Abstract
Curcumin (CUR), a natural product of turmeric, from rhizomes of Curcuma longa, is a known agent of reversal of drug resistance phenotypes in cancer cells overexpressing ATP-binding cassette (ABC) transporters, viz., ABCB1, ABCG2, and ABCC1. In the present study, we evaluated whether CUR could also modulate multidrug transporters of yeasts that belong either to the ABC family or to the major facilitator superfamily (MFS). The effect of CUR on multidrug transporter proteins was demonstrated by examining rhodamine 6G (R6G) efflux in Saccharomyces cerevisiae cells overexpressing the Candida albicans ABC transporters Cdr1p and Cdr2p (CaCdr1p and CaCdr2p, respectively) and the MFS transporters CaMdr1p and S. cerevisiae Pdr5p. CUR decreased the extracellular concentration of R6G in ABC transporter-expressing cells but had no effect on methotrexate efflux mediated through the MFS transporter CaMdr1p. CUR competitively inhibited R6G efflux and the photolabeling of CaCdr1p by [125I]iodoarylazidoprazosin, a drug analogue of the substrate prazosin (50% inhibitory concentration, 14.2 μM). Notably, the mutant variants of CaCdr1p that displayed abrogated efflux of R6G also showed reduced modulation by CUR. Drug susceptibility testing of ABC protein-expressing cells by spot assays and checkerboard tests revealed that CUR was selectively synergistic with drug substrates such as R6G, ketoconazole, itraconazole, and miconazole but not with fluconazole, voriconazole, anisomycin, cycloheximide, or FK520. Taken together, our results provide the first evidence that CUR modulates only ABC multidrug transporters and could be exploited in combination with certain conventional antifungal drugs to reverse multidrug resistance in Candida cells.
Overexpression of ATP-binding cassette (ABC) multidrug transporters, including P-glycoprotein (ABCB1), multidrug resistance protein (ABCC1), and mitoxantrone resistance protein (ABCG2), plays a major role in the development of multidrug resistance (MDR) in cancer cells (19). Among the various strategies to combat MDR, blocking the functioning of MDR transporters represents an attractive approach (11). Notably, several functional inhibitors of MDR proteins have been tested, but thus far none are clinically successful, due to the dose-limiting toxic effect of the modulators.
To circumvent this problem, extensive efforts have been under way in recent years to identify natural inhibitors of MDR exporters, since natural products have the potential to yield a large number of new drugs. Curcuminoids, from the rhizomes of Curcuma longa, have been reported to reverse the drug resistance phenotype in cancer cells overexpressing ABC transporters, viz., ABCB1, ABCG2, and ABCC1 (2, 4, 5). Curcuminoids blocked the efflux of fluorescent substrates calcein AM, rhodamine 123, and bodipy-FL-vinblastine in MDR cervical carcinoma cell lines overexpressing ABCB1 and the efflux of mitoxantrone and pheophorbide A, mediated by ABCG2, in HEK293 cells (3, 4).
In yeasts, including species of the pathogenic genus Candida, upregulation of multidrug transporter genes belonging either to the ABC family or to the major facilitator superfamily (MFS) is frequently observed in cells exposed to drugs and leads to the phenomenon of MDR (29, 31, 32). For clinical isolates of Candida albicans, it has been established that the ABC transporters C. albicans Cdr1p (CaCdr1p) and CaCdr2p and the MFS transporter CaMdr1p are major MDR transporters that contribute to azole resistance. There are compounds, such as FK506, enniatins, milbemycins, synthetic d-octapeptides, cyclosporine, isonitrile, disulfiram, ibuprofen, and unnarmicins (12, 30), that inhibit fungal ABC transporters. Such inhibitors or chemosensitizers probably act directly by affecting substrate binding and transport mediated by MDR efflux proteins.
Notably, the effect of curcuminoids on fungal ABC transporters is not known. However, due to functional and structural similarities between ABCB1 and ABC transporters in yeasts, it is very likely that the curcuminoids could act as “reversal agents” of drug resistance in yeasts as well. In this study, we have examined the potency of curcumin (CUR) in modulating the efflux activity of CaCdr1p and have compared it with those of CaCdr2p and Saccharomyces cerevisiae Pdr5p (ScPdr5p). Our results demonstrate that CUR behaves as a specific modulator of rhodamine 6G (R6G) efflux mediated by CaCdr1p, CaCdr2p, and ScPdr5p in S. cerevisiae cells overexpressing these transporters. Notably, CUR had no impact on efflux activity mediated by the MFS transporter CaMdr1p. Furthermore, CUR reversed drug resistance by displaying synergism with selected drugs.
MATERIALS AND METHODS
Materials.
R6G, a commercial-grade mixture of curcuminoids (commonly known as CUR), protease inhibitors (phenylmethylsulfonyl fluoride, leupeptin, aprotinin, pepstatin A, Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK], and N-tosyl-l-phenylalanine chloromethyl ketone [TPCK]), a bicinchoninic acid protein determination kit, miconazole (MCZ), ketoconazole (KTC), itraconazole (ITC), anisomycin (ANISO), cycloheximide (CYH), FK520, oligomycin, dinitrophenol, deoxyglucose, 3-(4,5-dimethyl thiazol-2-yl)-2,5 diphenyl tetrazolium bromide (MTT), and other molecular-grade chemicals were obtained from Sigma Chemical Co. (St. Louis, MO). [3H]-labeled fluconazole (FLC; specific activity, 19 Ci/mmol) was custom synthesized by Amersham Biosciences, United Kingdom, and [3H]methotrexate (MTX; specific activity, 8.60 Ci/mmol) was procured from Amersham Biosciences, United Kingdom. Radiolabeled [125I]iodoarylazidoprazosin (IAAP) (2,200 Ci/mmol) was purchased from Perkin-Elmer Life Sciences (Boston, MA).
Yeast strains and growth media.
The strains used in this study are listed in Table 1. The yeast strains were cultured in yeast extract-peptone-dextrose (YEPD) broth (Bio 101, Vista, CA). For agar plates, 2.5% (wt/vol) Bacto agar (Difco, BD Biosciences, NJ) was added to the medium. All strains were stored as frozen stocks with 15% glycerol at −80°C. Before each experiment, cells were freshly revived on YEPD plates from the stock.
TABLE 1.
