Abstract
Vancomycin (VAN)-intermediate Staphylococcus aureus (VISA) and heterogeneous VISA (hVISA) isolates are considered to have emerged from VAN-susceptible S. aureus (VSSA) by spontaneous mutation during VAN exposure. We previously reported that laboratory mutant H14, obtained from VSSA strain ΔIP by exposure to imipenem (IPM), showed overexpression of the vraSR two-component system and a typical hVISA phenotype. In the present study, to elucidate the mechanism of VSSA conversion to hVISA, we further characterized strain H14 by determining its whole-genome sequence, morphology, cell wall synthetic activity, and gene expression. Genome sequencing revealed that H14 harbored a mutated vraS (designated vraSH14) that caused an amino acid substitution (S329→L). This mutation is different from the VraS mutation (N5→I) identified in representative clinical hVISA strain Mu3. However, H14 exhibited a phenotype similar to that of Mu3, including heterogeneous resistance to VAN, enhanced cell wall synthetic activity, and vraSR overexpression. Replacement of the vraS gene of ΔIP with the mutated vraSH14 gene confirmed that the S329→L substitution was responsible for both the upregulation of vraSR and conversion to the hVISA phenotype. This conversion was also achieved by using the vraS gene of Mu3, which carries a mutation (N5→I), but not with the native vraS gene of strain N315. Finally, we carried out a study to analyze the appearance of hVISA from VSSA by exposure of ΔIP to selective concentrations of VAN and beta-lactam antibiotics. A total of 8 and 5 hVISA isolates were detected among 50 isolates selected with VAN and IPM, respectively. Among the 13 hVISA mutants, mutation in vraSR was detected only in mutant strain H14, suggesting that additional mutational mechanisms can be responsible for evolution to the hVISA phenotype. We conclude that exposure not only to VAN but also to beta-lactams may select for reduced glycopeptide susceptibility in S. aureus.
Methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) is a major cause of serious nosocomial infections, and the emergence of virulent MRSA strains in the community is of particular concern (6). Vancomycin (VAN) still serves as the main therapeutic agent for infections caused by multiresistant MRSA strains (17). However, MRSA strains with various degrees of reduced susceptibility to glycopeptides, vancomycin-intermediate S. aureus (VISA) and heterogeneous VISA (hVISA) strains, have emerged among multidrug-resistant MRSA clinical strains (9, 16).
Recently, we identified several genes that are overexpressed in VISA strain Mu50 and hVISA Mu3 compared to their levels of expression in their isogenic VAN-susceptible S. aureus (VSSA) strain, strain Mu50Ω (13); and among these, we found the overexpression of the vraSR two-component system (TCS), an upregulator of the S. aureus cell wall biosynthesis pathway (12, 13). We also demonstrated that the vraS gene is overexpressed in ΔIP-H14 (H14), a laboratory-derived hVISA strain obtained by selecting VSSA strain N315ΔIP (ΔIP) with 8 mg/liter of imipenem (IPM) (12) and showed that a single amino acid substitution in VraS was present in H14, Mu3, and Mu50 (10a).
In the study described here, we further characterized hVISA strain H14, investigated the role of the vraS mutation on the phenotype of H14, and evaluated the rates of selection of hVISA from VSSA ΔIP following exposure to VAN and beta-lactam antibiotics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth condition.
