Abstract
We evaluated the potency of garenoxacin in selecting resistant Streptococcus pneumoniae mutants by determining its mutant prevention concentration, using strains with and without topoisomerase gene mutations, and compared its potency to that of other quinolones. Garenoxacin had a significantly greater potency against pneumococci, including strains containing topoisomerase mutations. Genetic analysis of the S. pneumoniae mutants created by garenoxacin revealed that the gyrA gene was a primary target of garenoxacin.
The emergence of Streptococcus pneumoniae strains with resistance to β-lactams and macrolides has complicated the treatment of pneumococcal respiratory tract infections and created a need for new antibiotic agents. Recently developed compounds within the quinolone group have demonstrated enhanced potency against S. pneumoniae. In particular, agents such as moxifloxacin (MXF), gatifloxacin (GAT), and levofloxacin (LVX) have been recommended and used for therapy (17). However, S. pneumoniae strains with quinolone resistance have been observed in several countries (3, 9, 11, 16, 19). Furthermore, evidence suggests that increased usage of these compounds could lead to the development of further resistance and treatment failures (5, 7, 20). Quinolone resistance in S. pneumoniae is mediated by amino acid substitutions within the quinolone resistance-determining regions (QRDRs) of DNA gyrase (GyrA or GyrB) and/or topoisomerase IV (ParC or ParE), sometimes in combination with efflux (1, 12, 14). The mutant prevention concentration (MPC) of a drug is the concentration that prohibits the growth of mutants from a susceptible population of more than 1010 cells, and determining the MPC is a novel approach for evaluating quinolone potency (2, 6). Additionally, the mutant selection window (MSW), which is defined as the range between the MIC and MPC, provides a means for defining the ability of an antibiotic to prevent the emergence of mutants (22). Garenoxacin (GAR) is a novel des-F(6) quinolone with a broad spectrum of activity against respiratory tract pathogens with elevated or resistant-level fluoroquinolone MICs, including S. pneumoniae (4, 10, 13, 21). The aim of this study was to evaluate the potency of GAR in selecting for resistant S. pneumoniae and to compare GAR to other quinolones by determining the MPC and the MSW, using strains with and without mutations in the QRDRs of topoisomerase genes. Additionally, to determine the targets of GAR, we examined the intrinsic development of resistant S. pneumoniae mutants that were created by GAR exposure, with detailed evaluation of additional QRDR mutations and efflux.
A total of eight S. pneumoniae clinical isolates were used in this study. The QRDR genetic backgrounds and quinolone MICs of the isolates are summarized in Table 1. All strains were wild type for GyrB and ParE. Isolates W001 and W002 were quinolone-susceptible isolates with wild-type ParC and GyrA. Isolate S001 had an Asp83Asn mutation in ParC and wild-type GyrA. Isolate S002 had a Ser79Phe mutation in ParC and wild-type GyrA. Isolates S003 and S004 had single GyrA mutations (Ser81Phe for S003 and Gly85Asn for S004). Isolates D001 and D002 had both GyrA and ParC mutations (for D001, Ser79Phe in ParC and Gly85Lys in GyrA; for D002, Ser79Phe in ParC and Ser81Phe in GyrA). All the strains were exposed to 2, 4, 8, 16, 32, and 64× the MICs for ciprofloxacin (CIP), LVX, GAT, MXF, and GAR for 48 to 72 h at 37°C in 5% CO2. The MPCs were measured using a procedure previously described (2). Briefly, 200 μl of a culture containing 10 log10 CFU/ml was applied to Mueller-Hinton II agar plates containing 