Strains used in this study
| Strain no. | Strain name or mutation | Genotype or descriptiona | Source |
|---|---|---|---|
| 1 | AD1-8u− | MATα pdr1-3 his1 ura Δyor1::hisG Δsnq2::hisG Δpdr5::hisG Δpdr10::hisG Δpdr11::hisG Δycf1::hisG pdr3::hisG Δpdr15::hisG | 7 |
| 2 | AD-CDR1 | AD1-8u− cells harboring the CaCDR1-GFP ORF integrated at the PDR5 locus | 27 |
| 3 | A1346G | CDR1-GFP cells carrying an A1346G mutation in the CDR1 ORF and integrated at the PDR5 locus | 24 |
| 4 | A1347G | CDR1-GFP cells carrying an A1347G mutation in the CDR1 ORF and integrated at the PDR5 locus | 24 |
| 5 | T1351A | CDR1-GFP cells carrying a T1351A mutation in the CDR1 ORF and integrated at the PDR5 locus | 24 |
| 6 | F1360A | CDR1-GFP cells carrying an F1360A mutation in the CDR1 ORF and integrated at the PDR5 locus | 24 |
| 7 | G1362A | CDR1-GFP cells carrying a G1362A mutation in the CDR1 ORF and integrated at the PDR5 locus | 24 |
| 8 | L1358A | CDR1-GFP cells carrying an L1358A mutation in the CDR1 ORF and integrated at the PDR5 locus | 24 |
| 9 | T1355A | CDR1-GFP cells carrying a T1355A mutation in the CDR1 ORF and integrated at the PDR5 locus | 24 |
| 10 | G682A | CDR1-GFP cells carrying a G682A mutation in the CDR1 ORF and integrated at the PDR5 locus | Puri et al., unpublished data |
| 11 | AD-CaMDR1 | AD1-8u− cells harboring the CaMDR1-GFP ORF integrated at the PDR5 locus | 22 |
| 12 | AD-CDR2 | AD1-8u− cells harboring the CaCDR2-GFP ORF integrated at the PDR5 locus | 17 |
| 13 | AD-PDR5 | AD1-8u− cells harboring the ScPDR5-GFP ORF integrated at the PDR5 locus | 17 |
| 14 | CAI4 | Δura3::imm434/Δura3::imm434 | 8 |
ORF, open reading frame.
Efflux of R6G.
The efflux of R6G was determined essentially using a previously described protocol (27). Briefly, approximately 1 × 106 yeast cells from an overnight-grown culture were transferred to YEPD medium and allowed to grow for 5 h. Cells were pelleted, washed twice with phosphate-buffered saline (PBS) (without glucose), and resuspended as a 2% cell suspension, which corresponds to 108 cells (wt/vol) in PBS without glucose. The cells were then de-energized for 45 min in deoxyglucose (5 mM) and dinitrophenol (5 mM) in PBS (without glucose). The de-energized cells were pelleted, washed, and then resuspended as a 2% cell suspension (wt/vol) in PBS without glucose, to which R6G was added at a final concentration of 10 μM and incubated for 40 min at 30°C. The equilibrated cells with R6G were then washed and resuspended as a 2% cell suspension (wt/vol) in PBS without glucose. Samples with a volume of 1 ml were withdrawn at the indicated time and centrifuged at 9,000 × g for 2 min. The supernatant was collected, and absorption was measured at 527 nm. Energy-dependent efflux (at the indicated time) was measured after the addition of glucose (2%) to the cells resuspended in PBS (without glucose). Glucose-free controls were included in all the experiments. For competition assays, CUR (100 μM) was added to the de-energized cells 5 min before the addition of R6G and allowed to equilibrate.
Measurement of drug accumulation.
The accumulation of [3H]FLC (specific activity, 19 Ci/mmol) and [3H]MTX (specific activity, 8.60 Ci/mmol) was determined essentially by the methods described previously (22). Briefly, cells from mid-log phase (5 × 106) were centrifuged at 3,000 × g for 3 min and resuspended in PBS as a 2% cell suspension. For accumulation studies, 100 nM FLC and 25 μM MTX were routinely used (22). CUR at 100 μM was added 5 min before the addition of drugs and was allowed to equilibrate. A 100-μl volume of the cell suspension containing drugs alone or drugs plus CUR was incubated at 30°C for 40 min, filtered rapidly, and washed twice with PBS (pH 7.4) on a Millipore manifold filter assembly using a 0.45-μm-pore size cellulose nitrate filter (Millipore). The filter discs were dried and put in cocktail “O,” and the radioactivity was measured in a liquid scintillation counter (Beckman). Accumulation was expressed as picomoles per milligram (dry weight).
Photoaffinity labeling with IAAP.
The crude membrane proteins (50 μg) prepared from AD-CDR1 cells (27) were incubated with CUR or with R6G for 10 min at 37°C in 0.1 ml of 50 mM Tris-HCl (pH 7.5). The samples were brought to room temperature, and 3 to 6 nM [125I]IAAP (2,200 Ci/mmol) was added and incubated for an additional 5 min under subdued light. The samples were then illuminated with a UV lamp assembly (PGC Scientifics, Gaithersburg, MD) fitted with two black-light (self-filtering) UVA long-wavelength F15T8BLB tubes (365 nm) for 10 min at room temperature (21 to 23°C). One milliliter of radioimmunoprecipitation assay buffer was added to the samples, and CaCdr1p cross-linked with [125I]IAAP was immunoprecipitated with 10 μg of a monoclonal antibody (BD Biosciences, Palo Alto, CA) against green fluorescent protein (GFP) (27). The samples were then separated on a 7% Tris-acetate gel at a constant voltage, and the gels were dried and exposed to Bio-Max MR film (Eastman Kodak, Rochester, NY) at −80°C for 12 to 24 h. The radioactivity incorporated into the CaCdr1p band was quantified using a Storm 860 PhosphorImager system (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software as described previously (27).
ATPase assay.
The ATPase activity of the plasma membrane (PM) fractions was measured as oligomycin-sensitive release of inorganic phosphate either alone, as described previously (27), or in the presence of CUR (100 μM) and varying concentrations of ATP (0.5 mM to 7 mM).
Immunodetection of ABC proteins.
PMs were prepared from S. cerevisiae cells overexpressing ABC transporters as described previously (27) or in the presence of CUR (100 μM). The PM protein concentration was determined by a bicinchoninic acid assay using bovine serum albumin as the standard. Western blot analysis was conducted using an anti-GFP monoclonal antibody (1:5,000) as described previously (27). Proteins on immunoblots were visualized using the enhanced chemiluminescence assay system (ECL kit; Amersham Biosciences, Arlington Heights, IL).
Drug susceptibility assay.
The sensitivities of yeast cells to different drugs in the presence of CUR were determined by spot assays as described previously (22). The interaction of CUR with KTC, MCZ, ITC, R6G, FLC, ANISO, CYH, FK520, or MTX was evaluated by the checkerboard method recommended by the CLSI (formerly NCCLS) and was expressed as the fractional inhibitory concentration (FIC) index, the sum of the FICs for each agent. The FIC of each agent is calculated as the MIC of the agent in combination divided by the MIC of the agent alone (21). A range of concentrations were tried: 0.202 to 208 μM for FLC or voriconazole (VORI), 0.007 to 3.6 μM for KTC, 0.004 to 8.32 μM for MCZ, 0.002 to 5.6 μM for ITC, 0.200 to 103.5 μM for R6G, 0.177 to 92.5 μM for ANISO, 0.066 to 35 μM for CYH, 0.122 to 63 μM for FK520, and 1.05 to 540 μM for CUR. Each checkerboard test generates many different combinations, and by convention the FIC of the most effective combination is used in calculating the FIC index.
