Abstract
Lacticin Q is a pore-forming bacteriocin produced by Lactococcus lactis QU 5, and its antimicrobial activity is in the nanomolar range. Lacticin Q induced calcein leakage from negatively charged liposomes. However, no morphological changes in the liposomes were observed by light scattering. Concomitantly with the calcein leakage, lacticin Q was found to translocate from the outer to the inner leaflet of the liposomes, after it initially bound to the membrane within 2 s. Lacticin Q also induced lipid flip-flop. These results reveal that the antimicrobial mechanism of lacticin Q can be described by the toroidal pore model. This is the first report of a bacteriocin of gram-positive bacteria that forms a toroidal pore. From liposomes, lacticin Q leaked fluorescence-labeled dextran with a diameter of 4.6 nm. In addition, lacticin Q caused the leakage of small proteins, such as the green fluorescent protein, from live bacterial cells. There are no other reports of antimicrobial peptides that exhibit protein leakage properties. The proposed pore formation model of lacticin Q is as follows: (i) quick binding to outer membrane leaflets; (ii) the formation of at least 4.6-nm pores, causing protein leakage with lipid flip-flop; and (iii) the migration of lacticin Q molecules from the outer to the inner membrane leaflets. Consequently, we termed the novel pore model in the antimicrobial mechanism of lacticin Q a “huge toroidal pore.”
Bacteriocins are antimicrobial peptides or proteins produced by bacterial strains (7). Small bacteriocins (<10 kDa) produced by lactic acid bacteria (LAB) are potential food preservatives and alternatives to antibiotics (5, 7, 13). The antimicrobial mechanisms of the LAB bacteriocins nisin, pediocin PA-1, and lacticin 3147 are well characterized (3, 5, 9, 11, 13, 34). These LAB bacteriocins show strong activity against specific gram-positive bacteria in nanomolar concentration ranges. These bacteriocins cause ion efflux via pore formation with the aid of initial receptors, the so-called docking molecules (15). A peptidoglycan precursor, lipid II, is used as the docking molecule for nisin and lacticin 3147. These bacteriocins do not exhibit pore-forming activities against liposomes without the docking molecule (34, 35). In addition, other killing mechanisms that use lipid II are known to involve the inhibition of peptidoglycan biosynthesis (34, 35). Similarly, a membrane protein, MptD, is the docking molecule for pediocin PA-1 and its homologs, and a region inserted in MptD observed only in sensitive species such as Listeria and Enterococcus is the specific recognition site (11, 14). These types of bacteriocins do not demonstrate pore-forming activity against liposomes in the absence of MptD or against bacteria containing no specific region in MptD (8, 26). In conclusion, docking molecules play essential roles in the antimicrobial activities of these LAB bacteriocins.
Multicellular eukaryotes also produce pore-forming antimicrobial peptides to prevent microbial invasion (6, 38). Some antimicrobial peptides inhibit both gram-positive and gram-negative bacteria in the micromolar range through a membrane permeation mechanism such as mechanisms involving the barrel stave, carpet, and toroidal pore. One of the most-characterized peptides involved in antimicrobial activities is magainin 2, produced by Xenopus laevis (18). Initially, the cationic peptide magainin 2 binds to negatively charged bacterial membranes and forms an amphiphilic α-helical structure. Magainin 2 then forms a pore, which is accompanied by rapid lipid transbilayer movement, the so-called lipid flip-flop. The peptide-lipid supramolecular complex pore is called the toroidal pore. The diameter of the magainin 2 toroidal pore, which can leak water-soluble substances, is estimated to be 2 to 3 nm. The pore causes the leakage of calcein (molecular weight, 622) but not that of dextran, which has an average molecular weight of about 4,400 (19). Some magainin 2 molecules are translocated from the outer membrane leaflets to the inner membrane leaflets when the pore closes. While the peptide-lipid supramolecular complex pores caused by peptides from some multicellular eukaryotes have been well characterized, those created by LAB bacteriocins are poorly understood.
Previously, we discovered a novel LAB bacteriocin, lacticin Q, produced by Lactococcus lactis QU 5 (10). Lacticin Q, composed of 53 amino acids without intramolecular bridges, is a cationic membrane-permeating peptide that has a wide bactericidal activity against a wide variety of gram-positive bacteria in the nanomolar range. The antimicrobial activity, pH stability, and heat tolerance of lacticin Q are comparable to those of nisin, which shows a higher degree of stability than other LAB bacteriocins. Unlike typical LAB bacteriocins, such as nisin, the pore-forming activity of lacticin Q does not require a docking molecule, as confirmed by its high level of activity against negatively charged liposomes (37). Lacticin Q forms an amphiphilic α-helix, which is frequently observed in antimicrobial peptides (37, 38).
In the present study, we characterized the features of the antimicrobial mechanism of lacticin Q. These features were found to be more similar to those of magainin 2 than to those of typical LAB bacteriocins. On the other hand, lacticin Q exerted strong antimicrobial activity in the nanomolar range, and the activity of magainin 2 was in the micromolar range. We hypothesized that lacticin Q possesses unique features in its antimicrobial mechanism. Here, we report on a new lacticin Q-mediated antimicrobial mechanism termed the “huge toroidal pore” (HTP).
MATERIALS AND METHODS
Antimicrobial peptides.
Lacticin Q and nisin were purified from an L. lactis QU 5 culture supernatant and a commercial preparation (Sigma, St. Louis, MO), respectively, as described previously (10). Magainin 2 was chemically synthesized by standard solid-phase peptide synthesis (27). The purities of the peptides were checked by electrospray ionization-time-of-flight mass spectrometry on a JMS-T100LC instrument (Jeol, Tokyo, Japan).
Liposome preparation.
A zwitterionic phospholipid, egg yolk l-α-phosphatidylcoline (PC), and an anionic phospholipid, l-α-phosphatidyl-dl-glycerol (PG), were purchased as the highest grade from Sigma. Liposomes, large unilamellar vesicles (LUVs), were prepared as follows. A lipid film (PC-PG at a 1:1 molar ratio) was hydrated with buffer A (10 mM Tris-HCl, 75 mM NaCl, 1 mM EDTA, pH 7.4). For the leakage experiments, 70 mM calcein (Dojindo Laboratories, Kumamoto, Japan) was added to buffer A when the lipid film was hydrated. The suspension underwent 10 freeze-thaw cycles and was subsequently extruded through a 0.1-μm-pore-size polycarbonate filter. For the calcein-entrapping LUVs, untrapped calcein was removed by gel filtration chromatography with a 4% plain agarose bead column (Agarose Bead Technologies, Madrid, Spain). The phosphorus-based lipid concentration (of the lipid stocks and LUVs) was determined by the method of Bartlett (2). All other LUVs used in this study contained a negatively charged lipid (PG) at a molar ratio of 50%.