5% sheep blood and a drug at various concentrations. MPCs were recorded as the lowest concentration of the antibiotic that prevented bacterial colony formation after 48 h. All determinations were done in duplicate, and the results were identical. Genomic DNA was extracted from mutants growing on the plates (a maximum of eight mutants per plate were cultured individually) (Table 2) by using a Qiagen blood minikit (Qiagen, Hilden, Germany). All mutant DNA extracts were screened for QRDR mutations of the parC and gyrA genes, using a relatively new PCR-melting curve analysis (PCR-MCA) method, which we reported previously (8). Briefly, probes labeled with LC-Red 640 and fluorescein were used with designated primers (8) that targeted four QRDR positions (Ser79 and Asp83 of the parC gene product and Ser81 and Gly85 of the gyrA gene product). PCR was performed in a 20-μl volume containing 5 μl of DNA template, 4 μl of LightCycler 480 genotyping master (Roche Diagnostics, Basel, Switzerland), 3 mM MgCl2, 0.2 μM each probe, and 0.5 μM each primer. Thermal cycling was performed with an initial hold for 10 min at 95°C, followed by 35 cycles of 5 s at 95°C, 10 s at 55°C, and 12 s at 72°C. A melting curve was generated by cooling to 40°C for 30 s, followed by heating to 80°C at a rate of 2.0°C/s. The PCR-MCA assay was performed using LightCycler 480 analysis software (Roche Diagnostics, Basel, Switzerland). The total assay time was approximately 1 h. Nucleotide mismatches between the sequence and the hybridization probe resulted in a lower melting temperature for the mutant than for the wild type. The assay made it possible to quickly and easily differentiate a mutant strain from a wild-type strain. The QRDR sequences for the topoisomerase genes (parC, parE, gyrA, and gyrB) of the mutant strains were confirmed at the nucleotide level by direct sequencing, using a BigDye Terminator version 3.1 sequencing standard kit and an ABI PRISM 310 genetic analyzer (both by Applied Biosystems, CA) with published primers (18). The QRDR DNA sequencing results were compared with those for the R6 strain (GenBank accession no. NC_003098). MIC determination was done in parallel, both with and without an efflux inhibitor (10 μg of reserpine/ml). Efflux was considered to be present when a ≥2-fold reduction in the MIC was observed.
TABLE 1.
QRDR genetic backgrounds and quinolone MICs
| S. pneumoniae strain | QRDR genetic backgrounda
|
MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| parC | gyrA | CIP | LVX | GAT | MXF | GAR | |
| W001 | wt | wt | 0.5 | 0.5 | 0.12 | 0.06 | 0.015 |
| W002 | wt | wt | 0.25 | 0.25 | 0.12 | 0.06 | 0.015 |
| S001 | Asp83Asn | wt | 2 | 1 | 0.25 | 0.12 | 0.03 |
| S002 | Ser79Phe | wt | 2 | 1 | 0.12 | 0.25 | 0.06 |
| S003 | wt | Ser81Phe | 0.5 | 0.5 | 0.12 | 0.06 | 0.06 |
| S004 | wt | Gly85Asn | 0.5 | 1 | 0.25 | 0.12 | 0.06 |
| D001 | Ser79Phe | Gly85Lys | 32 | 16 | 4 | 4 | 0.25 |
| D002 | Ser79Phe | Ser81Phe | 8 | 16 | 8 | 4 | 0.5 |
wt, wild type.
TABLE 2.
Numbers of cultured mutants per plate
| S. pneumoniae strain | GAR MIC (μg/ml) | No. of cultured mutants (n = 231) for indicated GAR concna
|
Total no. of cultured mutants | ||||
|---|---|---|---|---|---|---|---|
| 2× MIC | 4× MIC | 8× MIC | 16× MIC | 32× MIC | |||
| W001 | 0.015 | 8 | 8 | 8 | —b | — | 24 |
| W002 | 0.015 | 8 | 8 | 8 | — | — | 24 |
| S001 | 0.03 | 8 | 8 | 8 | 7 | — | 31 |
| S002 | 0.06 | 8 | 8 | 8 | 8 | 1 | 33 |
| S003 | 0.06 | 8 | 8 | 8 | 8 | — | 32 |
| S004 | 0.06 | 8 | 8 | 8 | 8 | — | 32 |
| D001 | 0.25 | 8 | 8 | 8 | 7 | — | 31 |
| D002 | 0.5 | 8 | 8 | 8 | — | — | 24 |
There were no mutants cultured at 64× MIC.
There were no mutants cultured.
