Abstract
Although methicillin (meticillin)-resistant Staphylococcus aureus (MRSA) strains with reduced susceptibility to vancomycin (RVS-MRSA; including vancomycin-intermediate S. aureus [VISA] and heterogeneous VISA [hVISA]) have been linked with vancomycin treatment failure, it is unclear whether they are more pathogenic than vancomycin-susceptible MRSA (VS-MRSA). We prospectively assessed patients with clinical MRSA isolates during a 10-month period to determine clinical status (infection versus colonization) and therapeutic outcome before correlating these findings with the results of detailed in vitro assessment of vancomycin susceptibility, including population analysis profile (PAP) testing. hVISA and VISA were defined by standard PAP criteria (area-under-the-curve ratio compared to that of the reference hVISA strain Mu3 [≥0.9]) and routine CLSI criteria (vancomycin MIC, 4 to 8 μg/ml), respectively. Among the 117 patients assessed, 58 had RVS-MRSA isolates (56 hVISA and 2 VISA) and 59 had VS-MRSA isolates; the patient demographics and comorbidities were similar. RVS-MRSA was associated with a lower rate of infection than VS-MRSA (29/58 versus 46/59; P = 0.003), including a lower rate of bacteremia (3/58 versus 20/59, respectively; P < 0.001). The cure rates in RVS-MRSA and VS-MRSA groups were not statistically different (16/26 versus 31/42; P = 0.43), but the post hoc assessment of treatment regimes and study size made detailed conclusions difficult. The results of the macro method Etest correlated well with the PAP results (sensitivity, 98.3%, and specificity, 91.5%), but broth microdilution and our preliminary RVS-MRSA detection method correlated poorly. All isolates were susceptible to linezolid and daptomycin. These data suggest that detailed prospective laboratory identification of RVS-MRSA isolates may be of limited value and that, instead, such in vitro investigation should be reserved for isolates from patients who are failing appropriate anti-MRSA therapy.
Staphylococcus aureus strains with reduced susceptibility to vancomycin (RVS) have been recognized increasingly since their initial description in 1997 (13) and have generally arisen from methicillin (meticillin)-resistant S. aureus (MRSA) strains. In particular, strains of vancomycin-intermediate S. aureus (homogenous [VISA] and heterogeneous [hVISA]) have been associated with vancomycin treatment failures, including prolonged bacteremia, endocarditis, osteomyelitis, and prosthetic-joint sepsis (3, 9, 16), suggesting that these strains may be more pathogenic than routine strains of MRSA, although such reports have been retrospective in nature.
We aimed to prospectively assess the relative clinical importance of RVS-MRSA and vancomycin-susceptible MRSA (VS-MRSA) in terms of their rates of clinical infection and to compare the relative accuracies of various laboratory methods in identifying RVS-MRSA strains. Although not the primary purpose of the study, treatment outcomes for infections due to RVS-MRSA and VS-MRSA were compared.
(This work has been presented in part at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC], San Francisco, CA, 27 to 30 September, 2006 [15].)
MATERIALS AND METHODS
Patient selection.
During a 10-month period from March to December of 2005, inpatients at our institution (Austin Hospital) from whom MRSA was isolated from clinical (nonscreening) cultures which had been ordered by the patients' managing clinicians as part of an infection workup were identified and prospectively assessed to determine whether the isolate was associated with clinical infection or simply “colonization” (see definition below), using standard definitions of infection (1, 14). Two approaches to clinical assessment were used. During the initial 5-month period (March to July; period A), a case-control assessment was undertaken in which patients with presumptive RVS-MRSA isolates were identified by our laboratory's preliminary hVISA identification method (see below) and matched with patients who had presumptive VS-MRSA isolated from an identical clinical site during the same time period. All patients were reviewed within 72 h of the reporting of detailed confirmatory vancomycin susceptibility results (see below). Due to concerns that delays in the laboratory confirmation of RVS-MRSA isolates were influencing the immediacy of the clinical assessments and that results from our preliminary hVISA identification method were occasionally discordant with those from our confirmatory testing, we amended our approach to simply assess all patients with MRSA isolates within 72 h of MRSA identification, before completion of any in vitro assessment of vancomycin susceptibility (5-month period from August to December of 2005; period B). We then correlated the clinical impact of each isolate with the patients' demographic details and clinical features. The study was conducted as a quality assurance initiative under the Austin Health Ethics Committee Quality Program.
Clinical assessment and definitions.