Calcein leakage.
Calcein leakage from LUVs was examined at an excitation wavelength (EX) of 490 nm and emission wavelength (EM) of 520 nm on an F-7000 spectrofluorometer (Hitachi High-Technologies, Tokyo, Japan). The peptides were maintained in buffer A at 30°C with agitation, prior to the addition of LUVs to the solution. Leakage of 100% was determined by the addition of 0.1% (vol/vol) Triton X-100 (Wako Pure Chemical Industries, Osaka, Japan).
Light scattering.
Peptide-induced morphological changes in the LUVs with PC-PG at a ratio of 1:1 were monitored in buffer A by using 90° light scattering on the F-7000 spectrofluorometer at 30°C. Both EX and EM were set at 400 nm.
Peptide translocation.
A dansyl-labeled lipid, N-(5-dimethylaminonaphthalene-1-sulfonyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (DNS-DHPE), was purchased from Invitrogen (Carlsbad, CA). Dansyl-labeled LUVs (PC-PG-DNS-DHPE, 4:5:1) and unlabeled LUVs (PC-PG, 1:1) were prepared by the aforementioned procedures. Peptide migration from the outer to the inner membrane leaflets (translocation) was detected by fluorescence resonance energy transfer (FRET) from the tryptophan residues of lacticin Q to dansyl-labeled LUVs, as reported previously (21). Dansyl-labeled LUVs were added to the peptides in buffer A on the F-7000 spectrofluorometer at an EX of 280 nm and an EM of 336 nm at 30°C. After incubation, excess unlabeled LUVs were added for the desorption of untranslocated peptides from the dansyl-labeled LUVs. On the basis of the fluorescence intensities before and after the addition of unlabeled LUVs, the ratios of peptide translocation were calculated, as reported previously (21).
Calcein leakage from the dansyl-labeled LUVs (PC-PG-DNS-DHPE, 4:5:1) was monitored by the method described above.
Flip-flop.
A pyrene-labeled lipid, 1-lauroyl-2-(1′-pyrenebutyroyl)-sn-glycero-3-phosphocholine (pyPC), was purchased from Invitrogen. The fluorescence of pyPC depends on the distance between the molecules (17, 32). Symmetrically pyrene-labeled LUVs show monomer fluorescence (Im) at 397 nm (exited at 341 nm). In the case of the asymmetrically pyrene-labeled LUVs, excimer fluorescence (Ie) at 479 nm is observed, whereas Im is lower than that of the symmetrical LUV. The Ie/Im ratio is the indicator of transbilayer lipid movement.
Asymmetrically and symmetrically pyrene-labeled LUVs (PC-PG-pyPC, 47:50:3) were prepared as described previously (25). Briefly, to prepare asymmetrically pyrene-labeled LUVs, dried pyPC was dissolved in 50 μl of ethanol, and 1 ml of buffer A was added to form a pyPC micelle. The pyPC micelle (3% of the final product) and unlabeled LUVs (PC-PG, 47:50) were mixed at 37°C with agitation for a few minutes, and the mixture was kept overnight at room temperature. Peptide-mediated lipid flip-flop in buffer A was detected on the F-7000 spectrofluorometer at an EX of 341 nm and an EM of 350 to 500 nm. The fluorescence spectra of symmetrically and asymmetrically pyrene-labeled LUVs served as positive and negative controls, respectively. The extent of flip-flop was calculated from the spectrum Ie/Im ratio, as reported previously (25).
Calcein leakage from the pyrene-labeled LUVs (PC-PG-pyPC, 47:50:3) was monitored by the method described above.
Dextran leakage.
Fluorescein isothiocyanate-labeled dextrans (FITC-dex) with average molecular weights of about 10,000, 20,000, and 40,000 (FD-10K, FD-20K, and FD-40K, respectively) were purchased from Sigma. FITC-dex-entrapping dansyl-labeled LUVs (PC-PG-DNS-DHPE, 45:50:5) were prepared by the method described above. FITC-dex (2.5 mM) was added to buffer A when the lipid film was hydrated. Untrapped FITC-dex was removed by gel filtration chromatography, as described above.
FITC-dex leakage was monitored as described previously (21). Briefly, dansyl-labeled LUVs (50 μM lipids) harboring FITC-dex were incubated with peptides in buffer A at 30°C. After 10 min, an excess of unlabeled LUVs (500 μM lipids; PC-PG, 1:1) was added to stop the leakage. Leaked FITC-dex was eliminated by gel filtration chromatography, and the LUV fraction was collected. After addition of 0.1% Triton X-100, the excitation spectrum of the fraction was measured on the F-7000 spectrofluorometer at an EX of 300 to 500 nm and an EM of 520 nm. The spectrum was normalized at 490 nm. The extent of FITC-dex leakage was determined by measurement of the fluorescence intensity at 340 nm (21).
GFP leakage.
A green fluorescent protein (GFP)-encoding plasmid, pGreen (23), was a kind gift from the National Institute of Genetics (Mishima, Japan). The gfp gene and two PstI sites located immediately upstream and downstream of gfp were amplified by PCR with the DNA polymerase KOD plus (Toyobo, Osaka, Japan) and oligonucleotide primers pGreen-F (5′-CAGCTATGGTACCGGTAGA-3′) and pGreen-R (5′-CAGGTCGACTCTAGAGGAT-3′). The PCR product and a lactococcal expression vector, pNZ8048 (22), were treated with PstI (Nippon Gene, Toyama, Japan) and were ligated by the use of Ligation high (Toyobo). The plasmid that was constructed, pNZgfp, was introduced into L. lactis NZ9000 by electroporation by standard procedures (1). The transformant, L. lactis NZgfp, was grown in M17 medium (Merck, Whitehouse Station, NJ) supplemented with 0.5% (wt/vol) glucose (GM17) and 50 μg/ml chloramphenicol at 30°C.
For the induction of GFP in L. lactis NZgfp, 10 ng/ml nisin was added 2 h after the beginning of culture, and the cells were grown for an additional 2.5 h. The cells were harvested by centrifugation (6,000 × g, 10 min, 4°C) and washed twice with cold buffer A. The cells were added to the peptide solution in buffer A (1 ml) to adjust the absorbance at 600 nm (A600) to 0.25 and were incubated for 10 min at 30°C with agitation. The cells were then harvested by centrifugation (6000 × g, 10 min, 4°C) and resuspended in 20 μl of buffer A. The remaining GFP was monitored by fluorescence microscopy (Eclipse 80i/D-FL microscope with a GFP-B filter; Nikon, Tokyo, Japan). The fluorescent image was contrasted with the bright-field view.
