Abstract
Treatment of acute malaria caused by Plasmodium falciparum may include long-half-life drugs, such as the antifolate combination sulfadoxine-pyrimethamine (SP), to provide posttreatment chemoprophylaxis against parasite recrudescence or delayed emergence from the liver. An unusual case of P. falciparum recrudescence in a returned British traveler who received such a regimen, as well as a series of 44 parasite isolates from the same hospital, was analyzed by PCR and direct DNA sequencing for the presence of markers of parasite resistance to chloroquine and antifolates. The index patient harbored a mixture of wild-type and resistant pfdhfr and pfdhps alleles upon initial presentation. During his second malaria episode, he harbored only resistant parasites, with the haplotypes IRNI (codons 51, 59, 108, and 164) and SGEAA (codons 436, 437, 540, 581, and 613) at these two loci, respectively. Analysis of isolates from 44 other patients showed that the pfdhfr haplotype IRNI was common (found in 81% of cases). The SGEAA haplotype of pfdhps was uncommon (found only in eight cases of East African origin [17%]). A previously undescribed mutation, I431V, was observed for seven cases of Nigerian origin, occurring as one of two haplotypes, VAGKGS or VAGKAA. The presence of this mutation was also confirmed in isolates of Nigerian origin from the United Kingdom Malaria Reference Laboratory. The presence of the pfdhps haplotype SGEAA in P. falciparum parasites of East African origin appears to compromise the efficacy of treatment regimens that include SP as a means to prevent recrudescence. Parasites with novel pfdhps haplotypes are circulating in West Africa. The response of these parasites to chemotherapy needs to be evaluated.
The United Kingdom health system treats 1,000 to 2,000 cases of imported malaria each year, more than 70% of which are caused by Plasmodium falciparum (12). Infections from all over the globe are represented among these cases, but the majority are from sub-Saharan Africa. Parasite resistance to antimalarial drugs has an impact both on the effectiveness of chemoprophylaxis used by travelers (13) and on the efficacy of treatment regimens such as atovaquone-proguanil (AP), widely used to treat cases of malaria in hospitals in the United Kingdom (14).
Quinine remains an effective regimen for treating P. falciparum malaria and is still used in the United Kingdom for the treatment of both uncomplicated and severe malaria (9). At the Hospital for Tropical Diseases (HTD), patients with P. falciparum infections are treated with quinine until parasites are undetectable in peripheral blood; patients are then given a full dose of the fixed-combination antifolate sulfadoxine-pyrimethamine (SP). The long half-lives of both active components of SP in serum provide prophylaxis against the subsequent recrudescence of parasites surviving quinine therapy, or any parasites newly emerging from hepatic schizonts after the cessation of primary therapy (10, 15). This policy appears to have provided efficacious treatment for malaria patients, but there is no active follow-up of malaria patients at present.
The efficacy of quinine does not appear to have been significantly diminished over time by the evolution of parasite resistance, although studies of in vitro susceptibility of malaria parasites show some variability in sensitivity to quinine among South American isolates (6). In contrast, SP treatment failure due to high-level parasite resistance is widespread in Asia and common in East Africa (3), although SP retains good efficacy in West Africa (4, 17). Failure of SP therapy is associated with the accumulation of point mutations in two parasite genes, pfdhfr and pfdhps, encoding the folate biosynthesis pathway enzymes dihydrofolate reductase and dihydropteroate synthetase (DHPS), respectively. The continued efficacy of SP in West Africa is probably due to the absence or rarity of some of these mutations, particularly those at codon 164 of pfdhfr (widespread in Asia) and at codon 540 of pfdhps (widely reported from East and Southern Africa). Therefore, ongoing surveillance of the geographic distribution of mutations in pfdhfr and pfdhps is of great importance for informing treatment policy both in countries where malaria is endemic and in those where it is not, such as the United Kingdom, where a large number of imported P. falciparum malaria cases are treated each year.
