of flavivirus VLPs. Flavivirus replicon (Flavi-Rluc2A-Rep) contains a Renilla luciferase gene (Rluc) which substitutes for the deleted viral structural genes. The SFV vector (SFV-Flavi-CprME) was used to express flavivirus structural proteins. Double electroporations were performed to sequentially transfect Flavi-Rluc2A-Rep and SFV-Flavi-CprME into BHK-21 cells, leading to the production of VLPs in culture medium (see Materials and Methods for details). (B) Infection of CyP knockdown cells with flavivirus VLPs. Approximately 2 × 104 cells of each cell line were seeded per well in 96-well plates. At 24 h after seeding, the cells were infected with VLPs of WNV (1 FFU/cell), YFV (0.1 FFU/cell), or DENV-1 (0.01 FFU/cell). At 24 h p.i., the 96-well plates were assayed for luciferase activity. The percentages of the luciferase activities from the CyP knockdown cells versus on the sh-Fluc cells (set as 100%) are indicated. Average results, with standard deviations (error bars), of four independent experiments are shown.
The sh-AB cells are compromised in supporting the replication of a number of RNA viruses.
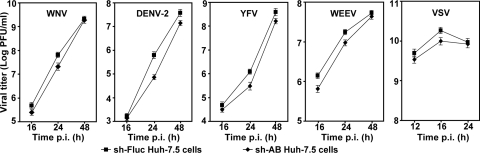
To validate the roles of CyPA and CyPB in flavivirus infection, we compared growth kinetics of authentic WNV, DENV-2, and YFV between the sh-AB cells and the control sh-Fluc cells (Fig. 3). After the cells were infected under an identical condition (MOI of 0.1), the sh-Fluc cells produced more viruses than the sh-AB did. The difference in virus titer was most apparent at 24 h p.i.; specifically, the sh-AB cells produced 67% (WNV), 89% (DENV-2), and 75% (YFV) less virus than the sh-Fluc cells did. However, at 48 h p.i., the difference became less apparent for DENV-2 and YFV; almost no difference was observed for WNV. Interestingly, the knockdown of CyPA and CyPB also reduced viral yields of two nonflaviviruses: a plus-strand RNA alphavirus (WEEV) and a negative RNA rhabdovirus (VSV) (Fig. 3). Similar to the flaviviruses, the difference in virus titer reduction was more dramatic at early time points (≤24 h p.i. for WEEV and ≤16 h p.i. for VSV) than those at late time points. The Student t test showed that the growth kinetics of the WNV between the two cell lines are not statistically significant, with a P value of 0.19. However, the growth curves of DENV-2, YFV, WEEV, and VSV between the two cell lines are statistically significant, with P values of 0.04, 0.03, 0.001, and 0.002, respectively. Overall, the results indicate that CyPA and/or CyPB function in both flavivirus and nonflavivirus replications.
FIG. 3.
Comparison of viral growth kinetics on sh-AB and sh-Fluc cells. The sh-AB and sh-Fluc cells were infected with WNV, DENV-2, YFV, WEEV, and VSV (MOI of 0.1). Culture fluids at the indicated time points were assayed for virus titers by a double-layer plaque assay. See details in Materials and Methods. Average results and standard deviations (n = 3) are presented.
Host CyPA interacts with WNV RNA and NS5.
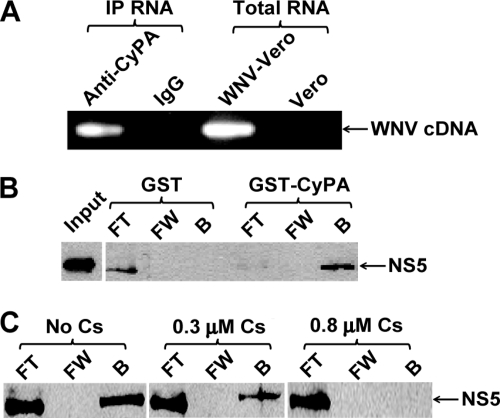
To explore the potential mechanism of CyPs in flavivirus replication, we performed Co-IP experiments with a monoclonal antibody of CyPA. Cell extracts prepared from the WNV replicon-containing Vero cells (Rluc-Neo-Rep; Fig. 4A) were immunoprecipitated with the CyPA antibody. Total RNA was extracted from the immunoprecipitated complexes and was subjected to virus-specific RT-PCR amplification. Agarose gel analysis of the RT-PCR showed an expected DNA fragment of viral sequence (Fig. 4A). In contrast, no RT-PCR product was detected when the same cell extract was immunoprecipitated using a rabbit control IgG. The results suggest that CyPA may participate in viral replication complex formation.
FIG. 4.