Standard information was collected for each patient, including demographic data (age, sex, and hospital admission date), presence of comorbidities, and diseases/medications associated with immune suppression. The investigating team (K.C.H. and M.G.) was not involved in patient care unless a formal referral to the Infectious Diseases Department was made. For the occasional instance where the patient was discharged before review, the relevant clinical and follow-up data were collected from the patient file. The clinical site from which MRSA (VS-MRSA or RVS-MRSA) was isolated was assessed using standard definitions to determine whether clinical infection was present. Centers for Disease Control definitions (14) were used for all sites, except for ventilator- and hospital-acquired pneumonia, when definitions from the guidelines of the American Thoracic Society and Infectious Diseases Society of America were used, since they were more specific (1). These definitions do not include the use of antibiotic therapy as criteria for presence of infection. Where these definitions were met, the site was deemed “infected”; where these definitions were not met, the site from which the RVS-MRSA or VS-MRSA strain were isolated was deemed to be “colonized.”
Where patients had MRSA isolates obtained from more than one site, a standard algorithm was used to identify which site was the most clinically significant based on site sterility, such that only one key isolate was assessed for each patient. The order of decreasing priority was as follows: blood cultures > sterile-site cultures (cerebrospinal fluid, joint fluid, and peritoneal fluid) > urine > sputum or wound swabs. The significance of sputum and wound swabs was influenced by the patient's clinical context—for instance, clinical signs of pneumonia or cellulitis, respectively, suggested infection. The most recent isolate from the most significant clinical site during the admission was the isolate selected for detailed in vitro susceptibility testing. All selected isolates were stored at −70°C prior to in vitro assessment.
Treatment outcomes for infected sites were assessed using standard clinical definitions of “cure,” “failure,” and “indeterminate” (12). Definitions were as follows. Cure was defined as the resolution of clinical signs and symptoms of infection in the absence of ongoing antibiotic therapy, failure as either unresolved signs or symptoms of infection at the time of completing a standard course of therapy or recurrence of infection requiring readmission within 1 month of ceasing therapy, and indeterminate as the clinical outcome or improvement being uncertain but the therapy ongoing at the time of assessment. Mortality data for patients were recorded 30 days after the isolate was collected.
Although the study was not primarily designed to assess potential factors related to the emergence of RVS-MRSA isolates, we reviewed patients' histories for the following information to identify any obvious associations: recent anti-MRSA therapy prior to admission to the study, including serum vancomycin levels (when tested), and the timing of such therapy in relation to the time the isolate was obtained; the duration of any hospitalization during the 3 months prior to admission; and the presence of laboratory markers of severe/chronic disease at the time of MRSA isolate collection (hemoglobin, hematocrit, leukocyte count, platelet count, and serum albumin) (10). Similarly, we recorded which antibiotic therapy was used to treat patients during the study, although interpretation of comparative drug efficacies was likely to be limited given the nonrandomized study design.
Laboratory methods.
Susceptibilities to routine antibiotics (methicillin [meticillin], tetracycline, erythromycin, clindamycin, co-trimoxazole, and vancomycin) were assessed by agar dilution according to CLSI criteria (4), and isolates were defined as multiresistant MRSA or non-multiresistant MRSA (resistant to fewer than three non-beta-lactam antibiotics) (5). These phenotypes were later correlated with the presence of RVS-MRSA or VS-MRSA. As required by our laboratory protocol, all nonmultiresistant strains of MRSA were assessed for the presence of Panton-Valentine leukocidin (PVL) by using standard PCR methods (18).
RVS-MRSA was defined by the results of population analysis profile (PAP) testing (22), using standard criteria in which the ratio of the area under the PAP curve (AUC) of the isolate compared with that of the reference hVISA strain Mu3 was required to be ≥0.9 for RVS-MRSA to be confirmed. All MRSA isolates were tested in duplicate by PAP test, and if the results were discrepant, a third test was performed and the isolate was classified according to the majority finding. Isolates with a ratio of <0.9 were defined as VS-MRSA (22). Consistent with CLSI guidelines, any MRSA isolate with an MIC of vancomycin of 4 to 8 μg/ml was defined as VISA (4), while isolates with an MIC of <4 μg/ml but a PAP result of ≥0.9 were defined as hVISA.
Susceptibility to vancomycin was also assessed by broth microdilution using Mueller-Hinton broth (4); the macro method Etest for vancomycin and teicoplanin (2.0 MacFarland inoculum) (21); and a new, potentially useful, preliminary hVISA identification method. This method used brain-heart infusion agar (Oxoid, Basingstoke, United Kingdom) with 20% horse serum plus 4 mg/liter teicoplanin (BHST4) and brain-heart infusion agar with 5% horse blood plus 2 mg/liter vancomycin (BHBV2). Any growth at 24 or 48 h was considered preliminary (but not confirmatory) evidence of RVS-MRSA (17).