The Staphylococcus strains and plasmids used in the present study are listed in Table 1. The cloning and transformation of Escherichia coli JM109 were carried out by standard techniques (http://catalog.takara-bio.co.jp/en/PDFFiles/9052_e.pdf; Takara-Bio Co., Ltd., Shiga, Japan). All S. aureus strains were cultivated in brain heart infusion (BHI) broth or agar (Difco Laboratories, Detroit, MI) with aeration at 37°C, unless indicated otherwise. The antibiotics tetracycline and chloramphenicol (Sigma Chemical Co., St. Louis, MO) were used for the selection of the S. aureus transformants. VAN (Sigma), teicoplanin (Astellas Pharma Inc., Osaka, Japan), ceftriaxone (CRO; Sigma), IPM (provided by Banyu Pharmaceutical Co., Tokyo, Japan), ampicillin-sulbactam (SAM; provided by Pfizer Pharmaceuticals Inc., Tokyo, Japan), and rifampin (RIF; rifampicin; Sigma) were used for antibiotic susceptibility tests. RIF-resistant (Rifr) strain ΔIP-rifR was selected by culturing ΔIP on BHI agar containing 1 mg/liter of RIF and was obtained at a frequency of 2.8 × 10−7. The isolates were evaluated for the presence of the His481→Tyr mutation in the rpoB gene of ΔIP-rifR, which confers RIF resistance, by DNA sequencing, as reported previously (26).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| N315 | Pre-MRSA strain carrying a functional mecI encoding mecA gene repressor, hetero-Metr, Nagasaki, Japan | 10 |
| N315P | Penicillinase plasmid-free strain derived from N315 | 10 |
| ΔIP | mecI null mutant derived from N315P (mecI::tetL), hetero-Metr Tetr | 15 |
| H14(or IPM8-14) | Mutant derived from ΔIP by selection with 8 mg/liter of IPM, VraS (S329→L) homo-Metr Tetr | 12 |
| ΔIP-KVR | vraSR null mutant from ΔIP (vraSR::cat), Mets Tetr Chlr | 12 |
| H14-KVR | vraSR null mutant from H14 (vraSR::cat), Mets Tetr Chlr | 12 |
| ΔIP-rifR | Mutant derived from ΔIP by selection with 1 mg/liter of RIF, RpoB (H481→Y) hetero-Metr Tetr Rifr | This study |
| ΔIP::vraSH14 | ΔIP in which vraS is replaced by vraSH14 (S329→L), homo-Metr Tetr | This study |
| ΔIP-rifR:: vraSH14 | ΔIP-rifR in which vraS is replaced by vraSH14, homo-Metr Tetr Rifr | This study |
| Mu3 | hVISA clinical strain, Tokyo, Japan | 9 |
| Mu50 | VISA clinical strain, Tokyo, Japan | 9 |
| NJ | VISA clinical strain, New Jersey | 23 |
| Plasmids | ||
| pKOR1 | S. aureus allele replacement vector | 2 |
| pKO- vraSH14 | pKOR1 harboring 1.0-kb PCR product of vraSH14 | This study |
Abbreviations: Mets, methicillin susceptible; Metr, methicillin resistant; Tetr, tetracycline resistant; Chlr, chloramphenicol resistant; Ampr, ampicillin resistant; Rifr, RIF resistant. Strains obtained by antibiotic selection are denoted “mutant.”
DNA methods.
DNA manipulations were performed by standard methods (11). Restriction enzymes were used as recommended by the manufacturer (Takara). Routine PCR amplification was performed with an Expand High Fidelity system (Roche, Mannheim, Germany).
Transmission electron microscopy.
The preparation for and the examination of S. aureus cells by transmission electron microscopy were performed as described previously (4).
Incorporation of 14C-labeled d-glucose or 14C-labeled N-acetyl-d-glucosamine into cells.
The preparation of the cells and the purification of peptidoglycan were performed as described previously (8), and the rate of incorporation was determined as described previously (8). These assays were performed in triplicate.
Whole-genome sequencing of H14 and identification of SNPs compared to the sequence of parent strain ΔIP.
Sequencing of the strain H14 genome was performed with a Genome Sequencer 20 system, a recently introduced highly parallel genome sequencer from 454 Life Sciences (Branford, CT). In a duplicate run with this instrument, 757,227 short reads, each of which had an average length of 108 bp, were collected. The short-read genome assembly generated by using the Genome Sequencer 20 system Assembler software contained 616 sequence contigs, including 95 contigs of >500 bp, for a total of 2,735,083 bp (average contig size, 28,790 bp). The chromosomal genome sequence of strain N315 (GenBank accession number BA000018) was used as a scaffold to assemble and orient the H14 contigs, and gap closing was carried out by long-range PCR primer walking. Sequencing of the PCR products was performed with a model 3730 capillary sequencer (Applied Biosystems Japan Ltd.). The resulting sequence of the H14 genome was then compared to that of N315, and the discovery of genome-wide single nucleotide polymorphisms (SNPs) was achieved by whole-genome alignments with the Mummer (version 3.20) software package (http://mummer.sourceforge.net/). The discrepancies identified between the two genomes were then verified by resequencing of the corresponding loci of N315 and H14 genomic DNAs that were as long as the corresponding loci of ΔIP, the parent strain of H14, with forward and reverse primers for each locus.