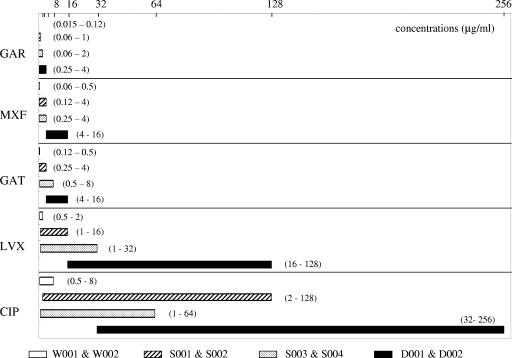
The MPC and MSW ranges of the S. pneumoniae isolates are summarized in Fig. 1. The MPCs that are shown were averaged between the W001 and W002, S001 and S002, S003 and S004, and D001 and D002 strains. The MPCs for the S. pneumoniae wild-type strains (from lowest to highest) were those for CIP (8 μg/ml), LVX (2 μg/ml), GAT (0.5 μg/ml) and MXF (0.5 μg/ml), and GAR (0.12 μg/ml). GAR potency was 16- to 64-fold greater than LVX or CIP potency and 4-fold greater than GAT or MXF potency. GAR also had a significantly narrower MSW than the other quinolones, even for strains with QRDR mutations. Among these strains, the MSWs (from lowest to highest) were those for the wild-type strains (W001 and W002), the strains with a single QRDR mutation (S001 to S004), and the strains with two QRDR mutations (D001 and D002). LVX, GAT, and GAR had narrower MSWs for strains with single parC mutations than for strains with single gyrA mutations, while CIP and MXF showed the opposite. The numbers of pneumococcal mutants that were created by GAR exposure and cultured individually are shown in Table 2. The QRDR wild-type strains generated mutants with GAR exposures of 1× to 8× MIC, while strains with QRDR mutations generated mutants from 1× to 16- to 32× MIC. The QRDR genetic changes in the S. pneumoniae mutants that were generated by GAR exposure are shown in Table 3. The PCR-MCA assays with the wild-type-strain mutants (W001 and W002) created by GAR exposure revealed a high frequency of additional gyrA mutations. In strains with single parC mutations (S001 and S002), a high percentage of additional gyrA mutations in codon 81 was seen. On the other hand, in the strains with single gyrA mutations (S003 and S004), additional parC mutations in codon 79 were seen at a high percentage. Strains with two QRDR mutations (D001 and D002) had additional gyrA mutations resulting from GAR exposure. None of the mutants created by GAR exposure showed the existence of efflux upon MIC determination with reserpine. DNA sequence analysis of the mutants without gyrA or parC mutations after GAR exposure showed two W001 mutants and a W002 mutant with an additional parE mutation (Asp435Asn). In addition, two W001 mutants, a W002 mutant, two S003 mutants, and an S004 mutant had an additional parC mutation (Lys137Asn). As for the gyrB gene, only one S001 mutant had an Asp435Asn mutation (data not shown).
FIG. 1.
MPCs and MSWs of S. pneumoniae strains. The horizontal axis represents concentrations of quinolones. Results are shown for the wild-type strains (W001 and W002), the parC single mutation-containing strains (S001 and S002), the gyrA single mutation-containing strains (S003 and S004), and the strains with both parC and gyrA mutations (D001 and D002). The left side, right end, and width of each bar represent the MIC, the MPC, and the MSW, respectively. The lower and higher values in each set of parentheses are quinolone MICs (μg/ml) and MPCs (μg/ml), respectively.
TABLE 3.
QRDR genetic changes in S. pneumoniae mutants generated by GAR exposure
| S. pneumoniae strain | QRDR genetic background
|
No. of mutants with indicated QRDR mutation/no. of isolated mutants (%) (n = 231)
|
||||
|---|---|---|---|---|---|---|
| parC | gyrA | ParC79 | ParC83 | GyrA81 | GyrA85 | |
| W001 | wta | wt | 0/24 (0) | 0/24 (0) | 12/24 (50) | 3/24 (12) |
| W002 | wt | wt | 0/24 (0) | 0/24 (0) | 16/24 (67) | 0/24 (0) |
| S001 | Asp83Asn | wt | 0/31 (0) | -b | 24/31 (77) | 0/31 (0) |
| S002 | Ser79Phe | wt | - | 0/33 (0) | 16/33 (48) | 3/33 (9) |
| S003 | wt | Ser81Phe | 21/32 (65) | 0/32 (0) | - | 0/32 (0) |
| S004 | wt | Ser81Phe | 18/32 (56) | 0/32 (0) | - | 0/32 (0) |
| D001 | Ser79Phe | Gly85Lys | - | 7/31 (23) | 22/31 (70) | - |
| D002 | Ser79Phe | Ser81Phe | - | 5/24 (21) | - | 12/24 (50) |
wt, wild type.