Susceptibilities to linezolid and daptomycin were assessed by Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. The interpretation criteria used for susceptibility to linezolid and daptomycin by Etest were as follows: for linezolid, an MIC of ≤4 mg/liter, and for daptomycin, an MIC of ≤1 mg/liter (4).
All blood and sterile-site isolates were assessed for molecular clonality by pulsed-field gel electrophoresis (PFGE) using SmaI DNA digestion as previously described (2, 19). Gels were analyzed with GelCompar II (Applied Maths, Saint-Martens-Latems, Belgium) using the Dice coefficient and the unweighted pair group method with arithmetic mean with settings for tolerance and optimization of 1% and 0.75%, respectively. Isolates were considered clonal if the band similarity was ≥85% or the number of band differences was three or fewer (11).
Statistical analysis.
A univariate analysis of patient demographic and clinical variables was undertaken to identify factors that might have an association with either RVS-MRSA or VS-MRSA. Variables with a P value of ≤0.2 in this analysis were considered suitable for inclusion in a subsequent multivariate analysis. Statistical significance was tested using the chi-square, Fisher's exact, Mann-Whitney, or t test, as appropriate, with a P value of ≤0.05 considered statistically significant. Where appropriate, given information obtained from the study, sample size calculations were undertaken for some key parameters (7).
RESULTS
Association between RVS-MRSA and clinical infection.
One hundred seventeen patients with MRSA isolates were assessed (period A, 21 patients, and period B, 96 patients), of whom 108 were assessed prospectively and 9 (8.3%) from the medical record (see the supplemental material for details of clinical data and laboratory results for all patients). A total of 58 patients had RVS-MRSA isolates (56 had hVISA and 2 VISA; n = 15, period A, and n = 43, period B) and 59 had VS-MRSA isolates (n = 6, period A, and n = 53, period B), with proportionally more RVS-MRSA patients being identified in period A (15/21 versus 43/96; P = 0.049). Overall, the majority of patients (62/117, 53%) had only one MRSA isolate identified. Among those who had more than one MRSA isolate identified, there were generally only 2 to 7 days between the collection of the first isolate and that of the index MRSA isolate used for in vitro assessment (for RVS-MRSA isolates, the median was 7 days and the range 0 to 70 days, and for VS-MRSA isolates, the median was 2 days and the range 0 to 78 days; P = 0.35).
Patient demographics and clinical assessment data regarding rates of infection versus rates of colonization are shown in Tables 1 and 2, respectively. The distributions of age, sex, and frequency of comorbidities were similar for patients with RVS-MRSA and those with VS-MRSA. Rates of infection with RVS-MRSA and VS-MRSA are shown in Table 2. Overall, RVS-MRSA was less likely to be associated with clinical infection than VS-MRSA (P = 0.003); this included the smaller number of patients who had RVS-MRSA bacteremia than VS-MRSA bacteremia (3/58 versus 20/59; P < 0.001). However, when blood culture and sterile-site isolates were excluded and specimens such as urine, sputa, and wound swabs were assessed, there was no difference in the rate of infection (21/50 RVS-MRSA isolates versus 16/29 VS-MRSA isolates; P = 0.37). The rates of deep-seated infection were low for both RVS-MRSA and VS-MRSA patients (see Table 2).
TABLE 1.
Univariate analysis assessing impact of patient demographics, including age, sex, and comorbidities, on likelihood of having RVS-MRSA versus VS-MRSA
| Parameter | RVS-MRSA (n = 58) | VS-MRSA (n = 59) | P value |
|---|---|---|---|
| Age [mean yr ± SD (range)] | 68 ± 17 (20-93) | 67 ± 16 (15-92) | 0.74 |
| No. (%) of patients with characteristic | |||
| Male | 36 (62.1) | 39 (66.1) | 0.70 |
| Diabetic | 20 (34.5) | 14 (23.7) | 0.23 |
| Dialysis patient | 3 (5.2) | 4 (6.7) | 1.00 |
| Transplant recipient | 2 (3.4) | 3 (5.1) | 1.00 |
| Malignancy | 14 (24.1) | 19 (32.2) | 0.41 |
| Immunosuppressiona | 12 (20.7) | 17 (28.8) | 0.39 |
| Chronic obstructive pulmonary disease | 11 (19.0) | 12 (20.3) | 1.00 |
| Laboratory parameters (mean or mean ± SD)b | |||
| Hemoglobin (g/dl) | 10.51 ± 1.6 | 10.55 ± 2.3 | 0.93 |
| Hematocrit | 0.32 | 0.32 | 0.59 |
| Leukocyte count | 11.2 | 9.5 | 0.14 |
| Platelet count | 268 | 262 | 0.83 |
| Serum albumin (g/dl) | 26.9 | 25.7 | |
| No. of days (mean ± SD) in hospital during the 3 mo before study admission | 9.3 ± 14.2 | 9.9 ± 16.6 | 0.58 |
| Previous vancomycin therapy | |||
| No. of patients who received vancomycin prior to isolation of index MRSA specimenc | 21/57 | 22/57 | 1.00 |
| Duration [median no. of days (range)] of vancomycin therapy during the 6 wks prior to collection of the index MRSA isolate | 0 (0-37) | 2 (0-39) | 0.78 |
Immunosuppression was primarily due to agents such as corticosteroids, cancer chemotherapy, agents to prevent transplant rejection, and hematological disorders.