Construction of ΔIP::vraSH14 strain.
For the replacement of the serine amino acid residue with the leucine at position 329 in vraS of ΔIP, we used the pKOR1 allele replacement system, as described previously (2). In brief, a 1.0-kb vraS insert DNA encompassing 1-kb flanking sequences of phage attachment sites was generated by PCR from the chromosomal DNA of strain H14 by using the following primers: attB1-vraS (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGTACTTACGTGAATGAAAAATCATATAAAGTTGAAAATAACAA T-3′) and attB2-vraS (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATGATCATCCACAAACAATACTTTAATCGTCATACGAATCCTC-3′). The PCR product was used for recombination with pKOR1, and then recombination products were transformed to E. coli DH5α, according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). The resulting plasmid, pKO-vraSH14 (where vraSH14 indicates the mutated vraS gene harbored by strain H14) was introduced into S. aureus ΔIP by electroporation, generating transformant ΔIP(pKO-vraSH14). Replacement of the vraSH14 gene allele with vraS in ΔIP was carried out by a two-step procedure, as described previously (1, 2). In this study, ΔIP-rifR was also used for the construction of a vraS mutant. Because it became apparent by whole-genome sequence determination that the nucleotide sequences of ΔIP and H14 did not differ except for the vraS mutation described above, it was necessary to introduce an artificial marker to exclude the unnoticed contamination of H14 in the culture of ΔIP::vraSH14. We utilized the amino acid substitution His481→Asn on rpoB, which confers RIF resistance, as a marker for mutant strain ΔIP-rifR::vraSH14 in which the vraS gene was replaced by vraSH14. In the final step of the procedure for mutant construction, 40 ΔIP-rifR(pKO-vraSH14) transformants were picked from tryptic soy agar plates containing 1 ng/liter of anhydrotetracycline, as described previously (1, 2), from which we identified the desired strains in which the vraS gene was replaced by vraSH14 by determination of the nucleotide sequence.
Northern blot analysis.
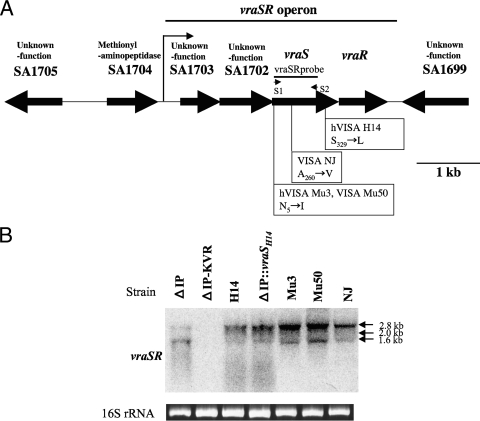
Preparation of total RNAs and Northern blot analysis for vraS and vraR were performed as described previously (12) (20). The vraSR probe was amplified by PCR with primers SR1 (5′-TCGATGAACCACTACATTAGAACA-3′) and SR2 (5′-CTGCGAATCTTGAACCATTTTCTCT-3′) (see Fig. 3A).
FIG. 3.
(A) Structural map of the vraSR operon in strains ΔIP, H14, Mu3, NJ, and Mu50 and the positions of the missense mutation in four strains. The vraSR probes used for Northern blotting analysis are illustrated by lines. The probe was amplified by PCR with the primers (short arrows) described in Materials and Methods. The arrows below the genes indicate the direction of transcription. The VraS histidine kinase has two amino-terminal transmembrane domains, located between residues 7 and 25 and between residues 49 and 67, as determined by the use of von Heijne's algorithm. The VraS kinase orthologs are similar in domain organization and contain a PAS (period clock protein, aryl hydrocarbon receptor nuclear translocator, and single-minded protein) family domain which has signaling modules essentially found in proteins involved with signal transduction. A major transcription start codon located in the 133 bp upstream of SA1703 is indicated by a thin arrow above SA1703 (27). The position of a single amino acid substitution in VraS in each VISA strain is shown on the map (10a, 19). (B) Northern blot analysis of vraSR in ΔIP, its derivative strains, hVISA strains, and VISA strains with a vraSR-specific DNA probe. Ethidium bromide staining of total RNA was used to demonstrate the equivalent loading of RNA. All strains were grown under noninducing conditions.