The amount of GFP that leaked into the supernatant was measured on the F-7000 spectrofluorometer. GFP expression in the L. lactis NZgfp cells and peptide treatment were performed by the procedures described above. The peptide-treated cells were eliminated by centrifugation. The supernatant (800 μl), 1 μM (glycine-treated) Alexa Fluor 430 (Invitrogen; 10 μl), and buffer A (1190 μl) were mixed together. The excitation spectrum of the mixture was recorded at 30°C. From the fluorescence intensities of 520 nm excited at 440 nm (FAF; Alexa Fluor 430) and 490 nm (FGFP; GFP), the ratio FGFP/FAF was calculated to determine the amount of GFP that leaked into the supernatant. As a positive control, the FGFP/FAF of whole cells was recorded. Fluorescence was seldom confirmed in cells in which GFP was not expressed.
Protein leakage.
L. lactis NZgfp and L. lactis IL1403, bacterial strains sensitive to lacticin Q, were cultured overnight in GM17 supplemented with and without 50 μg/ml chloramphenicol, respectively. Listeria innocua ATCC 33090T, a strain that is also sensitive to lacticin Q, was cultured in tryptic soy broth (Difco Laboratories, Detroit, MI) supplemented with 0.6% (wt/vol) yeast extract (Nacalai Tesque, Kyoto, Japan). Subsequently, the strains were inoculated onto their respective fresh media and grown to the logarithmic phase (A600 = 1.0) at 30°C. The cells were washed two times with cold buffer A and resuspended. The cells (1 ml, A600 = 0.5) were treated with the peptide at the indicated concentration for 10 min at 30°C with agitation. The cells were removed by two centrifugations (6,000 × g, 10 min, 4°C), and 900 μl of the supernatant was desalted and concentrated by use of a PAGEprep Advance kit (Pierce Biotechnology, Rockford, IL), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining with 2D-silver stain II (Cosmo Bio, Tokyo, Japan) were performed.
RESULTS
Membrane permeation of lacticin Q.
Lacticin Q-inducing calcein leakage from PC-PG LUVs was monitored (Fig. 1A). The calcein leakage started at a very low concentration of 31.3 nM.
FIG. 1.

Effects of lacticin Q on LUVs, calcein leakage, and morphological change. (A) Lacticin Q-induced leakage of calcein entrapped in PC-PG (1:1) LUVs. From the top, the traces show the leakage when lacticin Q was added at concentrations of 250, 125, 62.5, and 31.3 nM. The lipid concentration of the PC-PG LUVs was 50 μM. As controls, 0% leakage and 100% leakage were obtained by the addition of buffer and 0.1% Triton X-100, respectively. (B) Light scattering of PC-PG LUVs treated with lacticin Q. The arrow indicates the time of addition of PC-PG LUVs (50 μM lipids) to 250 nM lacticin Q (black line) and buffer (gray line). a.u., arbitrary units.
Next, we analyzed the LUVs for morphological changes by light scattering when lacticin Q was exhibiting membrane permeation of the LUVs. The absence of morphological changes in the LUVs was confirmed by treatment with 250 nM lacticin Q (Fig. 1B). In addition, a higher concentration of lacticin Q (2 μM) did not cause any morphological changes in the LUVs (data not shown). These results revealed that lacticin Q does not affect the size of the LUVs.
Characterization of lacticin Q pore.
We investigated peptide translocation by the detection of FRET. As shown in Fig. 2A, the tryptophan fluorescence energy of lacticin Q was transferred to DNS-DHPE when dansyl-labeled LUVs were added, resulting in a rapid reduction in the fluorescence intensity. After incubation, excess unlabeled LUVs were added. Lacticin Q molecules locating on the outer leaflet of the dansyl-labeled LUVs redistributed to the unlabeled LUVs, and an increase in the fluorescence intensity was observed. However, depending on the time of incubation with the dansyl-labeled LUVs, the fluorescence intensity recovered was lower than that for the control, to which both dansyl-labeled and excess unlabeled LUVs were added simultaneously. These results indicate that lacticin Q molecules translocated from the outer to the inner leaflets of LUVs. In addition, fast FRET showed quick binding. The ratio of translocated lacticin Q was calculated from the results shown in Fig. 2A, and the data are shown with the results of the calcein leakage experiment (Fig. 2B). This indicates that changes in the ratios of translocated lacticin Q were closely related to pore formation.
FIG. 2.
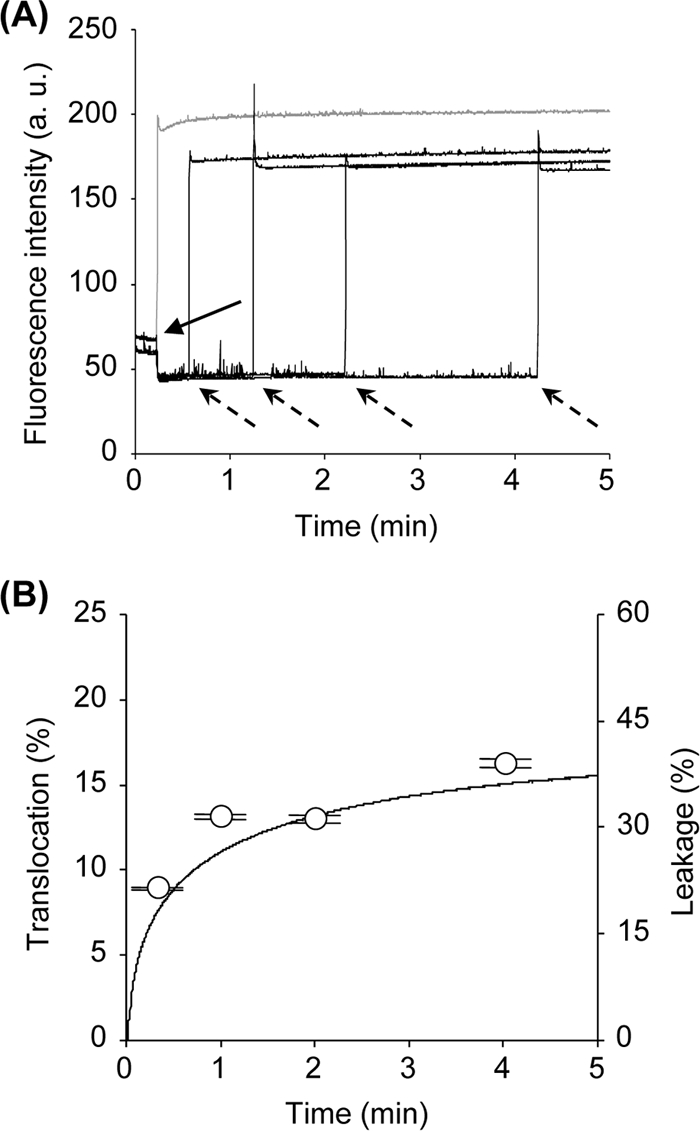
Transbilayer movement (translocation) of lacticin Q. (A) Detection of lacticin Q translocation by using FRET. The tryptophan fluorescence of 125 nM lacticin Q was monitored at an EM of 336 nm and an EX of 280 nm. Solid and dotted arrows show the times of addition of dansyl-labeled LUVs (PC-PG-DNS-DHPE, 4:5:1) and excess unlabeled LUVs (PC-PG, 1:1), respectively. As a control (gray line), the dansyl-labeled and unlabeled LUVs were added together. Lipid concentrations of dansyl-labeled and unlabeled LUVs at a large excess were adjusted to 50 μM and 350 μM, respectively. a.u., arbitrary units. (B) Co-occurrence of translocation of lacticin Q and calcein leakage. Open circles, translocated lacticin Q molecules (in percent) calculated from the data presented in panel A (see Materials and Methods). The line indicates the leakage of calcein entrapped in the dansyl-labeled LUVs (PC-PG-DNS-DHPE, 4:5:1). The leakage experiment was performed under the same conditions used for the detection of translocation (125 nM lacticin Q and 50 μM lipids).
