Abstract
New extended-spectrum β-lactamase GES-11 was detected in Acinetobacter baumannii BM4674. The enzyme conferred resistance to β-lactams, including aztreonam, and reduced susceptibility to carbapenems. The structural gene was part of a class 1 integron borne by self-transferable plasmid pIP847. GES-type β-lactamases have not been reported previously in A. baumannii.
Acinetobacter baumannii is a predominant species associated with outbreaks of nosocomial infections, such as pneumonia, urinary tract infections, septicemia, and meningitis. Its clinical significance is due to its ability either to upregulate indigenous efflux pumps (5) or to acquire numerous resistance mechanisms (7) that lead to therapeutic failure. A rapid, global emergence of A. baumannii strains resistant to all β-lactams, including carbapenems, aminoglycosides, quinolones, tetracyclines-glycylcyclines, polymyxins, and trimethoprim-sulfamethoxazole, has been observed (1). A. baumannii clinical specimens resistant to all known antibiotics, including polymyxins, have been reported, illustrating the genetic flexibility of this pathogen (13).
Resistance to β-lactams in A. baumannii is due mainly to the production of β-lactamases but can also result from several other mechanisms, including changes in outer membrane proteins, overexpression of multidrug efflux pumps, and alterations in the affinity or production of penicillin-binding proteins (9). β-Lactamases with carbapenemase activity, i.e., class D carbapenem-hydrolyzing oxacillinases or, less frequently, class B metallo-β-lactamases, represent the major clinical concern. To the best of our knowledge, Ambler class A carbapenemases KPC, GES, SME, NMC, and IMI have not yet been reported in A. baumannii (18).
A. baumannii BM4674 was isolated from the tibia fracture of a patient hospitalized at the Centre Hospitalier Universitaire in Nancy, France, in September 2008. MICs of antimicrobial agents for this strain were determined by Etest (AB Biodisk, Combourg, France) on Mueller-Hinton agar (bioMerieux, Marcy l'Etoile, France), and the breakpoints delivered by the Comité de l'Antibiogramme de la Société Française de Microbiologie were used for interpretations of results (3). A. baumannii BM4674 was resistant to all β-lactams, with decreased susceptibility to carbapenems (MICimipenem = 4 μg/ml; MICmeropenem = 8 μg/ml). It was also resistant to aminoglycosides, co-trimoxazole, quinolones, and chloramphenicol but remained susceptible to tetracyclines-glycylcyclines, colistin, and rifampin (rifampicin).
The transfer of β-lactam resistance from A. baumannii BM4674 to A. baumannii BM4652 was performed by conjugation on solid medium, as described previously (11). Transconjugants selected on agar containing apramycin (80 μg/ml) and ceftazidime (16 μg/ml) were obtained at a high frequency of ca. 1 × 10−3 per recipient cell. They exhibited a broad spectrum of resistance to β-lactams, including aztreonam, diminished susceptibility to imipenem (MIC = 0.75 μg/ml) and meropenem (MIC = 1.5 μg/ml) compared to that of the recipient (MICimipenem = 0.125 μg/ml; MICmeropenem = 0.094 μg/ml), and synergism between cefotaxime and clavulanic acid, suggesting production of an extended-spectrum β-lactamase (ESBL). The transconjugants were also resistant to amikacin, tobramycin, trimethoprim, and sulfonamides. Plasmid DNA extracted from A. baumannii BM4674 was electrotransformed into A. baumannii BM4454 and Acinetobacter radioresistens CIP1281. Transformants selected on agar containing ticarcillin (20 μg/ml) had a resistance phenotype to β-lactams and to the other drugs similar to that of the transconjugants. Analysis of plasmid DNA (2) from A. baumannii BM4674 and A. baumannii transformants by agarose gel electrophoresis following digestion with EcoRI or XbaI revealed the presence of plasmid pIP847 of ca. 90 kb (data not shown).