Antibiotic susceptibility tests.
Antibiotic susceptibility was examined by Etest (AB Biodisk, Solna, Sweden) and population analysis, as described previously (9). To detect hVISA strains among the 50 antibiotic-selected isolates, we determined the plating efficiency of each isolate on 4 mg/liter of VAN by population analysis. A VAN MIC of 4 mg/liter is the breakpoint for VISA (according to the criteria of the Clinical and Laboratory Standards Institute). hVISA status was defined for isolates that grew on VAN at 4 mg/liter at a frequency of greater than 10−6 when a total of 108 cells was inoculated.
Statistical analysis.
Two-by-two contingency tables were evaluated by Fisher's exact test. The statistical significance of the data was evaluated by Student's t test.
RESULTS AND DISCUSSION
Characterization of laboratory-derived hVISA mutant H14, which originated from VSSA strain ΔIP.
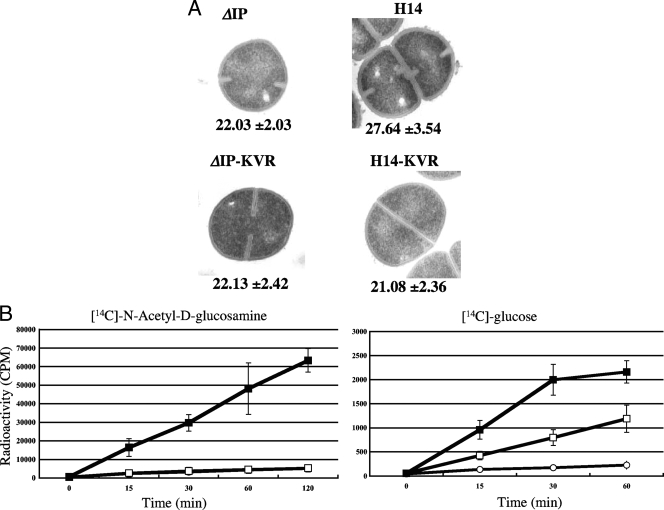
To investigate the phenotypes associated with reduced susceptibility to VAN, we analyzed the cell wall thickness and cell wall biosynthetic activity of H14, along with those of vraSR null mutant strain H14-KVR and parent strain ΔIP, which were used as controls. Electron microscopy analysis showed that H14 had a significantly thicker cell wall and a rougher cell surface than ΔIP, while H14-KVR had a cell wall much thinner than that of the parent strain (Fig. 1A). Student's t test demonstrated that the increase in cell wall thickness was statistically significant in all cases (P < 0.0001).
FIG. 1.
(A) Transmission electron micrographs of ΔIP derivative strains. The values given under each panel are the means and standard deviations of the cell wall thicknesses of the cells (in nanometers). Magnifications, ×30,000. (B) Incorporation of [14C]N-acetyl-d-glucosamine or [14C]d-glucose into the cell wall of the N315 derivative strains. Open squares, parent recipient ΔIP; open circles, H14-KVR; closed squares, H14. The counts per minute were measured at the indicated time points. The experiment was performed in triplicate on three independent occasions, and the results are shown as the mean values ± the standard deviations.
To test for cell wall biosynthetic activity, the rates of incorporation of 14C-labeled d-glucose and N-acetyl-d-glucosamine into the cell wall peptidoglycan fraction were evaluated by using a fixed number of nondividing cells. As shown in Fig. 1B, the incorporation of N-acetyl-d-glucosamine and d-glucose by H14 was significantly enhanced compared to that by ΔIP and H14-KVR. These data indicate that the vraS mutation could cause enhanced cell wall biosynthesis activity.
The vraS mutation is the only genetic event responsible for hVISA phenotype acquisition in H14.
To clarify the genetic mechanism of hVISA phenotype acquisition, genome sequencing of H14 was carried out. The 2,795,992-bp-long whole-genome sequence of H14 was determined. Since H14 is an in vitro derivative of N315, the sequence was practically identical to that of N315 (14), with the exceptions being 42 SNPs and 5 insertions or deletions between the two chromosomes (data not shown). PCR amplification and sequencing of ΔIP chromosomal DNA for the 45 differences found no additional mutations except for the previously reported vraS mutation, which causes the replacement of a single amino acid, Ser329 with Leu, in H14. This result confirms that the vraS mutation causing the replacement of Ser329 with Leu was the only genetic event responsible for the acquisition of the hVISA phenotype in H14.