In March 2007, a P. falciparum malaria patient treated at the HTD with quinine plus SP returned with recrudescent malaria and was confirmed as a case of therapeutic failure. It was hypothesized that quinine, a drug rapidly cleared from host circulation, had failed to eradicate all the parasites and that subsequent administration of long-lasting SP had selected a subpopulation of parasites resistant to antifolates that survived and subsequently caused a recrudescent infection. We report here a parasitological evaluation of this particular case and a concurrent survey of molecular markers of drug resistance in parasites from 44 HTD patients presenting with confirmed P. falciparum malaria over 12 months prior to the presentation of this case and in a further 39 isolates from P. falciparum malaria cases referred from all over the United Kingdom to the Health Protection Agency (HPA) Malaria Reference Laboratory (MRL) at the London School of Hygiene and Tropical Medicine.
MATERIALS AND METHODS
Patients and samples.
Venous blood samples from microscopy-confirmed P. falciparum malaria cases presenting at or referred to the HTD in London, United Kingdom, and blood samples sent to the HPA MRL for confirmation of diagnoses were used for this study. Patient identifiers were removed from sample aliquots given to research staff. The only information from patient records retained in the research milieu was the country in which malaria infection was considered to have occurred, if known. This study was pursued under the remit of the MRL to conduct surveillance of imported malaria cases for the presence of markers of antimalarial drug resistance. The study was approved by the Research Ethics Committee of University College London Hospitals.
qPCR.
Parasite DNA was extracted from 200 μl of peripheral blood using the Qiagen QIAamp blood extraction kit and was eluted in a volume of approximately 100 μl. An in-house quantitative real-time PCR (qPCR) assay (11) was employed to quantify parasite density. Primers PfalVL (GCCGAAAGGCGTAGGTAATC) and PfalVR (GTACAAAGGGCAGGGACGTA) were designed to amplify a 134-bp fragment of the 18S ribosomal subunit gene located on chromosome 5 of P. falciparum (sequence MAL5_18S, sourced from PlasmoDB [http://plasmodb.org/plasmo/; last accessed 21 April 2009]). An identical sequence is found on chromosome 7 (MAL7_18Sa). DNA (extracted as described below) was amplified in a 20-μl reaction volume, comprising 10 μl QuantiTect Sybr green mix (Qiagen, United Kingdom) and 2 μM each primer. Amplification was performed in a Rotorgene RG3000 thermocycler (Corbett, Sydney, Australia) with the following profile: 15 min at 95°C for activation of reagents, followed by 40 cycles of 94°C (30 s), 55°C (40 s), and 68°C (50 s). Products were stabilized at 68°C for 5 min before the following melt analysis was performed: 55°C for 45 s, followed by a temperature increase of 0.5°C per step with 5 s at each step of the gradient. Fluorescent data were collected on the 6-carboxyfluorescein/Sybr channel of the RG3000 thermocycler and were quantified against a serial dilution of the recently described WHO international standard for P. falciparum DNA (7).
Determination of parasite markers of drug resistance.
Resistance to the antimalarial chloroquine (CQ) is associated with the presence of mutations in the pfcrt gene, encoding the P. falciparum CQ resistance transporter (2). Alleles associated with resistance to CQ were identified by real-time double-labeled hydrolysis probe PCR in a Corbett RG3000 thermocycler, as described previously (13).
Allele sequences of pfdhfr were identified using a PCR amplification and direct-sequencing protocol designed specifically for this study. Briefly, 5 μl of DNA was subjected to PCR with forward primer FBR01 (AAGCAAAAATGAGGGGAAAAA) and reverse primer FBR02 (ACATCGCTAACAGAAATAATTTGA). The DNA was amplified under standard conditions with the following cycling program: 93°C for 5 min; 40 cycles of 93°C for 30 s, 56°C for 30 s, and 68°C for 75 s; and 68°C for 5 min.