Cytotoxicity assay.
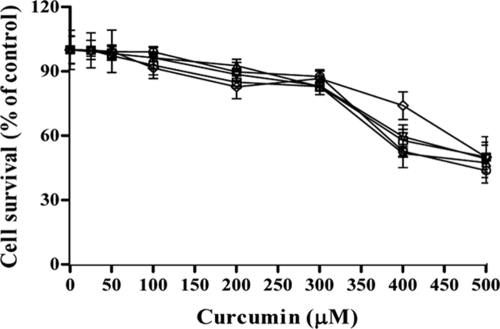
The cytotoxic effect of CUR was determined by an MTT assay (3, 4). Yeast cells (104) were seeded into 96-well plates in the absence and the presence of varying concentrations of CUR (25 to 500 μM) and were grown for 48 h at 30°C. One hundred microliters of an MTT solution was added to each well and incubated for 3 to 4 h, and 200 μl of isopropanol was added to stop the reaction. Absorbance was measured using a microplate spectrophotometer at 570 nm with a reference wavelength of 650 nm. Cell survival (as a percentage of the survival of control cells) was calculated as (mean absorbance in test wells)/(mean absorbance in control wells) × 100.
RESULTS
CUR inhibits R6G efflux.
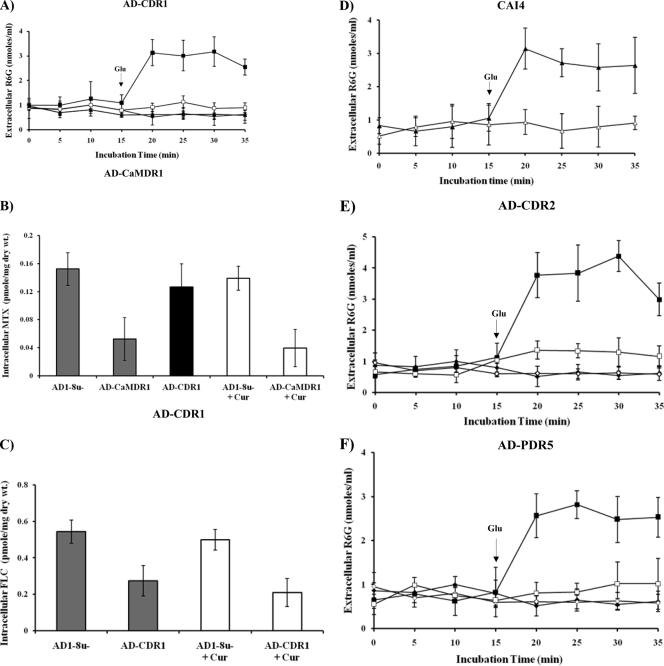
We used a commercial preparation of CUR in order to observe its effect on MDR efflux proteins of the pathogenic yeast C. albicans. For this purpose, we monitored the efflux of R6G in cells where GFP-tagged CaCdr1p (Cdr1p-GFP) was stably overexpressed from a genomic PDR5 locus in the S. cerevisiae mutant strain AD1-8u− (20). The host AD1-8u−, constructed by Goffeau's group (7), was derived from a Pdr1-3 mutant strain with a gain-of-function mutation in the transcription factor Pdr1p, resulting in constitutive hyperinduction of the PDR5 promoter (20). As shown in Fig. 1A, S. cerevisiae cells overexpressing CaCdr1p showed energy-dependent efflux of R6G, which was inhibited by CUR (100 μM). However, the addition of CUR had no effect on the leakage of preloaded R6G from de-energized S. cerevisiae cells. We tested if CUR could also affect a multidrug transporter belonging to the MFS, and we examined efflux mediated by CaMdr1p expressed in a similar heterologous background. As shown in Fig. 1B, the transport of [3H]MTX, a well-known substrate of CaMdr1p, remained unaffected by a fourfold excess of CUR. In the experiments for which results are shown in Fig. 1A, strain AD-CaMDR1 was used as a negative control for R6G transport, and in the experiments for Fig. 1B, strain AD-CDR1 was used as a negative control for MTX transport. The effect of CUR was also substrate specific, as evidenced by the fact that efflux of the well-known substrate FLC remained unimpeded in CaCdr1p-expressing S. cerevisiae cells, even though CUR was supplied in a 1,000-fold excess (Fig. 1C). Of note, CUR could also modulate R6G efflux in C. albicans cells (Fig. 1D); however, for subsequent studies, we used an S. cerevisiae strain overexpressing MDR transporters.
FIG. 1.
Effects of CUR on efflux of substrates in yeast cells. (A) Extracellular R6G concentrations in S. cerevisiae control cells (AD1-8u−) (diamonds) and in cells overexpressing CaCdr1p (AD-CDR1) (squares), incubated either with R6G (10 μM) alone (filled symbols) or with R6G plus CUR (100 μM) (open symbols). Filled triangles represent AD-CaMDR1 cells. Energy-dependent R6G efflux was initiated by adding 2% glucose (arrow) and was quantified by measuring the absorbance of the supernatant at 527 nm. Values are means and standard deviations (error bars) for three independent experiments. (B) [3H]MTX accumulation in S. cerevisiae control cells (AD1-8u−) and in cells overexpressing CaMdr1p (AD-CaMDR1). Cells were incubated either with [3H]MTX (25 μM; specific activity, 8.60 Ci/mmol) alone (shaded bars) or with [3H]MTX plus CUR (100 μM) (open bars). The solid black bar represents AD-CDR1 cells. The accumulated [3H]MTX was measured, 40 min after the initiation of efflux, using a liquid scintillation counter (Beckman). Values are means ± standard deviations (error bars) for three independent experiments. (C) [3H]FLC accumulation in S. cerevisiae control cells and in cells overexpressing CaCdr1p. Cells were incubated with either [3H]FLC (100 nM; specific activity, 19 Ci/mmol) alone (filled bars) or [3H]FLC plus CUR (100 μM) (open bars). The accumulated [3H]FLC was measured 40 min after the addition of glucose (2%). Values are means ± standard deviations (error bars) for three independent experiments. (D) Extracellular R6G concentrations in C. albicans strain CAI4. Cells were incubated either with R6G (10 μM) alone (filled triangles) or with R6G plus CUR (100 μM) (open triangles). Energy-dependent R6G efflux was initiated by adding 2% glucose (arrow) and was quantified by measuring the absorbance of the supernatant at 527 nm. Values are means and standard deviations (error bars) for three independent experiments. (E) Extracellular R6G concentrations in S. cerevisiae control cells (AD1-8u−) (diamonds) and in cells overexpressing CaCdr2p (AD-CDR2) (squares) incubated either with R6G (10 μM) alone (filled symbols) or with R6G plus CUR (100 μM) (open symbols). (F) Extracellular R6G concentrations in S. cerevisiae control cells (AD1-8u−) (diamonds) and in cells overexpressing ScPdr5p (AD-PDR5) (squares), incubated either with R6G (10 μM) alone (filled symbols) or with R6G plus CUR (100 μM) (open symbols). Energy-dependent R6G efflux was initiated by the addition of 2% glucose (arrows) and was quantified by measuring the absorbance of the supernatant at 527 nm. Values are means and standard deviations (error bars) for three independent experiments.