Phenotypic change conferred by vraS gene replacement.
To see if the single amino acid substitution in the sequence encoding VraSH14 is responsible for both the phenotypic expression of hVISA and the overexpression of vraSR, we constructed two vraS mutants from ΔIP and its RIF-resistant strain, ΔIP-rifR. The native vraS gene of the parent strains was replaced by vraSH14 to obtain ΔIP::vraSH14 and ΔIP-rifR::vraSH14. Of 40 transformants analyzed, 3 (7.5%) were found to carry vraSH14.
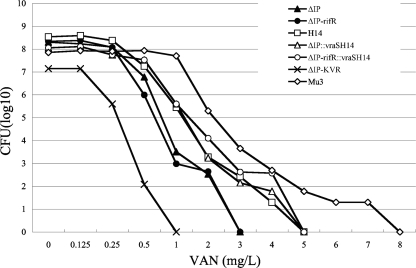
To evaluate the vraSH14-mediated phenotypic conversion, we determined the VAN susceptibilities of ΔIP::vraSH14 and ΔIP-rifR::vraSH14 (vraSH14 mutant strains) by the Etest method and population analysis. As shown in Table 2, the MICs of glycopeptides for the vraSH14 mutant were higher than those for the parent strain. The population analysis of ΔIP::vraSH14 and ΔIP-rifR::vraSH14 showed an increase in the proportion of VAN-resistant subpopulations compared with that for parent strain ΔIP (Fig. 2). The patterns of the population curves for H14 and ΔIP::vraSH14 were almost superimposable on each other. The same result was obtained by using ΔIP with the RIF resistance marker (the rpoB mutation), except that the population curve for ΔIP-rifR::vraSH14 was slightly deflected with 4 mg/liter of VAN. These data clearly demonstrate that the vraS mutation alone can confer the phenotypic expression of heterogeneous VAN resistance.
TABLE 2.
Antibiotic susceptibility profile of ΔIP and its derivative strains determined by Etest
| Strain | MIC (mg/liter)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| RIF | Oxacillin | IPM | VAN | Teicoplanin | Fosfomycin | Bacitracin | Daptomycin | Gentamicin | |
| ΔIP | 0.004 | 6 | 0.75 | 1 | 1 | 1 | 32 | 0.75 | 0.75 |
| H14 | 0.004 | >256 | >32 | 2 | 8 | 96 | 96 | 1.75 | 0.75 |
| ΔIP-KVR | 0.004 | 1.5 | 0.19 | 0.75 | 0.25 | 0.38 | 4 | 0.5 | 0.75 |
| H14-KVR | 0.004 | 1.5 | 0.19 | 0.75 | 0.25 | 0.38 | 4 | 0.5 | 0.75 |
| ΔIP-rifR | >32 | 5 | 1 | 1 | 1 | 1.5 | 24 | 0.5 | 1 |
| ΔIP::vraSH14 | 0.004 | >256 | >32 | 2 | 8 | 96 | 96 | 1.75 | 0.75 |
| ΔIP-rifR::vraSH14 | >32 | >256 | >32 | 3 | 12 | 96 | 96 | 2 | 1 |
| Mu3 | 0.006 | >256 | >32 | 3 | 24 | >1,024 | 256 | 2 | 384 |
FIG. 2.
Resistant subpopulation profiles of ΔIP, its derivative vraSR mutants, and hVISA Mu3. The numbers of cells (log10 CFU per milliliter) growing on BHI agar containing VAN are shown on the y axis; the VAN concentrations are shown on the x axis. The number of colonies that grew was counted after incubation at 37°C for 48 h.
The MICs were then determined, and the MICs of IPM, fosfomycin, bacitracin, and daptomycin for ΔIP::vraSH14 and ΔIP-rifR::vraSH14 were found to be increased compared with the MICs for ΔIP for which the MICs were similar to those for H14 (Table 2). Recently, Muthaiyan et al. reported that the inactivation of vraSR significantly increased susceptibility to daptomycin (18). It was concluded that the vraS mutation (Ser329→Leu) alone was responsible for the altered antibiogram of H14 compared to that of its parent, strain ΔIP.