Presented mutations with original isolates.
In the present study, the MIC results showed that GAR was potently active against pneumococci, including strains containing QRDR mutations, at a level significantly greater than those observed for other quinolones. In addition, the significantly narrower MSW and the low MPCs demonstrated that for pneumococci, it was more difficult to acquire resistance to GAR than to acquire resistance to other quinolones. We used a relatively new PCR-MCA assay to make a detailed genetic analysis of QRDR mutations from a vast number of mutants. Previous studies had difficulties analyzing large numbers of mutants, due to the time-consuming process of culturing them individually and analyzing their DNA sequences. A high proportion of the mutants that were derived from isolates with single parC mutations acquired secondary gyrA mutations and became highly resistant to all of the quinolones that we used in our study. With the wild-type pneumococcal strains, a high percentage (50% of mutants of strain W001 and 67% of mutants of strain W002) of additional GyrA mutations were seen in the mutants exposed to GAR. This may indicate that GAR has a more balanced affinity for the two target enzymes, with a slight initial preference for GyrA as an initial target. A total of 72 out of 231 mutants (31%) did not have additional detectable gyrA or parC mutations in this study. We expected that these remaining mutants would have acquired other additional QRDR mutations (parE or gyrB) or efflux; however, only 10 mutants (14%) were detected by sequencing the topoisomerase genes. Therefore, these mutants may have acquired other resistance mechanisms, such as plasmid-based resistance. Further studies will be needed to clarify the other mechanisms of resistance of pneumococci to GAR. Although quinolone resistance in S. pneumoniae isolates remains low, with susceptibility levels based on MICs at or below the susceptibility breakpoint, the findings of Lim et al. (15), which determined that a substantial percentage (60%) of S. pneumoniae isolates with LVX MICs of 2 μg/ml contained first-step QRDR mutations, may be of note. The opportunities to treat respiratory tract infections with quinolone have increased, and the potential for forming resistance should thus be considered when specific quinolones are selected for treatment. Including MPCs as part of a dosing strategy may be one method for limiting the selection of quinolone-resistant mutants and preserving this class of antibiotics.
In conclusion, the novel des-F(6) quinolone GAR showed a low MPC and a narrow MSW for QRDR mutation-containing pneumococcal strains, suggesting that GAR will be useful for minimizing the selection of quinolone-resistant mutants of S. pneumoniae.
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Bast, D. J., D. E. Low, C. L. Duncan, L. Kilburn, L. A. Mandell, R. J. Davidson, and J. C. S. de Azavedo. 2000. Fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae: contributions of type II topoisomerase mutations and efflux to levels of resistance. Antimicrob. Agents Chemother. 44:3049-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondeau, J. M., X. Zhao, G. Hansen, and K. Drlica. 2001. Mutant prevention concentrations of fluoroquinolones for clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:433-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, D. K., A. McGeer, J. C. de Azavedo, and D. E. Low. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N. Engl. J. Med. 341:233-239. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen, K. J., J. M. Bell, J. D. Turnidge, and R. N. Jones. 2004. Antimicrobial activities of garenoxacin (BMS 284756) against Asia-Pacific region clinical isolates from the SENTRY program, 1999 to 2001. Antimicrob. Agents Chemother. 48:2049-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson, R., R. Cavalcanti, J. L. Brunton, D. J. Bast, J. C. Azavedo, P. Kibsey, C. Fleming, and D. E. Low. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:747-750. [DOI] [PubMed] [Google Scholar]