Total DNA of A. baumannii BM4674, selected transconjugants, and transformants was screened by PCR for the presence of blaVEB, blaPER, blaKPC, blaGES-type, blaOXA-23, blaOXA-24, blaOXA-40, and blaOXA-58 genes using laboratory-designed sets of primers (Table 1) and for the presence of blaTEM, blaSHV, blaCTX-M-1-type, blaCTX-M-2-type, blaCTX-M-8-type, and blaCTX-M-9-type genes, as described previously (10). Results (not shown) indicated that A. baumannii BM4674 harbored both blaOXA-58 and blaGES-type genes, whereas the transconjugants and transformants carried only the blaGES-type gene as part of plasmid pIP847.
TABLE 1.
Oligonucleotide primers used for PCR amplification of β-lactam resistance genes
| bla gene | Primer (direction)a | Sequence (5′-3′) | Positionb | Size (pb) | GenBank accession no. |
|---|---|---|---|---|---|
| PER | per (+) | CCTGACGATCTGGAACCTTT | 157-176 | 715 | EF535600.1 |
| per (−) | GCAACCTGCGCAAT(GA)ATAGC | 872-853 | |||
| VEB | veb (+) | ATTTCCCGATGCAAAGCGT | 188-206 | 542 | AF010416 |
| veb (−) | TTATTCCGGAAGTCCCTGT | 730-712 | |||
| GES | ges (+) | ATGCGCTTCATTCACGCAC | 1-19 | 863 | AF156486 |
| ges (−) | CTATTTGTCCGTGCTCAGGA | 864-845 | |||
| KPC | kpc (+) | ATGTCACTGTATCGCCGTCT | 1-20 | 881 | AF297554 |
| kpc (−) | TTACTGCCCGTTGACGCCCA | 882-863 | |||
| OXA-23 | oxa23 (+) | ATGAATAAATATTTTACTTG | 1-20 | 821 | AJ132105 |
| oxa23 (−) | TTAAATAATATTCAGCTGTT | 822-803 | |||
| OXA-24 | oxa24 (+) | ATACTTCCTATATTCAGCAT | 13-32 | 809 | AJ239129 |
| oxa24 (−) | GATTCCAAGATTTCTAGCG | 822-803 | |||
| OXA-40 | oxa40 (+) | ATGAAAAAATTTATACTTCCTATA | 1-24 | 819 | AF509241 |
| oxa40 (−) | TTCCAAGATTTTCTAGCGAC | 820-801 | |||
| OXA-58 | oxa58 (+) | ATGAAATTATTAAAAATATTGAGT | 143-166 | 840 | AY570763 |
| oxa58 (−) | ATAAATAATGAAAAACACCCAA | 983-962 |
+, primer forward; −, primer reverse.
Refers to the first base of each β-lactamase gene.
The amplified blaGES-type gene was cloned in pCR-Blunt vector (Invitrogen, Leek, The Netherlands) under the control of the lac promoter, generating the plasmid pAT517, which was electrotransformed in Escherichia coli Top10 and E. coli HB4 deficient in porins OmpF and OmpC (12). Transformants E. coli Top10 (pAT517) and E. coli HB4 (pAT517) exhibited the ESBL phenotype and reduced susceptibility to carbapenems, as observed for transconjugants (Table 2). The MIC of imipenem for HB4 (pAT517) was 8 μg/ml, revealing that the expression of blaGES-11 in a porin-deficient E. coli strain could lead to imipenem resistance and likewise indicating the carbapenemase activity of GES-11.
TABLE 2.