Interaction of CyPA with WNV NS5 and RNA. (A) Co-IP of CyPA with WNV RNA. A monoclonal antibody of CyPA or a control IgG was incubated with WNV Rluc-Neo-Rep cell extracts. After being immobilized by protein G, the RNA (indicated as IP RNA) was extracted from the protein G complexes and subjected to virus-specific RT-PCR. The RT-PCRs were then analyzed on an agarose gel. As a positive control, total RNA, directly extracted from the Rluc-Neo-Rep Vero cells, was used for RT-PCR. (B) In vitro binding of WNV NS5 to CyPA. WNV Rluc-Neo-Rep cell extracts were incubated with GST or GST-CyPA. The mixtures were purified through glutathione beads, followed by Western blotting to detect viral NS5. FT, flowthrough; FW, final wash; B, bound. (C) Cs-mediated blockage of the WNV NS5-CyPA interaction. The GST pulldown assay, described in panel B, was performed in the presence of 0.3 or 0.8 μM Cs.
Next, we examined whether CyPA directly interacts with viral NS5 in the replication complexes. Recombinant GST-CyPA protein was incubated with cell extracts prepared from the WNV Rluc-Neo-Rep Vero cells. Viral NS5 was pulled down by GST-CyPA after the mixture was incubated with glutathione-Sepharose beads (Fig. 4B). As a negative control, no detectable level of NS5 was pulled down by GST. Inclusion of Cs in the binding reaction inhibited the binding of NS5 to GST-CyPA in a dosage-dependent manner (Fig. 4C; details are described below). The observed CyPA-NS5 interaction prompted us to test whether CyPA could affect the NS5 enzyme activities. Coincubation of recombinant NS5 of WNV with GST-CyPA at various stoichiometry ratios did not affect the methyltransferase and RdRp activities of NS5 (data not shown). The results indicate that, although CyPA interacts with WNV NS5, it does not directly interfere with the enzymatic activities of NS5, at least in vitro.
The PPIase activity of CyPA is critical in supporting the WNV replication.
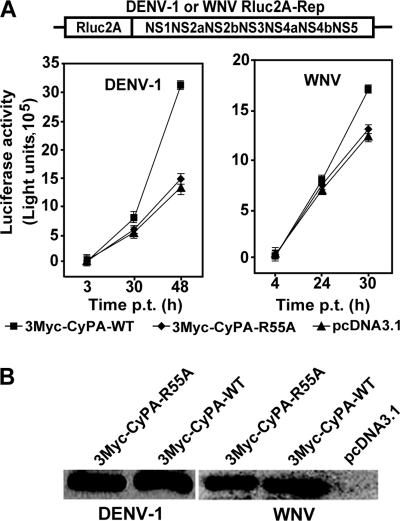
We performed a CyPA rescue experiment to determine whether the PPIase activity of CyPA is important for flavivirus replication. A cDNA of CyPA (N-terminally fused with three copies of Myc; 3Myc-CyPA-WT) that contained silent mutations in the recognition site of sh-A was cloned into a pcDNA3.1(+) vector. We also prepared a pcDNA3.1(+) plasmid containing an MT 3Myc-CyPA-R55A, which has an Ala substitution for Arg at amino acid position 55 in the PPIase domain of CyPA. The R55A mutation was previously shown to reduce the PPIase activity to <1% of the WT protein (58). The plasmid DNA (3Myc-CyPA-WT, 3Myc-CyPA-R55A, or empty pcDNA3.1) and flavivirus replicon RNAs (DENV-1 or WNV Rluc2A-Rep) were cotransfected into the sh-A cells. The effects of exogenous CyPA on viral replication were measured by the replicon-derived luciferase activities. As shown in Fig. 5A, equivalent levels of luciferase activities were observed at 3 or 4 h p.t., indicating similar transfection efficiencies. At later time points, luciferase signals were consistently higher in the WT CyPA-expressing cells than those in the MT CyPA-expressing cells. In the case of DENV-1 replicon, the luciferase signals derived from the WT CyPA-expressing cells were 32% (30 h p.t.) and 112% (48 h p.t.) greater than those derived from the MT CyPA-expressing cells; a similar, but less dramatic difference in luciferase signals was observed for the WNV replicon. The differences in luciferase signals between the WT and MT CyPA-expressing cells are statistically significant (student's t test P < 0.01) for both DENV-1 replicon (30 h and 48 h p.t.) and WNV replicon (30 h p.t.). As controls, Western blotting analysis showed that equivalent levels of WT and MT CyPA were expressed in the transfected cells at 48 h p.t. (Fig. 5B). The results demonstrate that exogenous expression of WT CyPA in the CyPA knockdown cells could rescue flavivirus replication; the PPIase activity of CyPA is essential for flavivirus replication.
FIG. 5.