Next, lipid flip-flop was detected by using symmetrically and asymmetrically pyrene-labeled LUVs. In the case of symmetrically pyrene-labeled LUVs, a high Im (397 nm) was observed, and a peak corresponding to Ie (479 nm) was hardly detected (Fig. 3A, trace 1). On the other hand, lower Im and higher Ie values were observed in the spectrum of the asymmetrically pyrene-labeled LUVs (Fig. 3A, trace 2). Treatment with lacticin Q led to the enhancement of Im and a reduction of Ie in the asymmetrically pyrene-labeled LUVs (Fig. 3A, trace 3). The changes in pyrene fluorescence indicate that lipid flip-flop was induced by lacticin Q. Time-dependent lipid flip-flop was analyzed from the ratio of Ie/Im, and these data are plotted along with the results of the calcein leakage experiments (Fig. 3B). The ratio of lipid flip-flop induced by lacticin Q was closely related to pore formation, as was observed in the case of peptide translocation. These features of the lacticin Q pore (calcein leakage, peptide translocation, and lipid flip-flop) corresponded to those observed in toroidal pores created by antimicrobial peptides from multicellular eukaryotes.
FIG. 3.
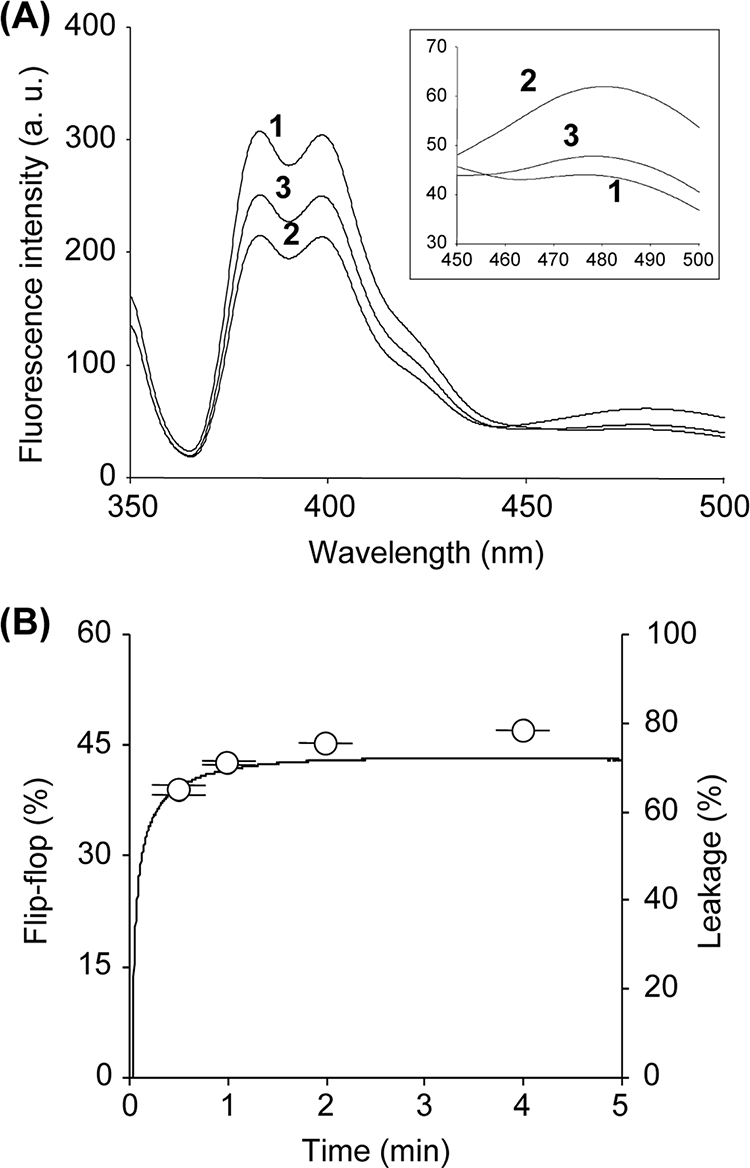
Lipid flip-flop induced by lacticin Q. (A) Fluorescence spectra of pyrene-labeled LUVs (PC-PG-pyPC, 47:50:3). Trace 1, the fluorescence spectra of the symmetrically labeled LUVs; traces 2 and 3, asymmetrically labeled LUVs and 250 nM lacticin Q-treated LUVs, respectively. (Inset) Enlarged view of the same spectra at 450 to 500 nm. The lipid concentration of the pyrene-labeled LUVs was 50 μM. a.u., arbitrary units. (B) Co-occurrence of lipid flip-flop and calcein leakage caused by lacticin Q. Open circles, lipid flip-flop (in percent) calculated from the ratio of Ie/Im (see Materials and Methods). The line indicates the leakage of calcein entrapped in the pyrene-labeled LUVs (PC-PG-pyPC, 47:50:3). The leakage experiment was performed under the same conditions used for lipid flip-flop (250 nM lacticin Q and 50 μM lipids).
Sizing of lacticin Q pore on LUVs.
We focused on the lacticin Q pore size, because lacticin Q exerts a high level of antimicrobial activity in the nanomolar range. Other antimicrobial peptides forming toroidal pores show activity in the micromolar range. Dansyl-labeled LUVs containing FITC-dex of various molecular weights were applied to determine the size of the lacticin Q pore. In the case of FD-10K, the concentration dependence of the leakage was similar to that of calcein (Fig. 4). However, the levels of FD-20K and FD-40K leakage were clearly lower than the level of FD-10K leakage, indicating that the average lacticin Q pore diameter is between the diameters of FD-10K and FD-20K.