- 6.Dong, Y., X. Zhao, B. N. Kreiswirth, and K. Drlica. 2000. Mutant prevention concentration as a measure of antibiotic potency: studies with clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2581-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endimiani, A., G. Brigante, A. A. Bettaccini, F. Luzzaro, P. Grossi, and A. Q. Toniolo. 2005. Failure of levofloxacin treatment in community-acquired pneumococcal pneumonia. BMC Infect. Dis. 5:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukushima, K. Y., Y. Hirakata, K. Sugahara, K. Yanagihara, A. Kondo, S. Kohno, and S. Kamihira. 2006. Rapid screening of topoisomerase gene mutations using a novel melting curve analysis for early warning of fluoroquinolone-resistant Streptococcus pneumoniae emergence. J. Clin. Microbiol. 44:4553-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldsmith, C. E., J. E. Moore, P. G. Murphy, and J. E. Ambler. 1998. Increased incidence of ciprofloxacin resistance in penicillin-resistant pneumococci in Northern Ireland. J. Antimicrob. Chemother. 41:420-421. [DOI] [PubMed] [Google Scholar]
- 10.Grohs, P., S. Houssaye, A. Aubert, L. Gutmann, and E. Varon. 2003. In vitro activities of garenoxacin (BMS 284756) against Streptococcus pneumoniae, viridans group streptococci, and Enterococcus faecalis compared to those of six other quinolones. Antimicrob. Agents Chemother. 47:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, P. L., R. W. Yung, D. N. Tsang, T. L. Que, M. Ho, W. H. Seto, T. K. Ng, W. C. Yam, and W. W. Ng. 2001. Increasing resistance of Streptococcus pneumoniae to fluoroquinolones: results of a Hong Kong multicentre study in 2000. J. Antimicrob. Chemother. 48:659-665. [DOI] [PubMed] [Google Scholar]
- 12.Janoir, C., V. Zeller, M. D. Kitzis, N. J. Moreau, and L. Gutmann. 1996. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob. Agents Chemother. 40:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, R. N., T. R. Fritsche, H. S. Sader, and M. G. Stilwell. 2007. Activity of garenoxacin, an investigational des-F(6)-quinolone, tested against pathogens from community-acquired respiratory tract infections, including those with elevated or resistant-level fluoroquinolone MIC values. Diagn. Microbiol. Infect. Dis. 58:9-17. [DOI] [PubMed] [Google Scholar]
- 14.Jorgensen, J. H., L. M. Weigel, M. J. Ferraro, J. M. Swenson, and F. C. Tenover. 1999. Activities of newer fluoroquinolones against Streptococcus pneumoniae clinical isolates including those with mutations in the gyrA, parC, and parE loci. Antimicrob. Agents Chemother. 43:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim, S., D. Bast, A. McGeer, J. de Azavedo, and D. E. Low. 2003. Antimicrobial susceptibility breakpoints and first-step parC mutations in Streptococcus pneumoniae: redefining fluoroquinolone resistance. Emerg. Infect. Dis. 9:833-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linares, J., A. G. de la Campa, and R. Pallares. 1999. Fluoroquinolone resistance in Streptococcus pneumoniae. N. Engl. J. Med. 341:1546-1548. [DOI] [PubMed] [Google Scholar]
- 17.Low, D. E. 2004. Quinolone resistance among pneumococci: therapeutic and diagnostic implications. Clin. Infect. Dis. 28(Suppl. 4):S357-S362. [DOI] [PubMed] [Google Scholar]
- 18.Morrissey, I., and J. George. 1999. Activities of fluoroquinolones against Streptococcus pneumoniae type II topoisomerase purified as recombinant proteins. Antimicrob. Agents Chemother. 43:2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pankuch, G. A., B. Bozdogan, K. Nagai, A. Tambic-Andrasevic, S. Schoenwald, T. Tambic, S. Kalenic, S. Plesko, N. K. Tepes, Z. Kotarski, M. Payerl-Pal, and P. C. Appelbaum. 2002. Incidence, epidemiology, and characteristics of quinolone-nonsusceptible Streptococcus pneumoniae in Croatia. Antimicrob. Agents Chemother. 46:2671-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Trallero, E., J. M. Marimon, L. Iglesias, and J. Larruskain. 2003. Fluoroquinolone and macrolide treatment failure in pneumococcal pneumonia and selection of multidrug-resistant isolates. Emerg. Infect. Dis. 9:1159-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhanel, G. G., L. Palatnick, K. A. Nichol, T. Bellyou, D. E. Low, and D. J. Hoban. 2003. Antimicrobial resistance in respiratory tract Streptococcus pneumoniae isolates: results of the Canadian Respiratory Organism Susceptibility Study, 1997 to 2002. Antimicrob. Agents Chemother. 47:1867-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, X., and K. Drlica. 2002. Restricting the selection of antibiotic-resistant mutant bacteria: measurement and potential use of the mutant selection window. J. Infect. Dis. 185:561-565. [DOI] [PubMed] [Google Scholar]