Restoration of flavivirus replication by exogenous expression of CyPA in sh-A cells. (A) Expression of 3Myc-CyPA-WT in CyPA knockdown cells rescued the replication of DENV-1 and WNV replicon in sh-A cells. DENV-1 and WNV Rluc2A-Rep (top panel) were used to monitor the effect of exogenous expression of CyPA on viral replication. The replicon RNA (5 μg) and plasmid DNA (expressing 3Myc-CyPA-WT, 3Myc-CyPA-R55A, or empty vector; 9 μg) were coelectroporated into sh-A cells. The transfected cells were assayed for luciferase activity at the indicated time points. Error bars represent the standard deviations from three independent experiments. (B) Western blotting with anti-Myc antibody. The electroporated cells, described in panel A, were lysed at 48 p.t. and monitored for CyPA expression by Western blotting with an anti-Myc antibody.
Cs inhibits a broad spectrum of RNA viruses at nontoxic concentrations.
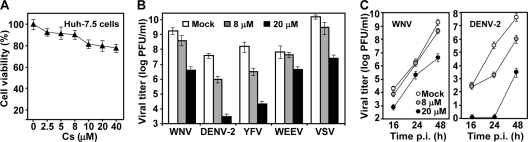
Since Cs binds to CyPs and inhibits the PPIase activity (40), we examined the antiviral activity of Cs against a panel of flaviviruses (WNV, DENV-2, and YFV) and two nonflaviviruses (WEEV and VSV). Initially, a cell proliferation-based MTT assay was performed to test the cytotoxicity of Cs. No substantial cytotoxicity (≤80% cell viability) was observed when Huh-7.5 cells were incubated with Cs at ≤40 μM for 48 h (Fig. 6A). Next, Huh-7.5 cells were infected with individual virus (MOI of 0.1); the infected cells were immediately treated with Cs (8 and 20 μM). Virus titers in culture medium were quantified by using plaque assays at 48 h p.i. for WNV, DENV-2, YFV, and WEEV and at 16 h p.i. for VSV (due to its short replication cycle). The Student t test indicates that, except for WNV and WEEV treated with 8 μM Cs (P > 0.05), the virus titer difference between the Cs-treated and mock-treated cells is statistically significant (P < 0.05). The results showed that both flaviviruses and nonflaviviruses were inhibited by Cs, among which DENV-2 and YFV exhibited the most sensitivity (Fig. 6B).
FIG. 6.
Antiviral activity of Cs. (A) Cytotoxicity of Cs in Huh-7.5 cells. Cytotoxicity was examined by incubation of Huh-7.5 cells with the indicated concentrations of Cs. Cell viability was measured by an MTT assay and is presented as the percentage of colorimetric absorbance derived from the compound-treated cells compared to that from the mock-treated cells (with 1% ethanol). (B) Virus titer reduction assays. Huh-7.5 cells were infected with indicated viruses (MOI of 0.1) and treated immediately with Cs at 8 and 20 μM. For WNV, DENV-2, YFV, and WEEV, culture medium were collected at 48 h p.i. For VSV, the culture medium was collected at 16 h p.i. The virus titers of all samples were determined by plaque assays on Vero cells. (C) Growth kinetics of WNV and DENV-2 on Huh-7.5 cells with or without Cs. Huh-7.5 cells were infected with WNV or DENV-2 (MOI of 0.1) and treated with Cs at 8 and 20 μM. Culture medium at indicated time points were quantified for viral yields by using plaque assays. Average results and standard deviations (n = 3) are presented.
These antiviral results were obtained at a single time point after infection. We also performed a growth kinetics of WNV and DENV-2 on Huh-7.5 cells in the presence or absence of Cs (Fig. 6C). The antiviral activity of Cs was consistently observed for both viruses at various time points. The Student t test shows that both 8 and 20 μM Cs significantly suppresses DENV-2 titer (P < 0.05), whereas only 20 μM Cs (P < 0.05) but not 8 μM Cs (P > 0.05) significantly suppresses the WNV titer. Overall, the results demonstrate that Cs inhibits flaviviruses and other RNA viruses in cell culture.
Cs inhibits WNV replication in CyPA knockdown cells.
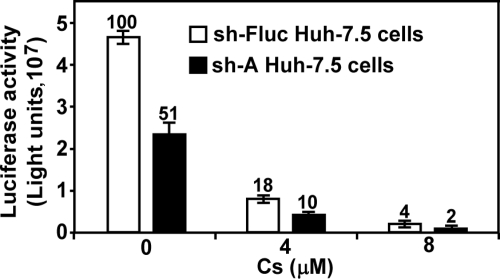
Since shRNA could not completely deplete target gene expression, treatment of the CyP knockdown cells with Cs is expected to further reduce flavivirus replication. We tested this possibility by treating infected sh-A cells with Cs. The sh-A cells were chosen because Cs was reported to primarily form a complex with CyPA (20). Both sh-A and sh-Fluc cells were infected with WNV VLP (1 FFU/cell), immediately treated with Cs (0, 4, or 8 μM), and assayed for luciferase activities at 24 h p.i. (Fig. 7). Without Cs treatment, the luciferase signal from the sh-A cells was ca. 51% of that from the sh-Fluc cells (set as 100%). Treatment of the infected sh-A cells with 4 and 8 μM Cs further reduced luciferase activities to 10 and 2%, respectively. In contrast, treatment of infected sh-Fluc cells with 4 and 8 μM Cs reduced the luciferase activities to 18 and 4%, respectively, of the mock-treated cells. The Student t test indicates that the luciferase differences between the sh-A and sh-Fluc cells at each concentration of Cs are statistically significant, with P values of <0.05. These results demonstrate that Cs can further inhibit the WNV replication in CyPA knockdown cells.