FIG. 4.
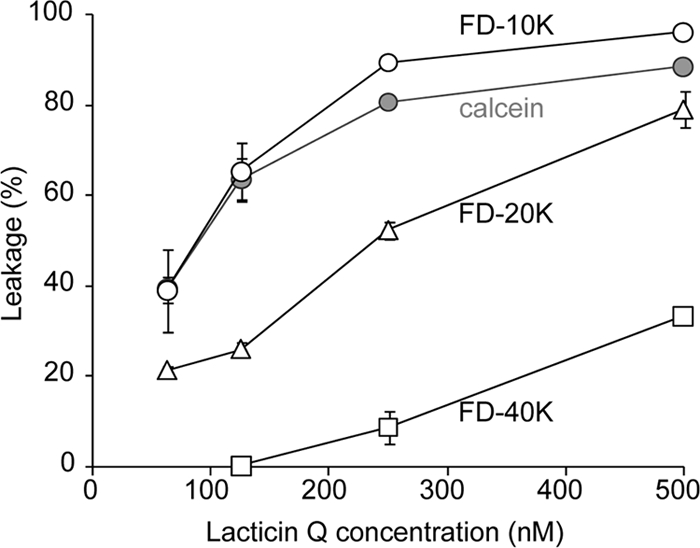
FITC-dex leakage by lacticin Q. Dansyl-labeled LUVs (PC-PG-DNS-DHPE, 45:50:5) entrapping FITC-dex were treated with various concentrations of lacticin Q for 10 min. Open circles, triangles, and squares, leakage (in percent) of FD-10K, FD-20K, and FD-40K, respectively; gray circles, leakage (in percent) of calcein (molecular weight, 622) determined by the method described in the legend to Fig. 1. The concentration of lipids was 50 μM.
Intracellular protein leakage by lacticin Q.
Next, we investigated whether lacticin Q could leak a small protein, GFP (27 kDa), from L. lactis NZgfp cells (Fig. 5). GFP-overexpressing L. lactis NZgfp cells were treated with lacticin Q. Nisin and magainin 2 were also applied as controls. To visualize a sufficient number of cells, the cell density used in this experiment (see Materials and Methods) was fivefold higher than that used for the MIC determinations described previously (36, 37). Only the lacticin Q treatment (5 μM) led to the loss of GFP fluorescence in the cells, while almost all cells used as negative controls (no peptide added) or treated with nisin or magainin 2 retained GFP (Fig. 5A). Conversely, the GFP released by the peptides into the supernatant was monitored with a standard fluorescence substance, Alexa Fluor 430 (Fig. 5B). Only lacticin Q caused an increase in the fluorescence of GFP, in agreement with the microscopic observations shown in Fig. 5A.
FIG. 5.
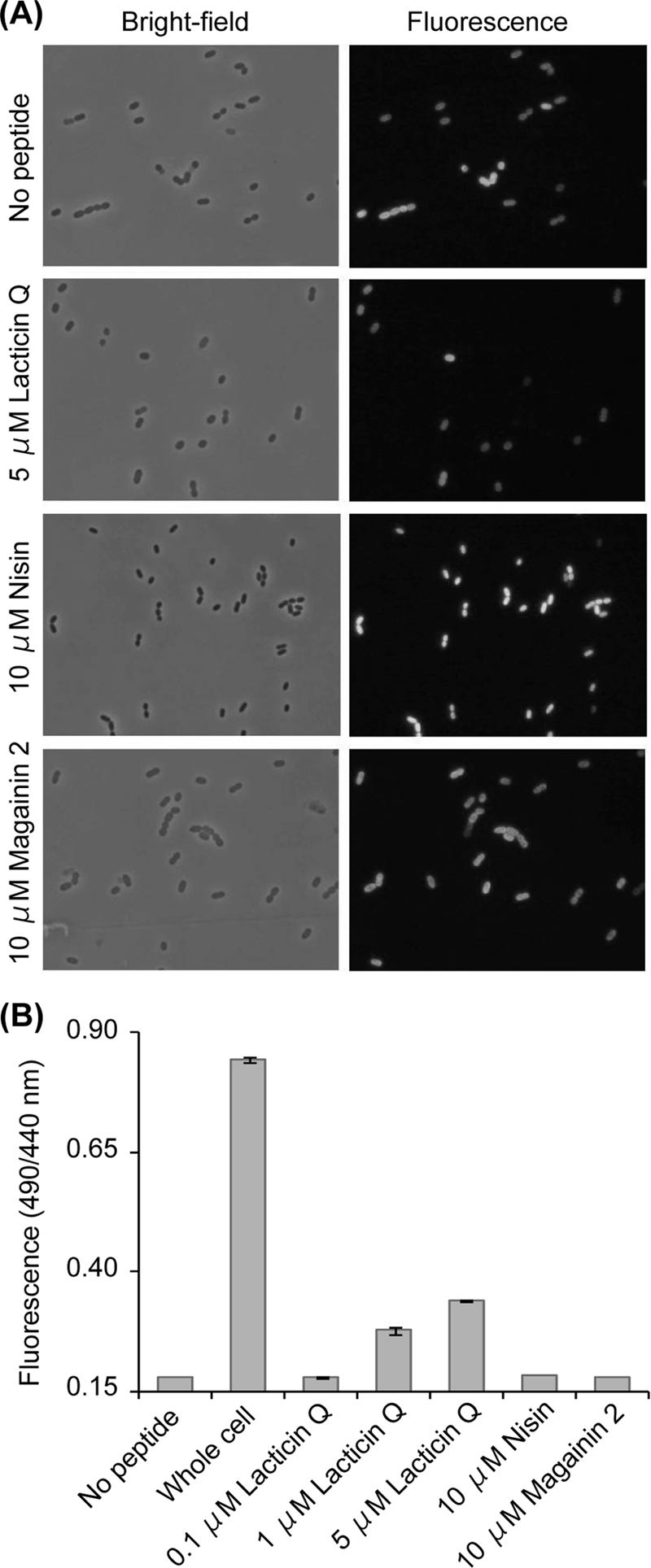
GFP leakage from L. lactis NZgfp. (A) Microscopic observation of GFP-overexpressed and peptide-treated cells. (B) Detection of GFP leaked into supernatant. The ordinate axis shows the ratio of the fluorescence intensity of 520 nm excited by 490 nm (GFP) and 440 nm (Alexa Fluor 430).