Assessment of vraS activation among VISA strains carrying a mutation in the vraSR operon.
By Northern blot analysis, we examined the transcriptional level of vraSR in the hVISA and VISA strains in which a single amino acid substitution in the vraSR operon has already been identified (12). The positions of mutations in the vraSR operons of different strains are shown in Fig. 3A. Northern blot analysis demonstrated the overexpression of vraS in ΔIP-rifR::vraSH14, which was indistinguishable from the level of expression by original mutant strain H14 (Fig. 3B). The fact that the whole genome of H14 has only a single mutation, in the vraS gene, compared to the sequence of ΔIP suggests that the VraSH14 S329→L substitution (on the ATP-binding domain [amino acids 248 to 340]) affects the activation of the VraS sensor histidine kinase by modulating its autophosphorylation (3). The overexpression of vraS was also observed in VISA strains NJ and Mu50 and hVISA strain Mu3 (Fig. 3B). The VraS A260→V substitution of NJ may also be associated with the activation of histidine kinase activity. These data are consistent with the findings of this study that implicate the increased expression of the vraSR system in the conversion of VSSA to hVISA. It has not previously been reported that amino acid mutations in a global regulator such as the vraSR TCS are involved in heterogeneous VAN resistance in S. aureus. Further investigation is needed to clarify the relationship between the function of TCS and the role of a mutation in vraS.
Recently, evidence of the evolution from hVISA to VISA in vivo during infection has been reported (5, 7, 21, 22, 24, 25). However, the present study described for the first time that a mutation in vraS is responsible for the increased level of transcription of vraS and the phenotypic expression of hVISA by the gene replacement experiment. It was also confirmed that this mutation could cause cell wall thickening and increase the rate of incorporation of N-acetyl-d-glucosamine and d-glucose into cells (Fig. 1B). Although the location of the VraS mutation identified in H14 is different from that identified in clinical hVISA strain Mu3, both mutations had the same effect on conferring VAN resistance in gene replacement experiments (data not shown).
Beta-lactam antibiotics can select hVISA mutants from VSSA strain ΔIP.
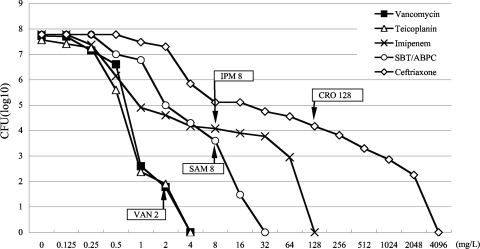
We exposed VSSA strain ΔIP to selective concentrations of three beta-lactams, IPM, SAM, and CRO, to determine the ability to select mutants with reduced VAN susceptibility. ΔIP is a derivative of N315 in which the mecI repressor was inactivated and from which the β-lactamase-carrying plasmid was cured (Table 1). The genotype of inactivated mecI and the absence of the β-lactamase-carrying plasmid are characteristic of hVISA strain Mu3 and VISA strain Mu50, as reported previously (9). Therefore, for this study we used ΔIP as a representative Japanese health care-associated MRSA strain from which VISA has emerged. We spread 108 CFU from an overnight culture on BHI agar plates containing several concentrations of antibiotics. As a control for this assay, we also spread them onto the agar plate containing VAN. We then picked 50 putative mutant colonies from each plate containing a selective concentration of antibiotic (VAN, 2 mg/liter; IPM, 8 mg/liter; SAM, 8 mg/liter; or CRO, 128 mg/liter) and established them as mutant strains by a colony purification procedure. A selective concentration was defined as the concentration which resulted in a reduction of the initial bacterial population by approximately 6 log units (with VAN) or 4 log units (with IPM, CRO, and SAM) (Fig. 4).
FIG. 4.
Profiles of antibiotic-resistant subpopulations. The numbers of cells (log10 CFU per milliliter) growing on antibiotic-containing BHI agar are shown on the y axis. The number of colonies of VSSA strain ΔIP growing was counted after incubation at 37° C for 72 h. The concentrations of VAN, IPM, CRO, and SAM are shown on the x axis. The numbers after the antibiotic designations indicate selective antibiotic concentrations (in mg/liter). SBT-ABPC, sulbactam-ampicillin.