Results available (see text) for 115 patients, except for leukocyte count (n = 114) and serum albumin (n = 98).
Insufficient detail was available for three patients (one with RVS-MRSA and two with VS-MRSA).
TABLE 2.
Rates of infection versus colonization for RVS-MRSA and VS-MRSA isolates
| Site colonized or infected or type of infection | No. of patients with indicated type of isolate
|
P value | |||
|---|---|---|---|---|---|
| RVS-MRSA (n = 58)
|
VS-MRSA (n = 59)
|
||||
| Infecteda | Colonizedb | Infecteda | Colonizedb | ||
| Sites | |||||
| Total no. | 29 | 29 | 46 | 13 | 0.003 |
| Blood | 3 | — | 20 | — | |
| Sterile sitec | 5 | — | 10 | — | |
| Urine | 0 | 7 | 3 | 2 | 0.045 |
| Sputum | 7 | 14 | 5 | 4 | 0.69 |
| Wound swab | 14 | 8 | 8 | 7 | 0.73 |
| Deep infections | |||||
| Prosthetic infection | 1f | 1g | |||
| Osteomyelitisd | 7 | 5 | |||
| Endocarditis | 1e | 3 | |||
| Total | 9 | 8 | |||
Among isolates associated with infection, 27/29 RVS-MRSA and 36/46 VS-MRSA isolates were multiresistant [5].
—, no colonization according to definition.
Sterile sites include operative specimens, intra-abdominal abscess fluid, and peritoneal fluid.
No prosthetic-joint infections.
Permanent pacemaker lead infection with subsequent peripheral emboli but no changes on echocardiogram.
Orthopedic pin site.
Silicone breast prosthesis.
Overall, we found no factors that were significantly associated with the presence of RVS-MRSA. Among the 114 patients whose preadmission drug therapy could be assessed in sufficient detail, there was no difference between patients with RVS-MRSA and those with VS-MRSA in the number who received vancomycin prior to isolation of the index MRSA specimen or in the duration of vancomycin therapy during the 6 weeks prior to specimen collection (see Table 1). Based on the results of our univariate analysis, no features reached the defined threshold to warrant a multivariate analysis.
The rates of treatment cure and failure were not statistically different between RVS-MRSA and VS-MRSA infections (16/26 versus 31/42 cured, respectively; Table 3), although when patients who received no effective therapy were excluded, the results approached statistical significance (cures numbered 14/24 for RVS-MRSA patients versus 25/31 for VS-MRSA patients; P = 0.08). Among the 114 patients followed to 30 days, there was no difference in the all-cause 30-day mortality (12/58 for RVS-MRSA patients versus 11/56 for VS-MRSA patients; P = 0.93).
TABLE 3.
Response to treatment in patients with assessable clinical infection outcome
| Treatment regimen | No. of patients with indicated type of isolatea
|
P value | |||
|---|---|---|---|---|---|
| RVS-MRSA (n = 26)
|
VS-MRSA (n = 42)
|
||||
| Cure | Failure | Cure | Failure | ||
| Glycopeptide aloneb | 8 | 6 | 21 | 5 | 0.22 |
| Vancomycin in combination with additional agents | 6c | 4d | 4e | 1 | 0.60 |
| No MRSA treatmentf | 2 | 0 | 6 | 5 | 0.49 |
| Total | 16 | 10 | 31 | 11 | 0.43 |
In addition, there were three RVS-MRSA and four VS-MRSA patients with clinical infection who had not completed their course of therapy at the time of final assessment and were therefore deemed indeterminate.
All patients received vancomycin alone, except for one patient with VS-MRSA who was treated with teicoplanin and cured.