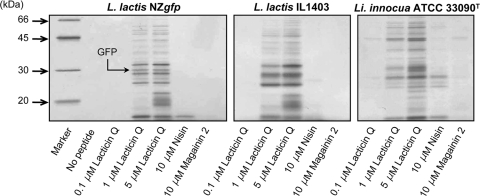
We also attempted to detect intracellular proteins that leaked into the supernatant from the peptide-treated cells, because no antimicrobial peptide causing protein leakage had previously been reported. As shown in Fig. 6, lacticin Q caused the leakage of intracellular proteins from two L. lactis strains and from L. innocua cells. In the case of L. lactis NZgfp, 1 μM lacticin Q caused protein leakage, while 5 μM lacticin Q caused further leakage. To detect a sufficient amount of protein by SDS-PAGE, the cell concentrations used in this experiment were 10-fold higher than those used for the MIC determinations described previously (36, 37). It made the concentrations of lacticin Q for protein leakage higher than the MIC (0.07 μM for L. lactis NZ9000), and 0.1 μM lacticin Q did not cause protein leakage. The sizes of the protein bands were confirmed to be in the range of 10 to 50 kDa. A band at about 30 kDa was observed when GFP was overexpressed, but that band was not detected without GFP expression (data not shown). These findings support the GFP leakage following the lacticin Q treatment described in Fig. 5. Similar results were obtained with other strains, L. lactis IL1403 and L. innocua ATCC 33090T. In all cases, we detected many protein bands, and the band patterns were manifold, depending on the indicator strains. The results reveal that lacticin Q is an antimicrobial peptide that causes random protein leakage.
FIG. 6.
SDS-PAGE of proteins leaked by peptide treatment. Indicator strains L. lactis NZgfp, L. lactis IL1403, and L. innocua ATCC 33090T were treated with lacticin Q, nisin, and magainin 2 in buffer A; and the proteins that leaked into the supernatant were detected by SDS-PAGE with a 12% polyacrylamide gel and silver staining.
DISCUSSION
In the study described in this paper, we show that lacticin Q is the first LAB bacteriocin reported to exert membrane permeation via the toroidal pore. In addition, the pore allows the leakage of macromolecules from both liposomes and cells. We name this novel antimicrobial mechanism of lacticin Q an HTP, and an HTP model is illustrated in Fig. 7.
FIG. 7.
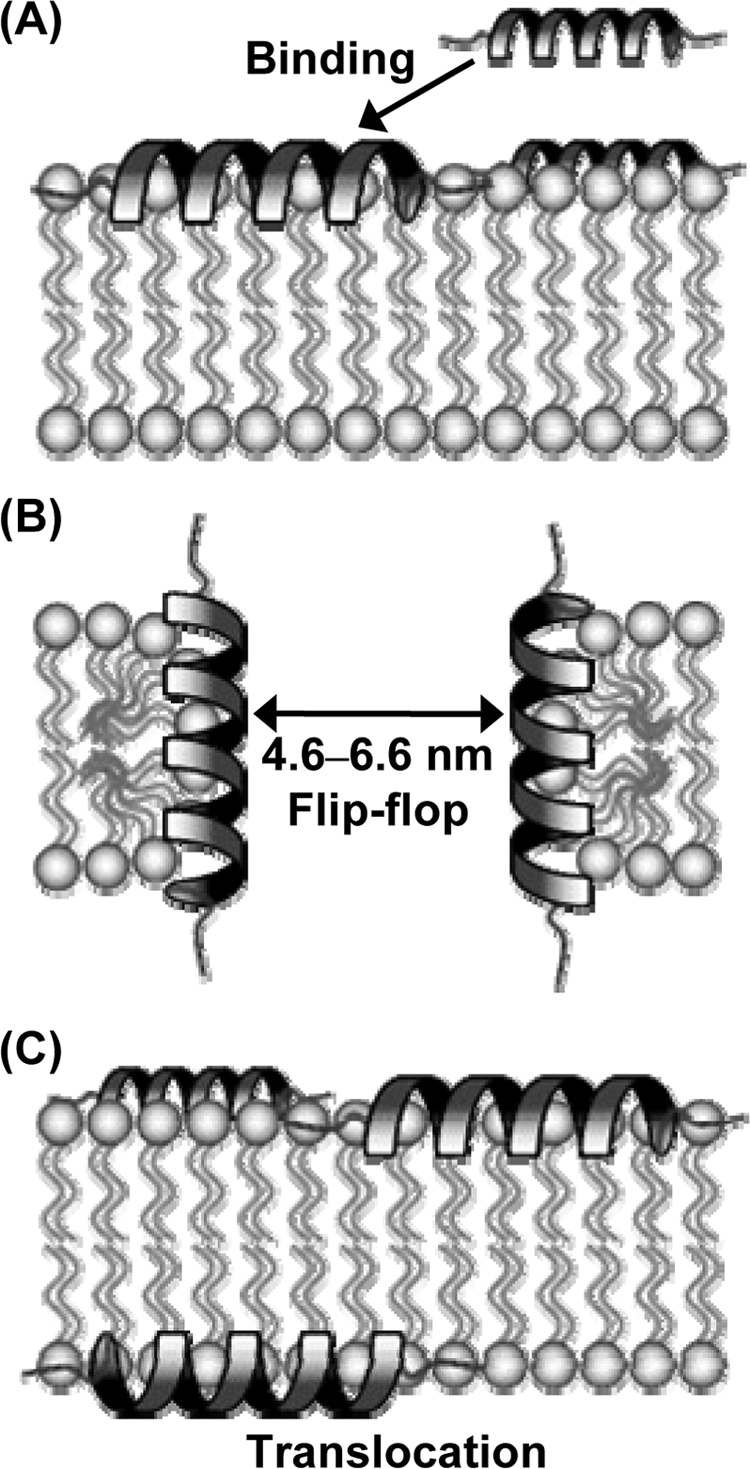
Model of HTP mediated by lacticin Q. (A) Lacticin Q rapidly binds to the negatively charged phospholipid bilayer. (B) Lacticin Q forms an HTP accompanied by lipid flip-flop. The average diameter of the peptide-lipid supramolecular complex pore is 4.6 to 6.6 nm, resulting in protein leakage. (C) Some lacticin Q molecules show transbilayer movement from the outer to the inner leaflet of the bilayer (translocation) when the pore closes.
First, lacticin Q rapidly binds to negatively charged membranes, forming an amphiphilic α-helical structure (Fig. 7A). Previously, we reported that lacticin Q forms an amphiphilic α-helical structure under aqueous conditions and in the presence of small unilamellar vesicles (37). As shown in Fig. 2A, the fluorescent energy from tryptophan residues in lacticin Q was transferred to the DNS-DHPE on the LUVs within 2 s, and an increase in fluorescence also occurred rapidly upon the redistribution to unlabeled LUVs. The observed FRET and its cancellation by excess unlabeled LUVs can be explained by the quick binding between lacticin Q and the LUVs. In addition, the rapid calcein leakage shown in Fig. 1A supports the existence of rapid binding.