CUR selectively modulates ABC transporters.
Before investigating whether CUR affects drug transporters, we examined whether it affected the viability of cells. For this purpose, control cells and transporter-overexpressing cells were exposed to various concentrations of CUR for 48 h, and cytotoxicity was determined by an MTT assay (3, 4). The percentage of viable cells was calculated in order to determine the 50% inhibitory concentrations (IC50s) (Fig. 2). As shown in Table 2, the IC50s for control cells (AD1-8u−) and those for cells expressing various transporters (AD-CDR1, AD-CDR2, AD-PDR5, and AD-CaMDR1) were not very different, ranging from 410.6 ± 9.4 μM to 498 ± 5.5 μM. Further, our transport assays confirmed that CUR is not a substrate of ABC or MFS proteins, since the extracellular concentration of CUR remained the same even after the initiation of efflux (data not shown). Our data suggest that CUR interacts with the yeast transporters, but these multidrug transporters may not transport it, since the IC50s and relative resistance factors were similar whether cells were overexpressing a transporter or not. We investigated whether the effect of CUR is specific to ABC transporters, and we examined R6G efflux mediated by CaCdr1p homologues, such as CaCdr2p and ScPdr5p, which were expressed in similar backgrounds. It was observed that CUR could inhibit the efflux of R6G mediated by both the proteins (Fig. 1E and F). Our transport assays confirmed that CUR is not a substrate of ABC or MFS proteins, since the extracellular concentration of CUR remained the same even after the initiation of efflux (data not shown). This was further confirmed by a cytotoxicity assay (3, 4), where the IC50s were similar for cells that did and did not overexpress transporters (Table 2). It should be mentioned that the functionality of GFP-tagged versions of ABC and MFS transporters was similar to that of untagged proteins (22, 27). To further examine the effect of CUR, the ABC transporter CaCdr1p was selected for detailed functional analyses.
FIG. 2.
Effect of CUR on the viability of S. cerevisiae cells as determined by an MTT assay. Shown is the percentage of survival among control cells (AD1-8u−) (open circles) and among cells overexpressing ABC or MFS transporters: AD-CDR1 (open inverted triangles), AD-CDR2 (open triangles), AD-PDR5 (open diamonds), and AD-CaMDR1 (open squares) cells. The experiments were conducted in triplicate, and the values are means ± standard deviations for three independent experiments.
TABLE 2.
IC50s and relative resistance factors for fungal strains in the presence of CUR
| Strain | IC50 (μM) | Relative resistance factor |
|---|---|---|
| AD1-8u− | 421.6 ± 2.2 | 1 |
| AD-CDR1 | 492.8 ± 6.3 | 1.16 |
| AD-CDR2 | 410.6 ± 9.4 | 0.99 |
| AD-PDR5 | 498 ± 5.5 | 1.18 |
| AD-CaMDR1 | 454.6 ± 7.4 | 1.07 |
CUR competitively inhibits R6G efflux.
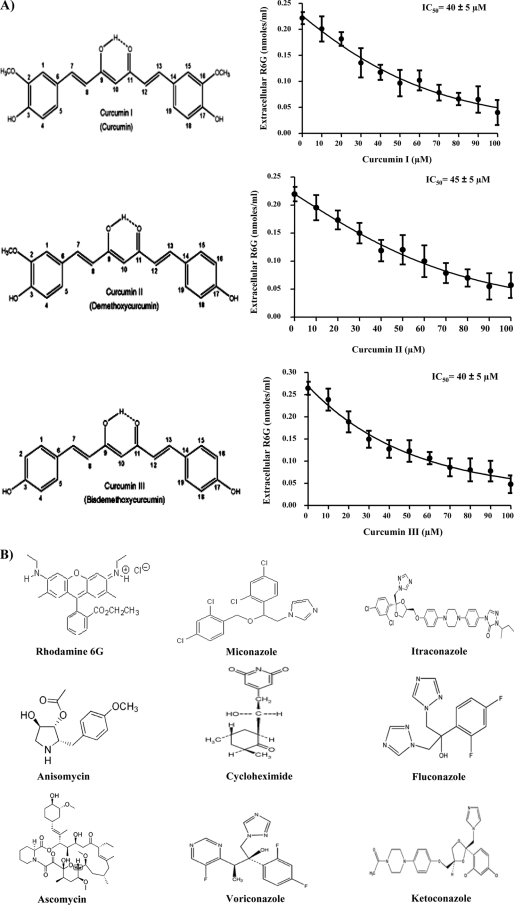
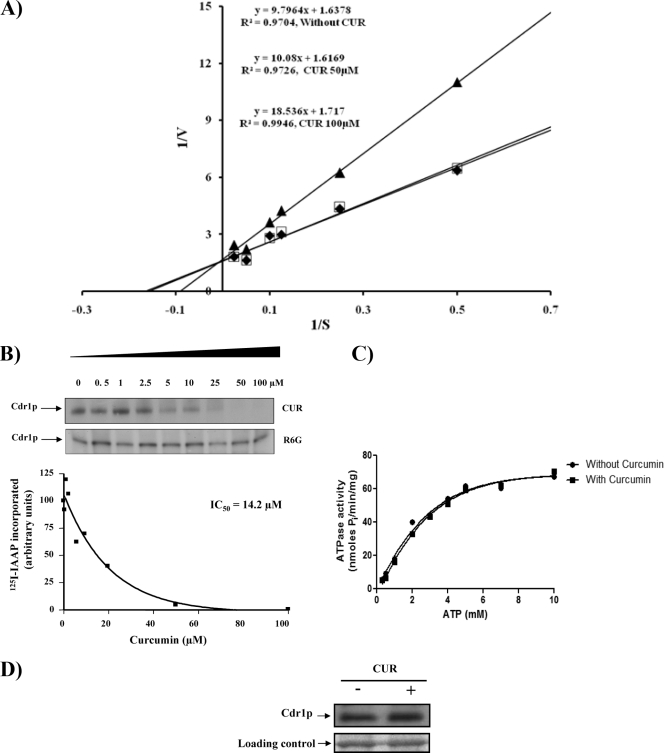
Commercially available CUR is a mixture of three major curcuminoids: curcumin, or diferuloylmethane (curcumin I), demethoxycurcumin (curcumin II), and bisdemethoxycurcumin (curcumin III). This mixture contains predominantly curcumin I (∼77%), followed by curcumin II (17%) and curcumin III (3%), which display a wide range of biological and pharmacological properties (1, 16). We used purified curcuminoids (curcumins I, II, and III) to see if these compounds showed any selectivity as modulators of R6G efflux. The efflux of R6G mediated by CaCdr1p was inhibited by all the pure forms of CUR in a concentration-dependent manner, with IC50s ranging from 40 ± 5 to 45 ± 5 μM (Fig. 3A). The Lineweaver-Burk plot revealed that CUR competitively inhibits R6G efflux, with an increase in apparent Km (5.87 to 11.83 μM) but no effect on the Vmax (Fig. 4A).