FIG. 7.
Antiviral effect of Cs on WNV VLP infection on sh-A cells. After incubation of sh-Fluc and sh-A cells for 24 h in a 96-well plate (2 × 104 per well), the cells were infected with WNV VLP (1 FFU/cell), immediately treated with Cs, and assayed for luciferase activities at 24 h p.i. The luciferase signals from 1% ethanol-treated sh-Fluc cells were set as 100%, and the percentages of luciferase activities derived from the Cs-treated cells are indicated. Error bars represent the standard deviations from three independent experiments.
Cs inhibits flaviviruses through suppression of viral RNA synthesis.
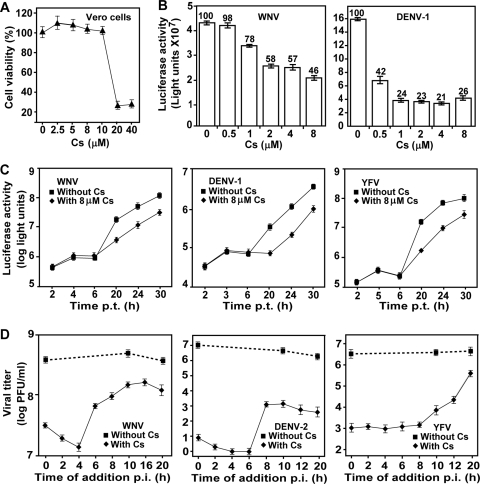
Inhibition of VLP infection by Cs indicates that the compound may block viral entry, translation, and/or replication. To define the step(s) at which Cs suppresses flavivirus infection, we analyzed the inhibitor in two replicon-based assays that have been validated for mode-of-action analysis (12, 42). Because some of the assays would be performed on Vero cells, we first examined the cytotoxicity of Cs on this cell type. No cytotoxicity was observed after treatment of Vero cells with Cs at ≤10 μM for 48 h (Fig. 8A).
FIG. 8.
Mechanism of Cs-mediated inhibition of flavivirus. (A) Cytotoxicity of Cs in Vero cells. Cytotoxicity was examined by incubation of Vero cells with the indicated concentrations of Cs for 48 h. Cell viability was determined by an MTT assay. The average results from three experiments are shown. (B) Antiviral activity of Cs in flavivirus replicon cells. Vero cells containing WNV or DENV Rluc-Neo-Rep were seeded in a 96-well plate (2 × 104 per well). At 24 h after seeding, the cells were treated with Cs at the indicated concentrations, and the luciferase activities were measured at 24 h posttreatment. The percentages of luciferase signals from the Cs-treated cells compared to those from the mock-treated cells (set as 100%) are indicated. Average results from three independent experiments are presented. (C) Analysis of Cs using transient replicon. Luciferase replicon (Rluc2A-Rep; 10 μg) of WNV, DENV-1, or YFV was electroporated into Huh-7.5 cells. The transfected cells were immediately incubated with 8 μM Cs and measured for luciferase activity at the indicated time points. Error bars indicate the standard deviations from three independent experiments. (D) Time-of-addition analysis of Cs in flavivirus infection. Huh-7.5 cells were infected with WNV, DENV-2 or YFV at an MOI of 3 at 4°C for 1 h. The infected cells were washed three times with cold PBS. At the indicated time points p.i., Cs was added to the infected cells at 20 μM for WNV and at 8 μM for DENV-2 and YFV. The supernatants were assayed to determine the virus titers at 24 h p.i. As controls, 1% ethanol was added to the infected cells at 0, 10, and 20 h p.i. to estimate its effect on viral production. Error bars represent the standard deviations from three independent experiments.
Based on the cytotoxicity result, we treated replicon-containing Vero cells with Cs at ≤8 μM (Fig. 8B). Both WNV or DENV-1 replicon cells were tested; each replicon (Rluc-Neo-Rep) contained a Renilla luciferase (replacing the viral structural genes) and a neomycin phosphotransferase (driven by an EMCV internal ribosome entry site engineered at the 3′ untranslated region of the replicon) (32, 42). The replicon cells allow for testing the effect of Cs on viral replication. As shown in Fig. 8B, Cs inhibited both WNV and DENV-1 replicons in a dose-responsive manner, with 50% effective concentrations of 8 and ≤0.5 μM, respectively. The Student t test shows that, except for the WNV treated with 0.5 μM Cs (P = 0.2), the suppression of luciferase activity by Cs in Fig. 8B is statistically significant (P < 0.005).