The membrane permeation by lacticin Q is due to pore formation and is not the result of micellization or the fusion of bilayers (Fig. 7B), because no morphological change in the LUVs or the cells caused by lacticin Q were observed (Fig. 1B and 5A). Changes in light scattering derived from peptide-induced micellization or the fusion of liposomes have been reported (39); however, pore-forming peptides do not affect the scattering intensity (31).
As shown in Fig. 3, lacticin Q forms pores coupled with lipid flip-flop (Fig. 7B). Lipid flip-flop, a main feature of the toroidal pore, has been characterized in several antimicrobial peptides from multicellular eukaryotes, and the behavior of lacticin Q was similar to that reported for magainin 2 (16). The pore created by lacticin Q corresponds to the toroidal pore model, but to our knowledge, lacticin Q is the first bacteriocin causing lipid flip-flop in gram-positive bacteria. Colicin E1, produced by Escherichia coli, is the sole bacteriocin reported to form a toroidal pore (30). However, colicin E1 is a protein-type bacteriocin composed of 522 amino acids. As described above, lacticin Q, which forms a toroidal pore, exerts its antimicrobial activity in the nanomolar concentration range (MICs, 5 to 750 nM), but other toroidal pore-type antimicrobial peptides exert their antimicrobial activities in the micromolar range (MICs, 1 to 20 μM) (37, 38). The strong activity of lacticin Q is probably due to the extraordinarily large size of the pores that it forms.
The diameter of the pore created by lacticin Q was determined by the leakage of FITC-dex of various sizes (Fig. 7B). On the basis of the supplier's information and a previous report (29), the average diameters of FD-10K and FD-20K were determined to be approximately 4.6 and 6.6 nm, respectively. Since the pore created by lacticin Q could similarly leak calcein and FD-10K (Fig. 4), the average size of the pores created by lacticin Q should be more than 4.6 nm. Furthermore, since that amount of FD-20K that leaked was less than the amount of FD-10K that leaked, the average size of the pore created by lacticin Q should be less than 6.6 nm. This is the largest among the pores created by antimicrobial peptides that have been reported. For example, the diameter of the pores created by magainin 2 was determined to be 2 to 3 nm (18). Among the lactococcal bacteriocins, the sizes of the pores created by nisin and lacticin 3147 were 2 to 2.5 and 0.6 nm, respectively (33, 34), while their antimicrobial activities were in the nanomolar range, as is the case for lacticin Q (4, 24). Although magainin 2 analogues, the magainin 2-peptidyl-glycylleucine-carboxyamide mixture, and melittin allow the leakage of FITC-dex with molecular weights of about 4,400 or lower, the peptide concentrations needed were higher than those needed for calcein leakage (12, 19, 21).
The huge pore created by lacticin Q was also demonstrated on the bacterial membrane (Fig. 5 and 6). We observed decreases in the level of GFP fluorescence in the bacterial cells and increases in the supernatant (Fig. 5). Since the size of GFP was determined to be approximately 3 by 4 nm by X-ray crystallography (28), the diameter of the lacticin Q pore on the biological membrane would be larger than the size of GFP. In addition, lacticin Q caused random proteins from the indicator strains to be leaked, which was rarely the case for nisin and magainin 2. Since the cell densities in the protein leakage experiments were 10-fold higher (in the GFP leakage experiment, 5-fold higher) than those in the MIC determinations, the protein leakage caused by lacticin Q in indicator strains at lower densities is expected even in the nanomolar range. Lacticin Q leaked proteins with higher molecular weights than FITC-dex (Fig. 4 and 6), which we speculate was due to the compact tertiary structures of some intracellular proteins. Regardless, there are no other reports of antimicrobial peptides that cause protein leakage. This type of protein leakage needs a huge pore; therefore, the pore created by lacticin Q was termed HTP.
Finally, some lacticin Q molecules migrate from the outer to the inner leaflet of the membrane (peptide translocation; Fig. 7C). The ratio of the translocation of lacticin Q was related to calcein leakage (Fig. 2B), and as shown in Fig. 1A, the rate of calcein leakage was initially fast and subsequently slowed. These results indicate that a large amount of lacticin Q molecules accumulated quickly on the outer leaflet of the membrane. After a toroidal pore was formed, which took a short time, the pore was closed concomitantly with peptide translocation. As a consequence, the lacticin Q molecules which had accumulated on the outer membrane leaflet were distributed to both leaflets, and the rate of pore formation was reduced in a time-dependent manner. Short-duration pores have been observed for various antimicrobial peptides (21, 33). Whereas peptide translocation has previously been reported in antimicrobial peptides from multicellular eukaryotes, such as magainin 2, melittin, and tachyplesin I (17, 20, 21), this is the first study of peptide translocation in a bacteriocin.
The pores created by nisin and lacticin 3147 are smaller than those created by lacticin Q, as described above. However, the former bacteriocins show strong antimicrobial activities in the nanomolar range. It has been reported that nisin and lacticin 3147 utilize lipid II as the docking molecule for their antimicrobial mechanisms, such as pore formation, inhibition of peptidoglycan biosynthesis, and lipid II segregation (13, 34, 35). On the other hand, lacticin Q forms an HTP without the presence of a docking molecule. The HTP of lacticin Q might be an important factor in achieving high levels of antimicrobial activity in the nanomolar range.
Acknowledgments
We thank S. Sugimoto of Kyusyu University, Fukuoka, Japan, for his technical support.
This work was partially supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science; a research project for utilizing advanced technologies in agriculture, forestry, and fisheries of the Ministry of Agriculture, Forestry and Fisheries of Japan; and the Takeda Science Foundation.