Additional agents were rifampin plus fusidic acid (two patients), linezolid plus rifampin plus fusidic acid (two patients), linezolid plus rifampin (one patient), and linezolid (one patient).
Additional agents were rifampin plus fusidic acid (one patient) and linezolid (three patients).
Additional agents were rifampin plus fusidic acid (two patients), linezolid plus rifampin plus fusidic acid (one patient), and clindamycin (one patient).
Reasons for no treatment in 13 cases were as follows: three patients died or were palliated prior to culture results becoming available; four patients had pneumonia and MRSA was isolated, but the managing clinicians did not consider the MRSA to be the causative organism; three patients were treated surgically or had prosthetic material removed and were considered to not require antibiotics; and in three cases, the managing clinicians decided not to treat.
We compared the degree of reduced vancomycin susceptibility, as measured by the PAP-AUC ratio, with the likelihood of clinical infection and subsequent treatment outcome (Fig. 1). There was no apparent correlation between the PAP-AUC ratio and the likelihood of either clinical infection or clinical treatment failure. However, for the two isolates with very high PAP-AUC ratio values, both were associated with infection, with one being cured and the other a treatment failure. Adjusting the definition of RVS-MRSA to a PAP-AUC ratio of >0.90 (e.g., 0.95, 1.0, or even 1.2) did not alter the relationship between RVS-MRSA and the likelihood of clinical infection or treatment failure (Fig. 1).
FIG. 1.
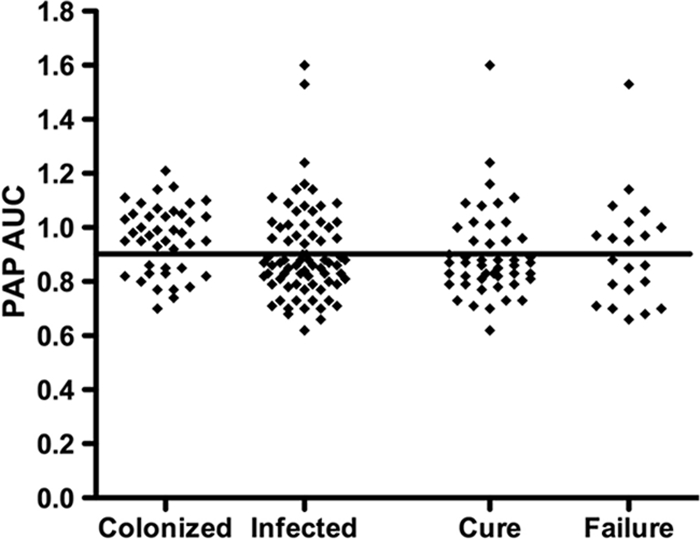
Comparison of the PAP-AUC ratio for each patient's MRSA isolate with the likelihood of clinical infection vs colonization, and, where infection was present, cure versus failure. No correlation was noted between PAP-AUC ratio and likelihood of the isolate causing infection or likelihood of treatment success. The horizontal line indicates a PAP-AUC ratio of 0.9. Each square represents the MRSA isolate for one patient in the study.
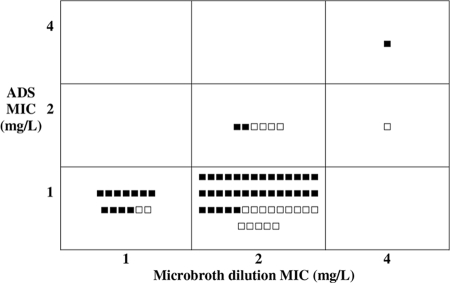
Similarly, we compared the likelihood of treatment failure with the MICs detected using each CLSI method (broth microdilution and agar dilution) among the 68 patients with infections for whom the treatment outcome was known (47 cures and 21 failures). In general, agar dilution identified MICs as one dilution lower than broth microdilution. Thus, the cure rates for patient whose isolates had MICs of 1, 2, and 4 mg/liter by broth microdilution were 11/13, 35/53, and 1/2, respectively, while the results using agar dilution were 44/60, 2/7, and 1/1, respectively (Fig. 2). Thus, an increasing MIC by either broth dilution or agar dilution was not associated with a reduced rate of cure (for broth microdilution, 11/13 had a MIC of 1 versus 36/55 with a MIC of 2 or 4 [P = 0.32, Fisher's exact test], and for agar dilution, 44/60 had a MIC of 1 versus 3/8 with a MIC of 2 or 4 [P = 0.096, Fisher's exact test]). An increasing MIC (mg/liter) was also not associated with an increased likelihood of the isolate being associated with infection (for broth microdilution, 15/23 isolates from infected patients had a MIC of 1 versus 60/94 with a MIC of 2 or 4 [P = 1.0], and for agar dilution, 66/101 isolates from infected patients had a MIC of 1 versus 9/16 with a MIC of 2 or 4 [P = 0.58]).