FIG. 3.
Effects of pure curcuminoids on R6G transport in S. cerevisiae cells overexpressing CaCdr1p. (A) Structures of curcumin I, curcumin II, and curcumin III and competition assays with R6G. CaCdr1p-overexpressing S. cerevisiae cells were incubated either with 10 μM R6G alone or with 10 μM R6G plus curcumin I, II, or III (10 to 100 μM). R6G efflux was monitored 40 min after the addition of glucose (2%). Extracellular R6G was quantified by measuring the absorbance at 527 nm. The data are plotted using GraphPad Prism. Values are means and standard deviations (error bars) for three independent experiments. (B) Structures of the various substrates used.
FIG. 4.
Biochemical analysis of CaCdr1p in the presence of CUR. (A) Lineweaver-Burk plot of CaCdr1p-mediated R6G efflux in the presence of CUR 5 min after the addition of 2% glucose. Filled diamonds, open squares, and filled triangles represent 0, 50, and 100 μM CUR, respectively. The rate of each reaction was calculated as nanomoles of R6G released per minute per 5 × 106 cells. (B) Effect of CUR or R6G on the photoaffinity labeling of CaCdr1p with [125I]IAAP. The autoradiogram represents the amounts of [125I]IAAP incorporated into CaCdr1p in the presence of the indicated concentrations of CUR or R6G. The graph represents the amounts of [125I]IAAP incorporated into CaCdr1p in the presence of the indicated concentrations of CUR. (C) Effect of CUR on the ATPase activity of CaCdr1p. PMs from cells overexpressing CaCdr1p were incubated with or without 100 μM CUR and varying concentrations of ATP (0.5 mM to 7 mM) in the ATPase buffer. The assay was performed essentially as described in Materials and Methods. The data are plotted using GraphPad Prism. (D) Effect of CUR (100 μM) on the expression of CaCdr1p. Western blot analyses were performed with an anti-GFP monoclonal antibody. Equal loading of protein was assessed by using a Coomassie-stained gel.
CUR inhibits drug binding and has no effect on ATPase activity or the expression of CaCdr1p.
We had shown previously that IAAP (a photoaffinity analogue of the human P-glycoprotein substrate prazosin) and azidopine (a dihydropyridine photoaffinity analogue of its modulator, verapamil) specifically bind to CaCdr1p (27). To monitor whether CUR affects drug binding, we labeled CaCdr1p with [125I]IAAP as described in Materials and Methods. Figure 4B demonstrates that CUR effectively inhibited the photoaffinity labeling of CaCdr1p with [125I]IAAP, with an IC50 of 14.2 μM. We also monitored [125I]IAAP labeling in the presence of R6G as described in Materials and Methods. Interestingly, R6G could not inhibit [125I]IAAP binding. In contrast to well-known inhibitors of CaCdr1p ATPase activity, such as vanadate, oligomycin, sodium azide, and N-ethylmaleimide (15, 26, 27), CUR at 100 μM had no effect on ATPase activity in the presence of varying concentrations of ATP (Fig. 4C). CUR also had no effect on the expression of CaCdr1p (Fig. 4D). Taken together, the data strongly indicate that although CUR is not transported, it acts as a competitive inhibitor at one of the transport sites used to transport clinically significant antifungal agents.
CUR displays synergism with selected azoles.
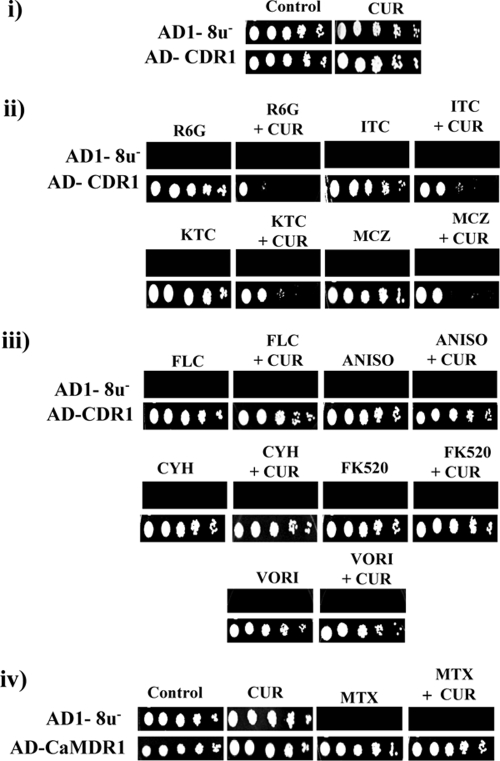
When the control (AD1-8u−) cells and the CaCdr1p-expressing cells were grown either in the presence of drugs alone (FLC at 6.52 μM, VORI at 5.72 μM, MCZ at 0.167 μM, KTC at 0.037 μM, ITC at 0.141 μM, ANISO at 2.97 μM, CYH at 0.28 μM, FK520 at 12.6 μM, R6G at 0.209 μM) or in the presence of both CUR (75.6 μM) and the indicated drug, it was observed that CaCdr1p-expressing cells displayed the expected drug resistance and thus were able to grow in the presence of drug alone. Similar results were obtained with CaCdr2p- and ScPdr5p-expressing S. cerevisiae cells (data not shown). However, the simultaneous presence of CUR with either R6G or azoles, viz., KTC, ITC, or MCZ, sensitized the cells, as evidenced by inhibition of the growth of the cells (Fig. 5ii). Interestingly, the presence of CUR along with noncompeting drugs, such as ANISO, CYH, FLC, VORI, and FK520, did not affect the level of resistance or the growth of cells expressing ABC proteins (Fig. 5iii). The observed inhibition of growth by CUR in the presence of drugs was not due to loss of viability, as determined by an MTT assay (Fig. 2). Notably, CUR (75.6 μM) alone did not inhibit the growth of control cells (AD1-8u−) or that of cells overexpressing CaCdr1p (Fig. 5i) or CaMdr1p (Fig. 5iv). The growth of CaMdr1p-overexpressing cells in the presence of MTX remained insensitive to CUR (Fig. 5iv). We performed checkerboard assays in the presence of CUR and various drugs. The FIC indices are below 0.5 for drugs such as KTC, ITC, MCZ, and R6G in AD-CDR1 cells, suggesting synergism with CUR (Table 3). Checkerboard analysis showed no synergism with CUR for FLC, VORI, ANISO, CYH, or FK520. Similar patterns of synergism between select drugs and CUR were observed with AD-CDR2 and AD-PDR5 cells (data not shown).
FIG. 5.