Amplification products were purified using Qiagen PCR Elute minicolumns, checked by agarose electrophoresis, and then sequenced in both directions using the ABI BigDye (version 3.1) sequencing kit according to the manufacturer's instructions, but all volumes were scaled down eightfold. Sequencing products were purified by alcohol precipitation and fractionated on an ABI 3770 sequencer. Chromatogram data were analyzed and manually curated using Chromas software. Good-quality DNA sequences from both strands were required in order for any sequence to contribute to analysis.
Allele sequences of pfdhps were amplified using the nested-PCR procedure of Pearce et al. (8) as described by these authors. PCR products were sequenced as for pfdhfr by using the following sequencing primers: AACCTAAACGTGCTGTTCAA (forward), AATTATTAAAAAAAAAAAAC (forward), AATTGTGTGATTTGTCCACAA (reverse), and TTTTAATAATTTTATAATTTC (reverse).
Estimating the multiplicity of infection.
The multiplicities of infection of different isolates from the index case were estimated using two polymorphic sequence tags with variable-length trinucleotide repeats, polyα on P. falciparum chromosome 4 and PfPK2 on chromosome 12, as previously described (1). Neither of these markers is closely linked to genetic loci associated with antifolate resistance; pfdhfr is located on chromosome 4 (220 kb from polyα), and pfdhps is on chromosome 8.
Statistical analysis was performed in Stata, version 8.0 (StataCorp, College Station, TX). Categorical variables were tested for associations by using the two-sided Fisher exact test.
RESULTS
Case history.
The primary case in this study had the following clinical and travel history.
A 37-year-old Caucasian male presented to the HTD as an outpatient on 21 February 2007 with a 3-day history of myalgia and a 1-day history of lethargy, headache, and fever. He had been traveling in Mozambique for 1 month, staying at a backpackers' hostel in a coastal resort, and had not used any malaria prophylaxis. He returned to the United Kingdom on 16 February 2007, and his first symptoms began 2 days later.
On examination, the patient was not acutely unwell; he was conscious and alert, with no signs of meningism. His temperature was elevated (39.9°C); there was no jaundice or lymphadenopathy; and his chest was clear. Sinus tachycardia was observed, but cardiac indicators were otherwise normal. There was no evidence of hepatosplenomegaly or rash. The hemoglobin level was normal, at 16.0 g/dl; the leukocyte count was 4.9 × 109 cells per liter; and the platelet count was 141 × 109/liter. Urea, electrolytes, and liver function tests were normal. Microscopy of a Giemsa-stained blood film revealed the presence of P. falciparum ring-stage trophozoites in 2.1% of red blood cells.
The patient was admitted to hospital and treated with intravenous quinine (one dose) followed by oral quinine (eight doses). Thick and thin blood films were examined daily for malaria parasites; asexual stages of P. falciparum were undetectable by microscopy after 3 days of treatment. The patient was given a single dose of SP and discharged the following day (26 February).
On 13 March 2007, 15 days after discharge, the patient returned to the HTD with fever, anorexia, and myalgia. He presented with a temperature of 38.7°C. A malaria blood film revealed late trophozoites of P. falciparum infecting 0.6% of erythrocytes. The patient was readmitted to the HTD as an inpatient and treated with AP daily for 3 days. He made a good recovery. On follow-up 2 months later, the patient reported being well, with no recurrence of symptoms.
Parasitological history.