FIG. 2.
Comparison of MIC results (broth microdilution versus agar dilution methods) and clinical outcome (cure [▪]/failure [□]) among the 68 patients with infections with assessable treatment outcomes. ADS, agar dilution susceptibility; L, liter.
Using isolates associated with infection, correlation between MRSA phenotype (multiresistant versus nonmultiresistant) and reduced vancomycin susceptibility revealed that multiresistant strains constituted 27/29 (93.1%) RVS-MRSA isolates versus 36/46 (78.3%) VS-MRSA isolates (P = 0.11) (Table 2).
Although detailed analysis of the comparative efficacy of various treatment regimens was not possible, the observed treatment outcomes for the 75 patients with clinical infections (29 RVS-MRSA and 46 VS-MRSA) were summarized (Table 3).
Laboratory results.
The results of initial and repeated PAP testing for all MRSA isolates were highly reproducible, with only 11/117 (9.4%) having two discordant results that required a third test (see Materials and Methods). The macro method Etest results correlated well with the PAP test interpretations, with 57/58 (98.3%) of RVS-MRSA and 54/59 VS-MRSA isolates being appropriately identified (sensitivity, 98.3%, and specificity, 91.5%). For the single RVS-MRSA isolate that was not identified by the macro method Etest, the two PAP-AUC ratios were 0.96 and 1.03 and the microbroth MIC was 2 mg/liter. Among the five isolates that were defined as VS-MRSA by PAP but found to have a positive macro method Etest, the PAP-AUC ratios were 0.82 to 0.93 for four isolates and a maximum of 0.71 for the fifth isolate; four isolates had a microbroth MIC of 2 mg/liter, and one had an MIC of 1 mg/liter to vancomycin.
Overall, the results of broth microdilution susceptibility analyses correlated poorly with the identification of RVS-MRSA by PAP, with only two RVS-MRSA isolates having an MIC to vancomycin of ≥4 mg/liter (thereby being defined as VISA), while the other 56/58 had MICs of 2 mg/liter. Among VS-MRSA isolates, 36/59 and 23/59 had MICs to vancomycin of 2 and 1 mg/liter, respectively. The sensitivity and specificity of broth microdilution for RVS-MRSA were 3.4% and 100%, respectively. Similarly, our proposed preliminary RVS-MRSA identification method performed poorly, correctly identifying only 39/58 RVS-MRSA isolates and incorrectly suggesting that 9/59 VS-MRSA strains were RVS-MRSA (sensitivity, 67.2%, and specificity, 84.7%).
A multiresistant MRSA phenotype was noted in 56/58 RVS-MRSA isolates and 47/59 VS-MRSA isolates (P = 0.008). All non-multiresistant MRSA isolates were negative for Panton-Valentine leukocidin. All 117 isolates were susceptible to linezolid and daptomycin as determined by Etest.
All 38 blood and sterile-site isolates were assessed by PFGE, with 10 PFGE clonal groups identified and RVS-MRSA isolates represented in five of these groups.
DISCUSSION
Previous clinical studies of RVS-MRSA (3, 9, 16) have generally identified such isolates after observing clinical treatment failures. This is the first study to assess the relative clinical importance of hVISA/VISA versus VS-MRSA isolates in terms of their propensity to be associated with infection and the likelihood of treatment failure.
Our findings suggest that RVS-MRSA isolates are less frequently associated with infections than VS-MRSA isolates (29/58 versus 46/59; P = 0.003). Indeed, among patients who had infections, RVS-MRSA was less likely than VS-MRSA to be associated with bacteremia (3/29 versus 20/46; P = 0.002). We have previously shown that prolonged fever and/or bacteremia, despite appropriate antibacterial therapy, is associated with a higher likelihood of isolating hVISA (3, 16). However, the results of this study suggest that the reverse association does not appear to be the case, namely, that the presence of RVS-MRSA in and of itself is not necessarily associated with a higher rate of bacteremia. This may have potentially important practical implications since, prior to this study, we believed that the early laboratory detection of RVS-MRSA might have important treatment and outcome (as well as infection control) consequences. Hence, considerable laboratory effort was expended attempting to develop a preliminary RVS-MRSA identification method and time-consuming PAP testing was prioritized on all MRSA bacteremia isolates, even when there was no evidence of clinical treatment failure. However, based on our current findings, we now support the previous speculative comments by others (8, 9) that laboratory efforts to diagnose RVS-MRSA should be guided by the clinical situation. The key trigger for detailed laboratory investigations should be treatment failure (as previously defined in references 3 and 16).