Synergistic effects of CUR on drug resistance. Control (AD1-8u−) and CaCdr1p-expressing (AD-CDR1) S. cerevisiae cells were grown overnight on YEPD plates and then resuspended in normal saline to an optical density at 600 nm of 0.1. The following stock solutions of drugs were used: R6G at 1 mg/ml in dimethyl sulfoxide, FLC at 1 mg/ml in water, VORI at 5 mg/ml in water, CYH at 0.1 mg/ml in water, MCZ at 1 mg/ml in methanol, KTC at 1 mg/ml in methanol, ANISO at 1 mg/ml in dimethyl sulfoxide, FK520 at 1 mg/ml in ethanol, MTX at 1 mg/ml in 10 mM Tris-Cl, and CUR at 11 mg/ml in dimethyl sulfoxide. Five microliters of a fivefold serial dilution of each strain was spotted onto YEPD plates as described previously (27) either in the absence (control) (i) or in the presence of antifungals, alone or in combination with CUR (ii through iv). (ii) R6G (0.209 μM), ITC (0.141 μM), KTC (0.037 μM), or MCZ (0.167 μM), alone or in combination with CUR (75.6 μM). (iii) FLC (6.52 μM), ANISO (2.97 μM), CYH (0.28 μM), FK520 (12.6 μM), or VORI (5.72 μM), alone or in combination with CUR (75.6 μM). (iv) MTX (11 μM) or CUR (75.6 μM), alone or in combination.
TABLE 3.
Interaction of CUR with KTC, MCZ, ITC, or R6G against AD-CDR1 cellsa
| Antifungal or dye | FIC of the antifungal or dye | FIC of CUR | FICI |
|---|---|---|---|
| KTC | 0.25 | 0.0054 | 0.255 |
| MCZ | 0.125 | 0.005 | 0.130 |
| ITC | 0.250 | 0.005 | 0.255 |
| R6G | 0.0026 | 0.0013 | 0.0039 |
Evaluated by the checkerboard method recommended by the CLSI and expressed as the FIC index (FICI). A FICI of ≤0.5 indicates synergistic interaction. FICI is calculated as the sum of the FICs of each agent. The FIC of each agent is calculated as the MIC of the agent in combination divided by the MIC of the agent alone (21).
CUR acts at the R6G transport site.
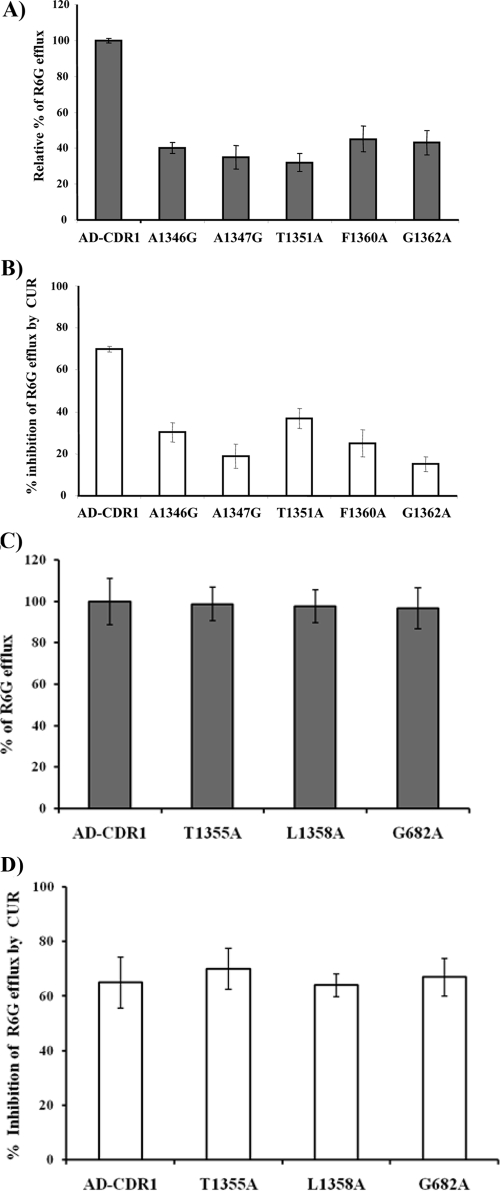
We had recently subjected transmembrane segment 11 of CaCdr1p to alanine scanning, replacing all 21 amino acid residues with alanine by site-directed mutagenesis (24). Out of 21 residues of transmembrane segment 11, substitution of 5 residues, namely, A1346G, A1347G, T1351A, F1360A, and G1362A, abrogated the efflux of R6G, while the rest of the mutant variants of CaCdr1p showed unaltered efflux (Fig. 6A). Since CUR selectively modulates R6G efflux, we argued that if R6G and CUR share binding sites, abrogation of R6G efflux should affect its modulation by CUR. Interestingly, we observed that the ability of CUR to modulate R6G efflux was indeed considerably reduced in those mutant variants that showed abrogated efflux of R6G. Figure 6B shows that the percentage of inhibition of R6G efflux by CUR was considerably lower in mutant variants than in native CaCdr1p-expressing cells. In addition, CUR inhibits R6G efflux even in those mutant variants that do not show abrogated efflux of the dye. This would suggest that in addition to common binding sites, CUR also has an independent binding site(s) in CaCdr1p (Fig. 6C and D). However, to resolve this issue, elaborate binding studies will be required.
FIG. 6.
R6G transport in S. cerevisiae cells overexpressing CaCdr1p or its mutant variants. (A and C) Extracellular R6G concentrations (expressed as percentages of those with wild-type CaCdr1p [AD-CDR1 cells]) in S. cerevisiae cells overexpressing CaCdr1p or mutant variants of CaCdr1p, incubated with 10 μM R6G alone. Energy-dependent R6G efflux was initiated by adding 2% glucose and was quantified by measuring the absorbance of the supernatant at 527 nm. (B and D) Percentages of inhibition of R6G efflux by CUR (100 μM), calculated by taking the level of R6G efflux with each mutant CaCdr1p variant in the absence of CUR as 100%. Values are means ± standard deviations (error bars) for three independent experiments.
DISCUSSION
Among the 28 putative ABC transporter genes and 95 putative MFS transporter genes identified in the C. albicans genome (9, 10), there is overwhelming clinical and experimental evidence that only ABC transporters, such as CaCdr1p and CaCdr2p, and the MFS transporter CaMdr1p are major determinants of azole resistance (23, 25). The reversal of the functionality of these multidrug efflux pump proteins represents an attractive strategy for combating azole resistance. In this study, we explored whether CUR, which inhibits the activities of the mammalian ABC multidrug transporter genes ABCB1, ABCG2, and ABCC1 (3, 4, 5), could be exploited as a modulating agent for multidrug transporters of C. albicans. Our study reveals that CUR inhibited R6G transport exclusively in S. cerevisiae cells overexpressing the ABC drug transporters CaCdr1p, CaCdr2p, and ScPdr5p and had no effect on efflux mediated by the MFS transporter CaMdr1p. All three pure forms of CUR—curcumins I, II, and III—showed similar levels of modulation of R6G efflux in S. cerevisiae cells expressing ABC transporters (Fig. 3A). The modulatory effect of CUR was restricted to R6G; it had no effect on the efflux of another substrate, FLC. We could observe a direct correlation between the modulatory effect of CUR and the status of R6G efflux. For instance, those mutant variants of CaCdr1p that show abrogated efflux of R6G also display decreased modulation by CUR.