Next, we used Rluc2A-Rep (Fig. 2A) of WNV, DENV-1, and YFV to differentiate the Cs-mediated inhibition between viral translation and RNA synthesis. Transfection of Huh-7.5 cells with this replicon reveals two Rluc peaks, one at 1 to 10 h p.t. and another at >10 h p.t., which represent viral translation and RNA replication, respectively (22, 31, 42). The replicon-transfected Huh-7.5 cells were immediately treated with 8 μM Cs and assayed for luciferase activities at the indicated time points. As shown in Fig. 8C, Cs did not suppress luciferase activities at ≤6 h p.t. In contrast, Cs inhibited luciferase activities by ≥70% at ≥20 h p.t.; statistical analysis indicated that the Cs-mediated luciferase suppression is significant, with P values of <0.05. These results suggest that Cs inhibits WNV, DENV-1, and YFV virus through suppression of viral RNA replication.
Time-of-addition experiments were performed to further characterize the mode-of-action for Cs. Huh-7.5 cells were synchronously infected with WNV, DENV-2, or YFV. It should be noted that, although all of the replicon experiments for DENV described above were derived from serotype 1 (DENV-1), we chose DENV-2 for the time-of-addition experiments because DENV-2 generates better plaques than DENV-1 does in our plaque assays. After the synchronous infection, Cs was added to the infected cells (without changing medium) at various time points p.i. Virus titers in the culture medium were determined at 24 h p.i. For mock treatment, 1% ethanol was added at 0, 10, and 20 h p.i. to estimate its effect on viral production. Because WNV was not very sensitive to Cs, we treated the WNV-infected Huh-7.5 cells with 20 μM Cs (80% cell viability; Fig. 6A); whereas the DENV-2- and YFV-infected cells were treated with 8 μM Cs. Consistent with the transient replicon results (Fig. 8C), a significant level of suppression in virus titer was observed when the compound was added during the initial 6 h (WNV) and 8 h (DENV-2 and YFV) of infection (Fig. 8D). The compound partially lost the antiviral activity when added between 6 and 20 h p.i. The Student t test indicates that the virus titer difference between the Cs-treated and the mock-treated cells in Fig. 8D is statistically significant (P < 0.001). It should be noted that the replication kinetics of raw viral infection are faster than those of the transfected replicon, possibly because the delivery of viral RNA through infection is more productive than that through electroporation. Specifically, during flavivirus infection, RNA synthesis could be detected at 6 p.i., with the release of infectious virus beginning at 12 h (10). Overall, the time-of-addition results were in agreement with the transient replicon results, indicating that Cs inhibits flavivirus at the step of viral RNA synthesis. However, in the case of DENV, treatment with 8 μM Cs reduced replicon RNA synthesis by ≤10-fold (Fig. 8C middle panel), whereas the same concentration of Cs (added at 20 h p.i.) suppressed the virus titer by ≥103-fold (Fig. 8D, middle panel). The results suggest that, in addition to RNA synthesis, Cs may also affect the assembly of DENV.
Cs does not inhibit WNV protease, methyltransferase, and RdRp.
Since the above results indicate that Cs inhibits viral RNA synthesis, we tested the compound in our previously established enzyme assays, including WNV protease (with NS2B), NTPase, methyltransferase, and RdRp (43, 55). None of the enzymatic activities were suppressed by the compound at concentrations up to 100 μM (data not shown), suggesting that Cs does not directly target the enzyme functions of viral NS3 or NS5 proteins.
Cs directly blocks the interaction between host CyPA and viral NS5.
To examine whether Cs could interfere with the interaction between CyPA and NS5, we performed the GST-CyPA pulldown experiments in the presence or absence of Cs. Different concentrations of Cs were incubated with GST-CyPA and WNV replicon cell extracts; the GST-CyPA were then immobilized onto glutathione beads, and viral NS5 (coimmobilized with GST-CyPA) was monitored by Western blotting. As shown in Fig. 4C, increasing concentrations of Cs decreased the mounts of NS5 that could be pulled down by GST-CyPA. No NS5 was detected when the experiment was performed with 0.8 μM Cs. These results demonstrate that Cs targets the association of CyPA with viral NS5.