To detect the presence of hVISA isolates among the selected isolates, all isolates were investigated for their VAN susceptibilities by population analysis. Eight and 5 hVISA isolatess were obtained from among the 50 isolates selected with VAN and IPM, respectively. No hVISA isolate was detected among the isolates selected with CRO or SAM. The rates of appearance of hVISA isolates among the 50 isolates selected with VAN and IPM isolates were 3.6 × 10−7 and 2.0 × 10−5, respectively.
We determined the nucleotide sequence of vraSR from 13 hVISA isolates, and a mutation was only in mutant strain H14. This suggests that different genetic mechanisms, other than vraS mutations, are used to acquire hVISA phenotypic expression. From these results, we also confirmed that hVISA can be selected not only by VAN but also by IPM. Since IPM was frequently used for the treatment of MRSA infections before the clinical introduction of VAN in 1991 in Japan, we suspect a possible role of IPM in the early emergence of hVISA in Japan, ahead of its emergence in other countries in 1996.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research on Priority Areas 16017294, Grant-in-Aid for Young Scientists (B) 17790677, and Grant-in-Aid for 21st Century COE Research from the Ministry of Education, Science, Sport, Culture and Technology of Japan.
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 101:12312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58-63. [DOI] [PubMed] [Google Scholar]
- 3.Belcheva, A., and D. Golemi-Kotra. 2008. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354-12364. [DOI] [PubMed] [Google Scholar]
- 4.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui, L., H. M. Neoh, M. Shoji, and K. Hiramatsu. 2009. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 53:1231-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deresinski, S. 2005. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin. Infect. Dis. 40:562-573. [DOI] [PubMed] [Google Scholar]
- 7.Finan, J. E., G. L. Archer, M. J. Pucci, and M. W. Climo. 2001. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 42:315-320. [DOI] [PubMed] [Google Scholar]
- 9.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu, K., K. Asada, E. Suzuki, K. Okonogi, and T. Yokota. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298:133-136. [DOI] [PubMed] [Google Scholar]
- 10a.Katayama, Y., et al. 2004. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-946.
- 11.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185:2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 15.Kuwahara-Arai, K., N. Kondo, S. Hori, E. Tateda-Suzuki, and K. Hiramatsu. 1996. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2′ production. Antimicrob. Agents Chemother. 40:2680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, C., and H. F. Chambers. 2003. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 47:3040-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moellering, R. C., Jr. 2005. The management of infections due to drug-resistant gram-positive bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 24:777-779. [DOI] [PubMed] [Google Scholar]
- 18.Muthaiyan, A., J. A. Silverman, R. K. Jayaswal, and B. J. Wilkinson. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mwangi, M. M., S. W. Wu, Y. Zhou, K. Sieradzki, H. de Lencastre, P. Richardson, D. Bruce, E. Rubin, E. Myers, E. D. Siggia, and A. Tomasz. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 104:9451-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neoh, H. M., L. Cui, H. Yuzawa, F. Takeuchi, M. Matsuo, and K. Hiramatsu. 2008. Mutated response regulator graR is responsible for phenotypic conversion of Staphylococcus aureus from heterogeneous vancomycin-intermediate resistance to vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 52:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neoh, H. M., S. Hori, M. Komatsu, T. Oguri, F. Takeuchi, L. Cui, and K. Hiramatsu. 2007. Impact of reduced vancomycin susceptibility on the therapeutic outcome of MRSA bloodstream infections. Ann. Clin. Microbiol. Antimicrob. 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pieper, R., C. L. Gatlin-Bunai, E. F. Mongodin, P. P. Parmar, S. T. Huang, D. J. Clark, R. D. Fleischmann, S. R. Gill, and S. N. Peterson. 2006. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 6:4246-4258. [DOI] [PubMed] [Google Scholar]
- 23.Roberts, R. B., M. Chung, H. de Lencastre, J. Hargrave, A. Tomasz, D. P. Nicolau, J. F. John, Jr., and O. Korzeniowski. 2000. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey, and Pennsylvania. Microb. Drug Resist. 6:245-251. [DOI] [PubMed] [Google Scholar]
- 24.Sieradzki, K., T. Leski, J. Dick, L. Borio, and A. Tomasz. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 26.Wichelhaus, T. A., V. Schafer, V. Brade, and B. Boddinghaus. 1999. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin, S., R. S. Daum, and S. Boyle-Vavra. 2006. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]