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Aso, Y., J. Nagao, H. Koga, K. Okuda, Y. Kanemasa, T. Sashihara, J. Nakayama, and K. Sonomoto. 2004. Heterologous expression and functional analysis of the gene cluster for the biosynthesis of and immunity to the lantibiotic, nukacin ISK-1. J. Biosci. Bioeng. 98:429-436. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett, G. R. 1959. Phosphorus assay in column chromatography. J. Biol. Chem. 234:466-468. [PubMed] [Google Scholar]
- 3.Breukink, E. 2006. A lesson in efficient killing from two-component lantibiotics. Mol. Microbiol. 61:271-273. [DOI] [PubMed] [Google Scholar]
- 4.Breukink, E., H. E. van Heusden, P. J. Vollmerhaus, E. Swiezewska, L. Brunner, S. Walker, A. J. Heck, and B. de Kruijff. 2003. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 278:19898-19903. [DOI] [PubMed] [Google Scholar]
- 5.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H.-G. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 6.Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, P. D., C. Hill, and R. P. Ross. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777-788. [DOI] [PubMed] [Google Scholar]
- 8.Dalet, K., Y. Cenatiempo, P. Cossart, and Y. Hechard. 2001. A σ(54)-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263-3269. [DOI] [PubMed] [Google Scholar]
- 9.Drider, D., G. Fimland, Y. Hechard, L. M. McMullen, and H. Prevost. 2006. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 70:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, K., S. Ichimasa, T. Zendo, S. Koga, F. Yoneyama, J. Nakayama, and K. Sonomoto. 2007. Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of gram-positive bacteria. Appl. Environ. Microbiol. 73:2871-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravesen, A., P. Warthoe, S. Knochel, and K. Thirstrup. 2000. Restriction fragment differential display of pediocin-resistant Listeria monocytogenes 412 mutants shows consistent overexpression of a putative β-glucoside-specific PTS system. Microbiology 146:1381-1389. [DOI] [PubMed] [Google Scholar]
- 12.Hara, T., H. Kodama, M. Kondo, K. Wakamatsu, A. Takeda, T. Tachi, and K. Matsuzaki. 2001. Effects of peptide dimerization on pore formation: antiparallel disulfide-dimerized magainin 2 analogue. Biopolymers 58:437-446. [DOI] [PubMed] [Google Scholar]
- 13.Hasper, H. E., N. E. Kramer, J. L. Smith, J. D. Hillman, C. Zachariah, O. P. Kuipers, B. de Kruijff, and E. Breukink. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636-1637. [DOI] [PubMed] [Google Scholar]
- 14.Hechard, Y., C. Pelletier, Y. Cenatiempo, and J. Frere. 2001. Analysis of σ(54)-dependent genes in Enterococcus faecalis: a mannose PTS permease (EII(Man)) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology 147:1575-1580. [DOI] [PubMed] [Google Scholar]
- 15.Hechard, Y., and H.-G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from gram-positive bacteria. Biochimie 84:545-557. [DOI] [PubMed] [Google Scholar]
- 16.Imura, Y., M. Nishida, and K. Matsuzaki. 2007. Action mechanism of PEGylated magainin 2 analogue peptide. Biochim. Biophys. Acta 1768:2578-2585. [DOI] [PubMed] [Google Scholar]
- 17.Imura, Y., M. Nishida, Y. Ogawa, Y. Takakura, and K. Matsuzaki. 2007. Action mechanism of tachyplesin I and effects of PEGylation. Biochim. Biophys. Acta 1768:1160-1169. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzaki, K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta 1462:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Matsuzaki, K., Y. Mitani, K. Y. Akada, O. Murase, S. Yoneyama, M. Zasloff, and K. Miyajima. 1998. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry 37:15144-15153. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1995. Translocation of a channel-forming antimicrobial peptide, magainin 2, across lipid bilayers by forming a pore. Biochemistry 34:6521-6526. [DOI] [PubMed] [Google Scholar]
- 21.Matsuzaki, K., S. Yoneyama, and K. Miyajima. 1997. Pore formation and translocation of melittin. Biophys. J. 73:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mierau, I., and M. Kleerebezem. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705-717. [DOI] [PubMed] [Google Scholar]
- 23.Miller, W. G., and S. E. Lindow. 1997. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene 191:149-153. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, S. M., P. M. O'Connor, P. D. Cotter, R. P. Ross, and C. Hill. 2005. Sequential actions of the two component peptides of the lantibiotic lacticin 3147 explain its antimicrobial activity at nanomolar concentrations. Antimicrob. Agents Chemother. 49:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller, P., S. Schiller, T. Wieprecht, M. Dathe, and A. Herrmann. 2000. Continuous measurement of rapid transbilayer movement of a pyrene-labeled phospholipid analogue. Chem. Phys. Lipids 106:89-99. [DOI] [PubMed] [Google Scholar]
- 26.Naghmouchi, K., D. Drider, and I. Fliss. 2007. Action of divergicin M35, a class IIa bacteriocin, on liposomes and Listeria. J. Appl. Microbiol. 102:1508-1517. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 28.Ormo, M., A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien, and S. J. Remington. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392-1395. [DOI] [PubMed] [Google Scholar]
- 29.Scherrer, R., and P. Gerhardt. 1971. Molecular sieving by the Bacillus megaterium cell wall and protoplast. J. Bacteriol. 107:718-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobko, A. A., E. A. Kotova, Y. N. Antonenko, S. D. Zakharov, and W. A. Cramer. 2006. Lipid dependence of the channel properties of a colicin E1-lipid toroidal pore. J. Biol. Chem. 281:14408-14416. [DOI] [PubMed] [Google Scholar]
- 31.Tachi, T., R. F. Epand, R. M. Epand, and K. Matsuzaki. 2002. Position-dependent hydrophobicity of the antimicrobial magainin peptide affects the mode of peptide-lipid interactions and selective toxicity. Biochemistry 41:10723-10731. [DOI] [PubMed] [Google Scholar]
- 32.Vanderkooi, J. M., and J. B. Callis. 1974. Pyrene. A probe of lateral diffusion in the hydrophobic region of membranes. Biochemistry 13:4000-4006. [DOI] [PubMed] [Google Scholar]
- 33.Wiedemann, I., R. Benz, and H.-G. Sahl. 2004. Lipid II-mediated pore formation by the peptide antibiotic nisin: a black lipid membrane study. J. Bacteriol. 186:3259-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiedemann, I., T. Bottiger, R. R. Bonelli, A. Wiese, S. O. Hagge, T. Gutsmann, U. Seydel, L. Deegan, C. Hill, P. Ross, and H.-G. Sahl. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61:285-296. [DOI] [PubMed] [Google Scholar]
- 35.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H.-G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 36.Yoneyama, F., M. Fukao, T. Zendo, J. Nakayama, and K. Sonomoto. 2008. Biosynthetic characterization and biochemical features of the third natural nisin variant, nisin Q, produced by Lactococcus lactis 61-14. J. Appl. Microbiol. 105:1982-1990. [DOI] [PubMed] [Google Scholar]
- 37.Yoneyama, F., Y. Imura, S. Ichimasa, K. Fujita, T. Zendo, J. Nakayama, K. Matsuzaki, and K. Sonomoto. 2009. Lacticin Q, a lactococcal bacteriocin, causes high-level membrane permeability in the absence of specific receptors. Appl. Environ. Microbiol. 75:538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, H., R. Sood, A. Jutila, S. Bose, G. Fimland, J. Nissen-Meyer, and P. K. Kinnunen. 2006. Interaction of the antimicrobial peptide pheromone plantaricin A with model membranes: implications for a novel mechanism of action. Biochim. Biophys. Acta 1758:1461-1474. [DOI] [PubMed] [Google Scholar]