DISCUSSION
A number of host proteins have been reported to be important for flavivirus replication, among which eEF1α and TIA-1 (T-cell intracellular antigen 1) are the two best characterized (11, 14). The eEF1α binds to the 3′ terminal stem-loop of flavivirus genomic; this interaction is critical for the synthesis of minus-strand RNA (11). In contrast, TIA-1 binds to the 3′-terminal stem-loop of WNV minus-strand RNA. This interaction negatively affects genomic RNA amplification (14). The present study demonstrated that, in addition to eEF1α and TIA-1, cellular CyPs also play an important role in flavivirus replication. We show that knockdown of different isoforms of CyP reduced flavivirus production in Huh-7.5 cells. However, the knockdown cells did not allow us to conclude which isoform(s) of CyP is most critical for flavivirus replication, because most cell lines with knockdown of one isoform exhibited enhanced expression of other isoforms (e.g., sh-A and sh-B; Fig. 1B). Only the double sh-AB knockdown cells specifically reduced the expression of CyPA (by 92%) and CyPB (by 84%), without a significant change in the CyPC expression (90% of the WT level). Compared to the parental cells, infection of the sh-AB cells with flavivirus VLPs produced 68 to 85% less luciferase signals at 24 h p.i. (Fig. 2B). Similarly, infection of the sh-AB cells with authentic flaviviruses yielded 67 to 89% less viruses at 24 h p.i. (Fig. 3). Furthermore, the low viral replication in sh-A cells could be restored by trans supplying of WT CyPA (Fig. 5A). These results clearly indicate that CyPA plays a role in flavivirus replication, but the potential function of CyPB and CyPC could not be excluded at this point.
Our results suggest that CyPA may serve as a component of the replication complex. An CyPA antibody was shown to coimmunoprecipitate WNV RNA from the replicon-containing cell extracts (Fig. 4A). Additionally, WNV NS5 in the replicon cell extracts could be pulled down by recombinant GST-CyPA protein (Fig. 4B). The latter results, together with the observation that only WT CyPA (but not the MT CyPA defective in PPIase activity) could rescue viral replication in sh-A cells (Fig. 4A), raised the possibility that CyPA may modulate the enzyme activities of WNV NS5 through the PPIase. However, biochemical analysis showed that recombinant CyPA affected neither the methyltransferase nor the RdRp activities of NS5 of WNV (data not shown). These data suggest that CyPA may not directly act on NS5, and an unidentified factor may mediate the CyPA-NS5 interaction. Alternatively, the conformation of recombinant CyPA and NS5 in vitro are different from that in the replication complex in vivo; in the context of replication complex, CyPA may directly bind to and enhance the NS5 activities. The fact that the PPIase activity of CyPA is required to facilitate viral replication suggests that CyPA serves as a molecular chaperone to keep the replication complex in an active conformation, leading to a more productive viral replication.
Consistent with the CyP knockdown results, Cs was shown to inhibit flavivirus replication in cell culture (Fig. 6). The compound could further suppress viral replication in the sh-A knockdown cells (Fig. 7). Mode-of-action analysis indicated that Cs inhibited viral replication at the step of RNA synthesis, possibly through direct blockage of the CyPA-NS5 interaction (Fig. 4C). It should be noted that, besides inhibiting the PPIase activity of CyPA (5, 40), Cs could also bind to and inhibit the PPIase activity of CyPB (21, 36), leading to the suppression of viral replication.
We found that sh-AB knockdown cells were less efficient in supporting nonflavivirus replication (WEEV and VSV; Fig. 3). A previous study reported that knockdown of CyPA did not affect the replication of VSV (56). Combining these results, one might conclude that CyPB contributes to the low replication of VSV in the sh-AB cells. However, care should be taken when interpreting these results, because the two studies used different methods to quantify the VSV yields. In the early study, equal amounts (PFU) of VSV were directly plaqued on parental and sh-A Huh-7.5 cells; the resulting numbers of plaques were compared to indicate the effect of CyPA on VSV replication. The incubation time of plaque assay was >24 h. In the present study, we infected parental and sh-AB cells at an MOI of 0.1. The culture media at different time points were measured for viral yields by plaque assay. The method used in the present study is expected to be more sensitive than the one described in the previous study. Nevertheless, a role of one or more of the CyPs in VSV and WEEV replication was further supported by the results that Cs inhibited both viruses in parental Huh-7.5 cells (Fig. 6B).
CyPs were previously reported to regulate HCV and HIV replication. Both CyPA and CyPB were initially shown to coprecipitate with HCV NS5B protein (53, 56) and with HIV Gag (8, 33), but RNA silencing experiments yielded conflicting results as to whether CyPA or CyPB, or both, are required for the viral replication cycle. CyPA was later demonstrated to be more important for HCV replication (56) and HIV infection (9). Moreover, CyPA was reported to be packaged into HIV-1 virion and to catalyze cis to trans isomerization of the viral capsid protein (7). The anti-HIV and anti-HCV actions of Cs was shown to rely on its capacity to bind to CyPs (33, 34, 48). Toward therapeutic development, two derivatives of Cs, NIM811 and DEBIO-025, are currently in clinical trials for HCV treatment (16, 35). Recent studies showed that the combination of NIM811 with HCV protease or RdRp inhibitors enhanced antiviral activity and suppressed the emergence of resistance (35) and that DEBIO-025 achieved proof-of-concept efficacy in HCV patients (16). It remains to be examined whether the two HCV clinical candidates could inhibit flaviviruses in cell culture and in animal models.