MICs of β-lactams for A. baumannii and E. coli strains
| β-Lactam | MIC (μg/ml) for indicated strain (plasmid):
|
||||||
|---|---|---|---|---|---|---|---|
|
A. baumannii
|
E. coli
|
||||||
| BM4674 | BM4652 | BM4652 (pIP847) | Top10 | Top10 (pAT517) | HB4 | HB4 (pAT517) | |
| Cefoxitin | 192 | 8 | 12 | 6 | 6 | 96 | >256 |
| Ceftazidime | >256 | 1.5 | >256 | 0.75 | >256 | 0.5 | >256 |
| Cefepime | >256 | 0.19 | 64 | 0.032 | 2 | 0.5 | 96 |
| Cefotaxime | >256 | 1 | >256 | 0.064 | >256 | 0.5 | >256 |
| Aztreonam | >256 | 1 | >256 | 0.125 | 24 | 0.75 | 48 |
| Imipenem | 4 | 0.125 | 0.75 | 0.19 | 0.25 | 0.125 | 8 |
| Meropenem | 6 | 0.064 | 1.5 | 0.016 | 0.047 | 0.25 | 4 |
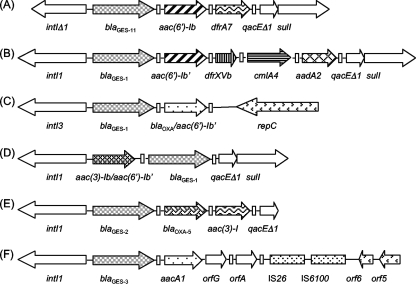
Sequence determination of purified blaGES-type amplicons revealed an 864-bp open reading frame. The deduced protein, designated GES-11, contained 287 amino acids and differed from GES-1 β-lactamase by a Gly-to-Ala substitution at Ambler position 243. A Gly-to-Ser change at this position has been previously reported in GES-9 (14). Substitution of the glycine at position 243 in GES-11 was associated with increased activity toward aztreonam, as had been observed for GES-9 (14). GES-11 did not have a substitution of the Gly170 residue that results in increased hydrolysis of imipenem as in GES-2, GES-4, GES-5, and GES-6 (17, 19, 20). The moles percent G+C content for blaGES-11 was 53.25%, a value higher than that of the genus Acinetobacter, which contains 38 to 39% G+C content (7). Using total DNA of A. baumannii BM4674 and of transformants BM4454 (pIP847) as a template and consensus primers for 5′-CS and 3′-CS ends of class 1 integrons (8), a 4,152-bp DNA fragment was obtained (Fig. 1). Sequence analysis revealed that the blaGES-11 gene was part of a class 1 integron containing, downstream, the aac(6′)-Ib gene encoding an aminoglycoside, 6′-N-acetyltransferase, type I, which modifies amikacin and tobramycin, and the dfrA7 trimethoprim resistance gene. A deletion of 83 bp in the class 1 integrase gene revealed that this integron alone can no longer acquire or lose any resistance genes. The cassette organization differed from those of other GES-containing integrons (4, 6, 15, 21).
FIG. 1.
Schematic representation of integrons containing blaGES-like genes. Arrows represent coding sequences and indicate the direction of transcription. Rectangles represent attC sites. (A) blaGES-11 containing a class 1 integron of pIP847; (B) blaGES-1 containing a class 1 integron of pTK-1 (15); (C) blaGES-1 containing a class 3 integron of p22K9 (4); (D) blaGES-1 containing a class 1 integron of pC23 (6); (E) blaGES-2 containing a class 1 integron of p22K9 (GenBank accession no. AF326355); and (F) blaGES-3 containing a class 1 integron of pKGB525 (21).
In addition to the finding of a novel GES-type variant as part of a class 1 integron, this work reports the emergence of GES-type ESBLs in A. baumannii. The occurrence of GES enzymes in this species is probably underestimated, since synergism between extended-spectrum cephalosporins and clavulanic acid may be masked by the presence of the intrinsic AmpC cephalosporinase and OXA-51-like oxacillinase that are frequently associated with other β-lactamases such as OXA-58 (16). Genotypic screening is therefore required to assess the prevalence of GES-type enzymes in A. baumannii.
Nucleotide sequence accession number.
The nucleotide sequence of the integron blaGES-11 is available in the GenBank nucleotide database under accession no. FJ854362.