Notably, R6G and FLC are both substrates of CaCdr1p, but only the former is competed with CUR (Fig. 1A and C). If the structure of CUR is compared with the structures of R6G and FLC, it is apparent that electronic factors, such as the number of π rings and an extended π surface, could be important for CUR and other substrates, such as ITC, KTC, and MCZ, which compete with R6G efflux (Fig. 3B). In this context, it is noteworthy that noncovalent π-π interactions have tremendous biological implications (13, 14). On the other hand, if one considers the structures of FLC, VORI, ANISO, and CYH, which do not compete with R6G, there are no such electronic factors but a good number of tetrahedral sites. Therefore, these subtle differences in properties between the structures of competing and noncompeting substrates could explain why CUR is a selective modulator.
The probability that the modulation of ABC transporter function would result in an increase in the intracellular concentrations of the drugs to toxic levels became apparent from the growth studies. When CUR was used in combination, it was synergistic with drugs in cells overexpressing ABC transporters. This synergism was restricted to those drugs whose efflux was modulated by R6G (24). Thus, the chemosensitization of cells by CUR was specific to competing drugs, such as KTC, ITC, and MCZ, and was not observed with noncompetitive drugs, such as FLC, VORI, ANISO, CYH, and FK520. The fact that the presence of CUR along with some drugs did not inhibit the growth of cells not only points to the selectivity of CUR for certain compounds but also suggests that R6G, KTC, ITC, MCZ, and CUR may share overlapping binding sites of ABC multidrug transporter proteins. The modulatory and synergistic effects of CUR confirm our previous observation that KTC, MCZ, and ITC share CaCdr1p binding sites with R6G (24).
A natural CUR mixture contains three major curcuminoids: curcumin I, curcumin II, and curcumin III. We tested these individual curcuminoids in our earlier studies with the mammalian ABC drug transporters P-glycoprotein, MRP1, and ABCG2 (3, 4, 5). We reported that these individual curcuminoids inhibited the function of these drug transporters with different efficiencies and that curcumin I was the most potent among them (3, 4, 5). In addition, we have also reported that tetrahydrocurcumin, a major metabolite of CUR, also inhibits these three mammalian ABC drug transporters (18). In this study, based on the initial data discussed above for the natural CUR mixture and its purified individual components with mammalian transporters, we evaluated the CUR mixture alone for its activity to synergize the activities of antifungal agents. There are several CUR derivatives that are synthetic analogues, and some of them may have better activity than the CUR mixture. Thus, these analogues merit further study.
There are reports to suggest that CUR can downregulate the expression of an MDR-linked transporter (ABCB1) and can even affect the function of several transcription factors (2, 6). For this reason, we tested the effect of CUR on the expression of an ABC transporter and observed that CUR did not affect the expression levels of CaCdr1p (Fig. 4D), implying that the modulation of R6G efflux by CUR is restricted to its direct effect on the functionality of ABC transporter proteins. The direct effect of CUR on CaCdr1p was confirmed by its ability to compete the photoaffinity labeling of CaCdr1p with [125I]IAAP (Fig. 4B) and by its competitive inhibition of R6G efflux (Fig. 4A). We excluded the possibility that CUR could be a preferred substrate of the ABC transporters studied (Fig. 2).
Our study shows that CUR, which is not a transport substrate of CaCdr1p, specifically modulates the efflux of R6G mediated by the transporter. This is not surprising, since it has been observed previously that curcuminoids can modulate drug transport without being a substrate of mammalian ABCG2 (4). Our cytotoxicity data (Fig. 2; Table 2) suggest that the presence or absence of efflux pump proteins did not affect the growth and viability of yeast cells, again pointing to the fact that CUR is not a substrate of these pumps. It is not clear, however, whether CUR modulates R6G efflux by binding to the substrate or to an allosteric site(s) of CaCdr1p. Considering the fact that the structures and substrate specificities of fungal ABC transporters such as CaCdr1p, CaCdr2p, and ScPdr5p are very different, our finding from this study that yeast transporters can be modulated by CUR is very significant.
It is reported that the poor bioavailability of CUR and its low concentrations in plasma decrease its effectiveness in modulating the function of ABC drug transporters in rodents and humans. However, recent studies indicate that the use of piperine to prevent the glucuronidation of curcumin, as well as the encapsulation of CUR in liposomes, can increase the absorption of CUR and its levels in plasma (28). It is, however, not known whether CUR is metabolized via glucuronidation in yeast cells or whether the intracellular level of CUR is lower than that in medium or plasma. These issues need to be resolved before CUR can be used as an effective in vivo or in vitro antifungal. In summary, the modulation of antifungal efflux by CUR is substrate and transporter specific. Nevertheless, curcuminoids are not toxic to the cell, nor are they transported by their target efflux pumps. Thus, their ability to sensitize cells to azoles opens up the possibility that they could be exploited in combination with conventional chemotherapy.
Acknowledgments
The work presented in this paper has been supported in part by grants to R.P. from Indo-DFG (INT/DFG/P-05/2005), the Department of Science and Technology (SR/SO/BB-12/2004), the Council of Scientific and Industrial Research [38(1122)/06/EMR-II], and the Department of Biotechnology (BT/PR9100/Med/29/03/2007, BT/PR9563/BRB/10/567/2009). S. Shukla and S. V. Ambudkar were supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. M.S. acknowledges the Department of Biotechnology, India, for the award of junior and senior research fellowships.
We are grateful to R. D. Cannon for providing the S. cerevisiae strains AD-CDR2 and AD-PDR5 and to R. Pasrija for providing strain AD-CaMDR1. We thank M. Darokar, CIMAP, India, for providing pure forms of curcuminoids. We are thankful to Pritam Mukhopadhyay for making suggestions during the preparation of the manuscript. We further thank Ranbaxy Laboratories Limited, India, for providing fluconazole and voriconazole.