Our study found that RVS-MRSA was not statistically more likely than VS-MRSA to be associated with clinical treatment failure (10/26 versus 11/42, respectively; P = 0.43). Similarly, we could detect no statistical association between an increasing vancomycin MIC among isolates associated with infections and clinical outcome, regardless of the CLSI susceptibility testing method used. However, these results should be interpreted with caution since it is possible, given our findings, that our study was of insufficient size to accurately detect anything other than a large difference in treatment outcomes. Based on our observed cure rate among actively treated patients (14/24 [58%] RVS-MRSA patients versus 25/31 [80.6%] VS-MRSA patients), sample size calculations suggest that to detect a ≥20% difference with 80% power and ≤0.05% significance (two sided), assuming a comparable rate of RVS-MRSA isolates (approximately 50% of MRSA isolates), a study would need to recruit 196 to 220 patients with MRSA—roughly twice the size of our study (7). Any reduction in the overall rate of RVS-MRSA isolates among MRSA isolates would require an even larger recruitment. Thus, our treatment outcome data are not definitive but set the parameters that allow appropriate planning for any future studies to address this specific issue. Given the large number of PAP analyses that would be required for such a study, careful planning and funding would be required.
The high rate of RVS-MRSA isolates (58/117, 50%) among our MRSA isolates instigated questions of either the accuracy of our PAP results or the study definition for hVISA and VISA (22). However, repeat PAP testing of all 117 isolates demonstrated highly reproducible results. Notably, there was excellent correlation between the PAP-AUC ratio of ≥0.9 and the results of the macro method Etest. This latter test may be a simple, more-practical diagnostic screening test for many routine microbiology laboratories when hVISA/VISA is suspected clinically, while the PAP test may be utilized for confirmation of any positive macro method Etest.
In this study, the results from routine agar dilution, microbroth dilution, and our preliminary RVS-MRSA detection method demonstrated poor sensitivity and specificity for detecting RVS-MRSA. In particular, given the recent recommendation by CLSI that VISA may be defined as any S. aureus isolate with an MIC to vancomycin of ≥4 mg/liter, we found it notable that only two RVS-MRSA isolates had an MIC of 4 mg/liter, with most (56/58) having an MIC of 2 mg/liter.
Our assessment of alternative antistaphylococcal agents suggests that linezolid and daptomycin have reliable (100%) in vitro activity against RVS-MRSA, as determined by Etest. Of note, we did not detect reduced daptomycin susceptibility in our RVS-MRSA isolates, which has recently been described in other VISA strains (6), although more-detailed testing may be required to detect subtle daptomycin heteroresistance.
This study has some limitations. First, perhaps not surprisingly given the initial study design, a higher proportion (but not a higher total number) of patients with RVS-MRSA were recruited during period A of the study than period B. Second, our study population did not include many patients with deep-seated infections (17/117, 14.5%) (Table 2), which appeared to be so prominent in previous reports of RVS-MRSA infections (3, 9, 16). In these published cases, however, the diagnosis of RVS-MRSA was first suggested by clinical treatment failure rather than a priori laboratory evidence of relative vancomycin resistance. Third, the fact that 50% of our MRSA isolates were RVS-MRSA was surprising, as our previous study of MRSA bacteremia suggested that approximately 10% of the MRSA blood culture isolates in our institution were due to RVS-MRSA (3). However, an assessment of only bacteremic patients in this study demonstrates that the rate of RVS-MRSA isolates was 3/23 (13%), a rate remarkably similar to our previous findings. It was only when we assessed MRSA isolates from all sources that the higher rate of RVS-MRSA isolates was noted. To our knowledge, there are no other studies that have assessed for RVS-MRSA in such a large number of prospectively collected clinical specimens; thus, our results may simply represent the day-to-day unrecognized ecology of MRSA in our institution. Lastly, the diversity of treatment regimens means that there may be unidentified confounders in our analysis of vancomycin treatment outcomes.
The results of our study suggest that reduced susceptibility to vancomycin among MRSA isolates may be more common than previously suspected and that the impact of this resistance phenotype only becomes important clinically when the infection occurs in a site where drug (especially vancomycin) penetration is limited, such as in cardiac vegetations, bone, or around prosthetic devices (20). Given these data, we believe steps to diagnose RVS-MRSA should be focused on patients with MRSA infections who demonstrate clinical evidence of vancomycin treatment failure, since reduced susceptibility to vancomycin per se currently appears to have limited relevance unless it is in the right clinical context. For this reason, a larger study will be needed to identify more-subtle associations, if present, among patients with prosthetic devices or who are at high risk of deep-seated infections.