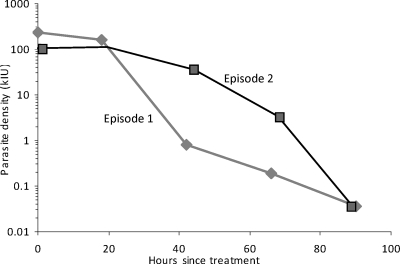
A total of 10 peripheral blood samples were taken from the patient for primary diagnosis and for monitoring of parasite clearance under treatment: 5 samples from the quinine- and SP-treated episode and 5 samples from the AP-treated episode. To test whether the rates of parasite clearance differed between the quinine- and AP-treated episodes, we used qPCR to estimate parasite clearance rates for the two episodes. The relative parasite density (measured against a serial dilution of P. falciparum-positive control DNA from a previous patient) was plotted over time for both episodes, with time zero representing the times of the first blood draws on 21 February and 13 March, respectively. The data are plotted against the newly established WHO international standard for P. falciparum DNA (7) in Fig. 1. Ninety percent parasite clearance (16) was estimated to have occurred by about 25 h under quinine treatment and 59 h under AP treatment. However, our sequential parasitological samples were taken 18 to 24 h apart, and the precise time of sampling was not always recorded. Therefore, although our data suggest that parasitological clearance was significantly faster under quinine than under AP therapy, more closely spaced and more accurately timed sampling would be required to verify this observation.
FIG. 1.
Quantitative PCR analysis of two sequential P. falciparum malaria episodes for the same patient. Episode 1 was treated with quinine; episode 2, with AP. Each point represents the mean estimate of parasite density from two replicates in a single experiment. The y axis shows parasite density, expressed in kilo-international units of P. falciparum DNA, on a logarithmic scale. The x axis shows the approximate time (in hours) after the first diagnostic blood sample was taken. Where the exact time was not recorded, an interval of 24 h between consecutive samples is assumed.
The apparent difference in clearance times observed between these two episodes led us to hypothesize that the parasites present in the second malaria episode in this individual were a drug-selected subset of those present during the first malaria episode and presumably represented a resistance-associated genotype or genotypes with enhanced survival under quinine and/or SP treatment. We therefore analyzed and compared parasite material collected during the two clinical episodes for a variety of molecular markers associated with drug resistance and for two microsatellites. This analysis is presented in Table 1; it clearly indicates that only parasites with resistant haplotypes of pfdhfr and pfdhps persisted after quinine and SP treatment and that these resistant parasites initiated the second malaria episode in this patient. This finding is supported by microsatellite typing at the polyα and PfPK2 markers, which indicated that at the time of first presentation in February 2007, our primary case was an infection with a minimum of four distinct parasite clones, but that these were reduced to one clone following treatment during the second episode.
TABLE 1.
Evaluation of resistance-associated markers in two malaria episodes in the same patient
| Episode (treatment) | Day | Haplotypea
|
No. of distinct clones
|
|||
|---|---|---|---|---|---|---|
| pfcrt (aa 72-76) | pfdhfr (aa 51, 59, 108, 164) | pfdhps (aa 436, 437, 540, 581, 613) | Polyα | PfPK2 | ||
| 1 (quinine) | 0 | CVIET | IRNI/ICSI | SAKAA/SGEAA | 4 | 2 |
| 1 | CVIET | IRNI/ICSI | SAKAA/SGEAA | 4 | 1 | |
| 2 | CVIET | IRNI/ICSI | SAKAA/SGEAA | ND | ND | |
| 3 | CVIET | ND | SAKAA/SGEAA | ND | ND | |
| 4 | ND | ND | SAKAA/SGEAA | ND | ND | |
| 2 (AP) | 0 | CVIET | IRNI | SGEAA | 2 | 2 |
| 1 | CVIET | IRNI | SGEAA | 1 | 1 | |
| 2 | CVIET | IRNI | SGEAA | ND | ND | |
| 3 | CVIET | IRNI | SGEAA | ND | ND | |
| 4 | ND | ND | SGEAA | ND | ND | |
Haplotypes are compiled using the one-letter amino acid (aa) code. Those haplotypes known to be associated with resistance to CQ (pfcrt) or SP (pfdhfr, pfdhps) are shown in boldface. ND, not determined. Note that only pfdhps was sequenced from nested PCR and thus consistently produced amplicons from days 3 and 4 after treatment, when parasite densities were very low. Microsatellites were determined only for the first two isolates of each episode.