One major advantage of targeting host factors for antiviral development is the higher genetic barrier to the emergence of viral escape mutants. Due to the error-prone nature of RdRp, drug resistance to virus-specific inhibitors can quickly emerge both in vitro and in patients. Since viral inhibitors usually bind to defined pockets of viral proteins, mutations in the viral genome that weaken or disrupt the binding would reduce the compound efficacy and lead to resistance. In this regard, host inhibitors do not directly bind to viral targets, creating a greater barrier for the emergence of resistant viruses. On the other hand, one major challenge for the development of host inhibitors is to identify a therapeutic dose that does not affect the normal function of host targets and, therefore, does not lead to significant side effect.
In summary, we found that host CyP plays a role in flavivirus replication. CyPA may function as a component of viral replication complex. The PPIase activity is essential for CyPA to function in WNV replication. Cs, a ligand and inhibitor of PPIase of CyP, inhibits flaviviruses in cell culture. Mechanism-of-action analysis showed that Cs directly blocks the interaction between cellular CyPA and WNV NS5 protein. The results suggest that CyPs represent a potential target for flavivirus antiviral development.
Acknowledgments
We thank the Molecular Genetics Core and the Cell Culture Facility at the Wadsworth Center for the DNA sequencing and maintenance of the BHK-21 and Vero cells, respectively.
This study was partially supported by federal funds from the National Institute of Allergy and Infectious Disease, National Institutes of Health (NIH), under contract N01-AI-25490, by NIH grants 1U01AI061193 and U54-AI057158 (Northeast Biodefense Center), and by the James Esther King Biomedical Research Program from Florida Department of Health.
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Bartelma, G., and R. Padmanabhan. 2002. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299:122-132. [DOI] [PubMed] [Google Scholar]
- 3.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billich, A., F. Hammerschmid, P. Peichl, R. Wenger, G. Zenke, V. Quesniaux, and B. Rosenwirth. 1995. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J. Virol. 69:2451-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borel, J. F., F. Di Padova, J. Mason, V. Quesniaux, B. Ryffel, and R. Wenger. 1990. Pharmacology of cyclosporine (sandimmune). I. Introduction. Pharmacol. Rev. 41:239-242. [PubMed] [Google Scholar]
- 6.Borowski, P., A. Niebuhr, O. Mueller, M. Bretner, K. Felczak, T. Kulikowski, and H. Schmitz. 2001. Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol. 75:3220-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosco, D. A., E. Z. Eisenmesser, S. Pochapsky, W. I. Sundquist, and D. Kern. 2002. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. USA 99:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten, D., H. Ansari, and J. Luban. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 11.Davis, W., J. Blackwell, P. Shi, and M. Brinton. 2007. Interaction between the cellular protein eEF1A and the 3′ terminal stem-loop of the West Nile virus genomic RNA facilitates viral RNA minus strand synthesis. J. Virol. 10172-10187. [DOI] [PMC free article] [PubMed]
- 12.Deas, T. S., I. Binduga-Gajewska, M. Tilgner, P. Ren, D. A. Stein, H. M. Moulton, P. L. Iversen, E. B. Kauffman, L. D. Kramer, and P.-Y. Shi. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 79:4599-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emara, M. M., H. Liu, W. G. Davis, and M. A. Brinton. 2008. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J. Virol. 82:10657-10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falgout, B., R. H. Miller, and C. J. Lai. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flisiak, R., A. Horban, P. Gallay, M. Bobardt, S. Selvarajah, A. Wiercinska-Drapalo, E. Siwak, I. Cielniak, J. Higersberger, J. Kierkus, C. Aeschlimann, P. Grosgurin, V. Nicolas-Metral, J. M. Dumont, H. Porchet, R. Crabbe, and P. Scalfaro. 2008. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology 47:817-826. [DOI] [PubMed] [Google Scholar]
- 17.Gubler, D., G. Kuno, and L. Markoff. 2007. Flaviviruses, p. 1153-1253. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 18.Guo, J., J. Hayashi, and C. Seeger. 2005. West nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 20.Handschumacher, R. E., M. W. Harding, J. Rice, R. J. Drugge, and D. W. Speicher. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544-547. [DOI] [PubMed] [Google Scholar]
- 21.Hasel, K. W., J. R. Glass, M. Godbout, and J. G. Sutcliffe. 1991. An endoplasmic reticulum-specific cyclophilin. Mol. Cell. Biol. 11:3484-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, C., C. Patkar, and R. Kuhn. 2005. Construction and applications of yellow fever virus replicons. Virology 331:247-259. [DOI] [PubMed] [Google Scholar]
- 23.Kramer, L., J. Li, and P.-Y. Shi. 2007. West Nile virus. Lancet Neurol. 6:171-182. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, H., S. Clum, S. You, K. E. Ebner, and R. Padmanabhan. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., H.-J. Thiel, and C. M. Rice. 2007. Flaviviridae: the virus and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 28.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807-815. [DOI] [PubMed] [Google Scholar]