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Ammon, H. P., and M. A. Wahl. 1991. Pharmacology of Curcuma longa. Planta Med. 57:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Anuchapreeda, S., P. Leechanachai, M. M. Smith, S. V. Ambudkar, and P. N. Limtrakul. 2002. Modulation of P-glycoprotein expression and function by curcumin in multidrug-resistant human KB cells. Biochem. Pharmacol. 64:573-582. [DOI] [PubMed] [Google Scholar]
- 3.Chearwae, W., S. Anuchapreeda, K. Nandigama, S. V. Ambudkar, and P. Limtrakul. 2004. Biochemical mechanism of modulation of human P-glycoprotein (ABCB1) by curcumin I, II, and III purified from turmeric powder. Biochem. Pharmacol. 68:2043-2052. [DOI] [PubMed] [Google Scholar]
- 4.Chearwae, W., S. Shukla, P. Limtrakul, and S. V. Ambudkar. 2006. Modulation of the function of the multidrug resistance-linked ATP-binding cassette transporter ABCG2 by the cancer chemopreventive agent curcumin. Mol. Cancer Ther. 5:1995-2006. [DOI] [PubMed] [Google Scholar]
- 5.Chearwae, W., C. P. Wu, H. Y. Chu, T. R. Lee, S. V. Ambudkar, and P. Limtrakul. 2006. Curcuminoids purified from turmeric powder modulate the function of human multidrug resistance protein 1 (ABCC1). Cancer Chemother. Pharmacol. 57:376-388. [DOI] [PubMed] [Google Scholar]
- 6.Choi, B. H., C. G. Kim, Y. Lim, S. Y. Shin, and Y. H. Lee. 2008. Curcumin down-regulates the multidrug-resistance mdr1b gene by inhibiting the PI3K/Akt/NFκB pathway. Cancer Lett. 259:111-118. [DOI] [PubMed] [Google Scholar]
- 7.Decottignies, A., A. M. Grant, J. W. Nichols, H. De Wet, D. B. McIntosh, and A. Goffeau. 1998. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273:12612-12622. [DOI] [PubMed] [Google Scholar]
- 8.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaur, M., D. Choudhury, and R. Prasad. 2005. Complete inventory of ABC proteins in human pathogenic yeast, Candida albicans. J. Mol. Microbiol. Biotechnol. 9:3-15. [DOI] [PubMed] [Google Scholar]
- 10.Gaur, M., N. Puri, R. Manoharlal, V. Rai, G. Mukhopadhayay, D. Choudhury, and R. Prasad. 2008. MFS transportome of the human pathogenic yeast Candida albicans. BMC Genomics 9:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman, M. M., T. Fojo, and S. E. Bates. 2002. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2:48-58. [DOI] [PubMed] [Google Scholar]
- 12.Holmes, A. R., Y. H. Lin, K. Niimi, E. Lamping, M. Keniya, M. Niimi, K. Tanabe, B. C. Monk, and R. D. Cannon. 2008. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob. Agents Chemother. 52:3851-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, C. A., and J. K. M. Sanders. 1990. The nature of π-π interactions. J. Am. Chem. Soc. 112:5525-5534. [Google Scholar]
- 14.Hunter, C. A., J. Singh, and J. M. Thornton. 1991. Pi-pi interactions: the geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J. Mol. Biol. 218:837-846. [DOI] [PubMed] [Google Scholar]
- 15.Krishnamurthy, S., U. Chatterjee, V. Gupta, R. Prasad, P. Das, P. Snehlata, S. E. Hasnain, and R. Prasad. 1998. Deletion of transmembrane domain 12 of CDR1, a multidrug transporter from Candida albicans, leads to altered drug specificity: expression of a yeast multidrug transporter in baculovirus expression system. Yeast 14:535-550. [DOI] [PubMed] [Google Scholar]
- 16.Kuo, M. L., T. S. Huang, and J. K. Lin. 1996. Curcumin, an antioxidant and anti-tumor promoter, induces apoptosis in human leukemia cells. Biochim. Biophys. Acta 1317:95-100. [DOI] [PubMed] [Google Scholar]
- 17.Lamping, E., B. C. Monk, K. Niimi, A. R. Holmes, S. Tsao, K. Tanabe, M. Niimi, Y. Uehara, and R. D. Cannon. 2007. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot. Cell 6:1150-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limtrakul, P., W. Chearwae, S. Shukla, C. Phisalphong, and S. V. Ambudkar. 2007. Modulation of function of three ABC drug transporters, P-glycoprotein (ABCB1), mitoxantrone resistance protein (ABCG2) and multidrug resistance protein 1 (ABCC1) by tetrahydrocurcumin, a major metabolite of curcumin. Mol. Cell. Biochem. 296:85-95. [DOI] [PubMed] [Google Scholar]
- 19.Litman, T., T. E. Druley, W. D. Stein, and S. E. Bates. 2001. From MDR to MXR: new understanding of multidrug resistance systems, their properties and clinical significance. Cell. Mol. Life Sci. 58:931-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura, K., M. Niimi, K. Niimi, A. R. Holmes, J. E. Yates, A. Decottignies, B. C. Monk, A. Goffeau, and R. D. Cannon. 2001. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob. Agents Chemother. 45:3366-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niimi, K., D. R. Harding, R. Parshot, A. King, D. J. Lun, A. Decottignies, M. Niimi, S. Lin, R. D. Cannon, A. Goffeau, and B. C. Monk. 2004. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a d-octapeptide derivative. Antimicrob. Agents Chemother. 48:1256-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasrija, R., D. Banerjee, and R. Prasad. 2007. Structure and function analysis of CaMdr1p, a major facilitator superfamily antifungal efflux transporter protein of Candida albicans: identification of amino acid residues critical for drug/H+ transport. Eukaryot. Cell 6:443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad, R., N. A. Gaur, M. Gaur, and S. S. Komath. 2006. Efflux pumps in drug resistance of Candida. Infect. Disord. Drug Targets 6:69-83. [DOI] [PubMed] [Google Scholar]
- 24.Saini, P., T. Prasad, N. A. Gaur, S. Shukla, S. Jha, S. S. Komath, L. A. Khan, Q. M. Haq, and R. Prasad. 2005. Alanine scanning of transmembrane helix 11 of Cdr1p ABC antifungal efflux pump of Candida albicans: identification of amino acid residues critical for drug efflux. J. Antimicrob. Chemother. 56:77-86. [DOI] [PubMed] [Google Scholar]
- 25.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 26.Shukla, S., V. Rai, D. Banerjee, and R. Prasad. 2006. Characterization of Cdr1p, a major multidrug efflux protein of Candida albicans: purified protein is amenable to intrinsic fluorescence analysis. Biochemistry 45:2425-2435. [DOI] [PubMed] [Google Scholar]
- 27.Shukla, S., P. Saini, Smriti, S. Jha, S. V. Ambudkar, and R. Prasad. 2003. Functional characterization of Candida albicans ABC transporter Cdr1p. Eukaryot. Cell 2:1361-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shukla, S., H. Zaher, A. Hartz, B. Bauer, J. A. Ware, and S. V. Ambudkar. 2009. Curcumin inhibits the activity of ABCG2/BCRP1, a multidrug resistance-linked ABC drug transporter in mice. Pharm. Res. 26:480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, W. L., and T. D. Edlind. 2002. Histone deacetylase inhibitors enhance Candida albicans sensitivity to azoles and related antifungals: correlation with reduction in CDR and ERG upregulation. Antimicrob. Agents Chemother. 46:3532-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanabe, K., E. Lamping, K. Adachi, Y. Takano, K. Kawabata, Y. Shizuri, M. Niimi, and Y. Uehara. 2007. Inhibition of fungal ABC transporters by unnarmicin A and unnarmicin C, novel cyclic peptides from marine bacterium. Biochem. Biophys. Res. Commun. 364:990-995. [DOI] [PubMed] [Google Scholar]
- 31.Vermitsky, J. P., and T. D. Edlind. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48:3773-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]