Supplementary Material
Acknowledgments
Some linezolid Etest strips were provided gratis by Pfizer.
Footnotes
Published ahead of print on 8 June 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.American Thoracic Society and Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 171:388-416. [DOI] [PubMed] [Google Scholar]
- 2.Ballard, S. A., K. K. Pertile, M. Lim, P. D. Johnson, and M. L. Grayson. 2005. Molecular characterization of vanB elements in naturally occurring gut anaerobes. Antimicrob. Agents Chemother. 49:1688-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448-451. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2007. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Coombs, G. W., G. R. Nimmo, J. M. Bell, F. Huygens, F. G. O'Brien, M. J. Malkowski, J. C. Pearson, A. J. Stephens, P. M. Giffard, and the Australian Group for Antimicrobial Resistance. 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui, L., E. Tominaga, H. M. Neoh, and K. Hiramatsu. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donner, A. 1984. Approaches to sample size estimation in the design of clinical trials—a review. Stat. Med. 3:199-214. [DOI] [PubMed] [Google Scholar]
- 8.Fridkin, S. K. 2001. Vancomycin-intermediate and -resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin. Infect. Dis. 32:108-115. [DOI] [PubMed] [Google Scholar]
- 9.Fridkin, S. K., J. Hageman, L. K. McDougal, J. Mohammed, W. R. Jarvis, T. M. Perl, and F. C. Tenover. 2003. Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin. Infect. Dis. 36:429-439. [DOI] [PubMed] [Google Scholar]
- 10.Gabay, C., and I. Kushner. 1999. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340:448-454. [DOI] [PubMed] [Google Scholar]
- 11.Goering, R. 2004. Pulsed-field gel electrophoresis, p. 185-196. In D. Persing, F. Tenover, J. Versalovic, Y. Tang, and E. Unger (ed.), Molecular microbiology diagnostic principles and practice. ASM Press, Washington, DC.
- 12.Grayson, M. L., G. W. Gibbons, G. M. Habershaw, D. V. Freeman, F. B. Pomposelli, B. I. Rosenblum, E. Levin, and A. W. Karchmer. 1994. Use of ampicillin/sulbactam versus imipenem/cilastatin in the treatment of limb-threatening foot infections in diabetic patients. Clin. Infect. Dis. 18:683-693. [DOI] [PubMed] [Google Scholar]
- 13.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 14.Horan, T., and R. Gaynes. 2004. Surveillance of nosocomial infections, p. 1659-1702. In G. Mayall (ed.), Hospital epidemiology and infection control. Lippincott Williams and Wilkins, Philadelphia, PA.
- 15.Horne, K. C., M. Graham, P. B. Ward, B. P. Howden, S. Xie, P. D. R. Johnson, B. Mayall, and M. L. Grayson. 2006. Vancomycin-intermediate Staphylococcus aureus (hVISA/VISA) and vancomycin-susceptible methicillin-resistant S. aureus (VSSA) have similar rates of clinical disease and overall outcomes when assessed prospectively, abstr. K-785, p. 326. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA.
- 16.Howden, B. P., P. B. Ward, P. G. Charles, T. M. Korman, A. Fuller, P. du Cros, E. A. Grabsch, S. A. Roberts, J. Robson, K. Read, N. Bak, J. Hurley, P. D. Johnson, A. J. Morris, B. C. Mayall, and M. L. Grayson. 2004. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin. Infect. Dis. 38:521-528. [DOI] [PubMed] [Google Scholar]
- 17.Howden, B. P., P. B. Ward, S. Xie, J. Wang, P. D. R. Johnson, P. G. P. Charles, and M. L. Grayson. 2004. Identification of a new agar dilution screening method for the accurate detection of heterogenous-vancomycin intermediate Staphylococcus aureus (hVISA), abstr. D-59, p. 140. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., Washington, DC.
- 18.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 19.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens, D. L. 2006. The role of vancomycin in the treatment paradigm. Clin. Infect. Dis. 42(Suppl. 1):S51-S57. [DOI] [PubMed] [Google Scholar]
- 21.Walsh, T. R., A. Bolmstrom, A. Qwarnstrom, P. Ho, M. Wootton, R. A. Howe, A. P. MacGowan, and D. Diekema. 2001. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J. Clin. Microbiol. 39:2439-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.