- 29.Liu, W., X. Wang, V. Mokhonov, P.-Y. Shi, R. Randall, and A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo, L., M. Tilgner, K. Bernard, and P.-Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus using a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo, L., M. Tilgner, and P.-Y. Shi. 2003. A potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 77:12901-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 34.Ma, S., J. E. Boerner, C. TiongYip, B. Weidmann, N. S. Ryder, M. P. Cooreman, and K. Lin. 2006. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 50:2976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathy, J. E., S. Ma, T. Compton, and K. Lin. 2008. Combinations of cyclophilin inhibitor NIM811 with hepatitis C Virus NS3-4A Protease or NS5B polymerase inhibitors enhance antiviral activity and suppress the emergence of resistance. Antimicrob. Agents Chemother. 52:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikol, V., J. Kallen, and M. D. Walkinshaw. 1994. X-ray structure of a cyclophilin B/cyclosporin complex: comparison with cyclophilin A and delineation of its calcineurin-binding domain. Proc. Natl. Acad. Sci. USA 91:5183-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patkar, C. G., and R. J. Kuhn. 2008. Yellow Fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J. Virol. 82:3342-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ptak, R. G., P. A. Gallay, D. Jochmans, A. P. Halestrap, U. T. Ruegg, L. A. Pallansch, M. D. Bobardt, M. P. de Bethune, J. Neyts, E. De Clercq, J. M. Dumont, P. Scalfaro, K. Besseghir, R. M. Wenger, and B. Rosenwirth. 2008. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 52:1302-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puig-Basagoiti, F., T. S. Deas, P. Ren, M. Tilgner, D. M. Ferguson, and P.-Y. Shi. 2005. High-throughput assays using luciferase-expressing replicon, virus-like particle, and full-length virus for West Nile virus drug discovery. Antimicrob. Agent. Chemother. 49:4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puig-Basagoiti, F., M. Tilgner, B. Forshey, S. Philpott, N. Espina, Wentworth, S. Goebel, P. S. Masters, B. Falgout, P. Ren, Ferguson, and P. Y. Shi. 2006. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob. Agents Chemother. 50:1320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray, D., A. Shah, M. Tilgner, Y. Guo, Y. Zhao, H. Dong, T. Deas, Y. Zhou, H. Li, and P.-Y. Shi. 2006. West Nile Virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robida, J. M., H. B. Nelson, Z. Liu, and H. Tang. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 81:5829-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, P. Y., E. B. Kauffman, P. Ren, A. Felton, J. H. Tai, A. P. Dupuis, 2nd, S. A. Jones, K. A. Ngo, D. C. Nicholas, J. Maffei, G. D. Ebel, K. A. Bernard, and L. D. Kramer. 2001. High-throughput detection of West Nile virus RNA. J. Clin. Microbiol. 39:1264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starzl, T. E., G. B. Klintmalm, K. A. Porter, S. Iwatsuki, and G. P. Schroter. 1981. Liver transplantation with use of cyclosporin A and prednisone. N. Engl. J. Med. 305:266-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinkasserer, A., R. Harrison, A. Billich, F. Hammerschmid, G. Werner, B. Wolff, P. Peichl, G. Palfi, W. Schnitzel, E. Mlynar, et al. 1995. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J. Virol. 69:814-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilgner, M., and P.-Y. Shi. 2004. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 78:8159-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waninger, S., K. Kuhen, X. Hu, J. E. Chatterton, F. Wong-Staal, and H. Tang. 2004. Identification of cellular cofactors for human immunodeficiency virus replication via a ribozyme-based genomics approach. J. Virol. 78:12829-12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warrener, P., J. K. Tamura, and M. S. Collett. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282-1288. [DOI] [PubMed] [Google Scholar]
- 53.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 54.Wengler, G., and G. Wengler. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707-715. [DOI] [PubMed] [Google Scholar]
- 55.Wong, S. J., R. H. Boyle, V. L. Demarest, A. N. Woodmansee, L. D. Kramer, H. Li, M. Drebot, R. A. Koski, E. Fikrig, D. A. Martin, and P.-Y. Shi. 2003. An immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections, and form flavivirus vaccination. J. Clin. Microbiol. 41:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, F., J. M. Robotham, H. B. Nelson, A. Irsigler, R. Kenworthy, and H. Tang. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 82:5269-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, Y., D. Ray, Y. Zhao, H. Dong, S. Ren, Z. Li, Y. Guo, K. Bernard, P.-Y. Shi, and H. Li. 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81:3891-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zydowsky, L. D., F. A. Etzkorn, H. Y. Chang, S. B. Ferguson, L. A. Stolz, S. I. Ho, and C. T. Walsh. 1992. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1:1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]