Acknowledgments
We thank L. Poirel for the gift of E. coli HB4, M. Galimand for helpful discussions, and P. E. Reynolds for reading of the manuscript.
This work was supported in part by the Institut National de la Veille Sanitaire (InVS).
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2009. Communiqué 2009. Société Française de Microbiologie, Paris, France. http://www.sfm.asso.fr.
- 4.Correia, M., F. Boavida, F. Grosso, M. J. Salgado, L. M. Lito, J. M. Cristino, S. Mendo, and A. Duarte. 2003. Molecular characterization of a new class 3 integron in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 47:2838-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Depardieu, F., I. Podglajen, R. Leclercq, E. Collatz, and P. Courvalin. 2007. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20:79-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubois, V., L. Poirel, C. Marie, C. Arpin, P. Nordmann, and C. Quentin. 2002. Molecular characterization of a novel class 1 integron containing bla(GES-1) and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:762-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnier, F., N. Raked, A. Gassama, F. Denis, and M. C. Ploy. 2006. Genetic environment of quinolone resistance gene qnrB2 in a complex sul1-type integron in the newly described Salmonella enterica serovar Keurmassar. Antimicrob. Agents Chemother. 50:3200-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giamarellou, H., A. Antoniadou, and K. Kanellakopoulou. 2008. Acinetobacter baumannii: a universal threat to public health? Int. J. Antimicrob. Agents 32:106-119. [DOI] [PubMed] [Google Scholar]
- 10.Guessennd, N., S. Bremont, V. Gbonon, A. Kacou-Ndouba, E. Ekaza, T. Lambert, M. Dosso, and P. Courvalin. 2008. Qnr-type quinolone resistance in extended-spectrum β-lactamase producing enterobacteria in Abidjan, Ivory Coast. Pathol. Biol. (Paris) 56:439-446. [DOI] [PubMed] [Google Scholar]
- 11.Lambert, T., G. Gerbaud, and P. Courvalin. 1988. Transferable amikacin resistance in Acinetobacter spp. due to a new type of 3′-aminoglycoside phosphotransferase. Antimicrob. Agents Chemother. 32:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mammeri, H., P. Nordmann, A. Berkani, and F. Eb. 2008. Contribution of extended-spectrum AmpC (ESAC) β-lactamases to carbapenem resistance in Escherichia coli. FEMS Microbiol. Lett. 282:238-240. [DOI] [PubMed] [Google Scholar]
- 13.Maragakis, L. L., and T. M. Perl. 2008. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 46:1254-1263. [DOI] [PubMed] [Google Scholar]
- 14.Poirel, L., L. Brinas, N. Fortineau, and P. Nordmann. 2005. Integron-encoded GES-type extended-spectrum β-lactamase with increased activity toward aztreonam in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:3593-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel, L., I. Le Thomas, T. Naas, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 17.Poirel, L., G. F. Weldhagen, T. Naas, C. De Champs, M. G. Dove, and P. Nordmann. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vourli, S., P. Giakkoupi, V. Miriagou, E. Tzelepi, A. C. Vatopoulos, and L. S. Tzouvelekis. 2004. Novel GES/IBC extended-spectrum β-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol. Lett. 234:209-213. [DOI] [PubMed] [Google Scholar]
- 20.Wachino, J., Y. Doi, K. Yamane, N. Shibata, T. Yagi, T. Kubota, and Y. Arakawa. 2004. Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A β-lactamase, GES-4, possessing a single G170S substitution in the Ω-loop. Antimicrob. Agents Chemother. 48:2905-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wachino, J., Y. Doi, K. Yamane, N. Shibata, T. Yagi, T. Kubota, H. Ito, and Y. Arakawa. 2004. Nosocomial spread of ceftazidime-resistant Klebsiella pneumoniae strains producing a novel class a β-lactamase, GES-3, in a neonatal intensive care unit in Japan. Antimicrob. Agents Chemother. 48:1960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]