Having established a probable role of resistant haplotypes of pfdhfr and pfdhps in the recrudescence of parasites in the primary case, we sought to determine the prevalence of these haplotypes, and that of other relevant haplotypes at pfcrt, pfdhfr, and pfdhps, among P. falciparum malaria patients presenting to the HTD. DNA was extracted from isolates collected at the HTD throughout 2006, and pfcrt genotypes in 66 of these isolates were determined by real-time PCR. Both pfdhfr and pfdhps genotypes were determined by direct sequencing of PCR amplicons in a subset of 44 isolates. No episodes of treatment failure were recorded among the patients contributing isolates used in the genotyping study. The results for pfcrt and pfdhfr are presented in Table 2. These results show that for both these loci, wild-type haplotypes are in the minority among our sample of isolates from United Kingdom travelers who have visited African destinations, and that the pfdhfr triple mutant (IRNI; associated with failure of the pyrimethamine component of SP) occurred in >80% of the patients tested. Parasites with the IRNI haplotype were found in isolates from Cameroon, Ghana, Guinea, Kenya, Liberia, Malawi, Mozambique, Nigeria, Sierra Leone, and Uganda; therefore, this haplotype shows no evidence of restricted dispersal across continental Africa. Similarly, parasites with the CVIET haplotype of pfcrt had a variety of origins, including Cameroon, Côte d'Ivoire, Ghana, Kenya, Mozambique, Nigeria, Sierra Leone, and Uganda. Therefore, the treatment failure experienced by this patient is unlikely to be due to the presence of either the pfcrt genotype CVIET or the pfdhfr genotype IRNI, since both of these are very common in our patient sample, in which no other quinine-plus-SP treatment failures were reported.
TABLE 2.
pfcrt and pfdhfr genotypes among P. falciparum isolates from malaria patients presenting to the HTD in 2006
| Genotype and haplotype | Frequency (no. of isolates) | Prevalence (%) |
|---|---|---|
| pfcrt (n = 66), codons 72-76 | ||
| CVMNK (wild type) | 19 | 28.8 |
| CVIET | 43 | 65.2 |
| SVMNT | 1 | 1.5 |
| CVMNK/CVIET, mixed | 3 | 4.5 |
| pfdhfr (n = 44), codons 51, 59, 108, 164 | ||
| NCSI (wild-type) | 5 | 11.4 |
| ICNI | 1 | 2.3 |
| IRTI | 2 | 4.5 |
| IRNI | 32 | 72.7 |
| IRNI/ICNI, mixed | 2 | 4.5 |
| IRNI/NCSI, mixed | 2 | 4.5 |
The results of the pfdhps sequencing were more complex and are presented in Fig. 2. Where more than one genotype was evident in the sequencing data, the dominant genotype only is shown. Eight different haplotypes, across six polymorphic codons, were identified in our sample of 46 isolates. Unexpectedly, a novel mutation was identified among these: substitution of valine for isoleucine at codon 431 (I431V) in seven of our patients. The other polymorphisms observed, S436G, A437G, K540E, A581G, and A613S, have been reported previously. The ISGEAA haplotype, which was found in the recrudescent infection observed in our case of quinine-plus-SP treatment failure, is rare among our patient samples but has been commonly reported in East and Southern Africa by previous studies. It is therefore possible that parasite persistence in this patient was due to the presence of this pfdhps allele.
FIG. 2.
Alignment of deduced PfDHPS amino acid sequences from 46 P. falciparum isolates. The eight different PfDHPS haplotypes encoded by DNA sequences determined in this study from patients at the HTD in 2006 are shown, and the number of isolates with each haplotype is given on the left (#). Haplotypes are arranged in increasing order of amino acid substitutions, and substitutions relative to the wild-type sequence are shaded. Amino acids 445 to 530 were invariant and are omitted for clarity. Amino acids 1 to 424 were not sequenced, because there are no previous reports of amino acid substitutions in this region of the polypeptide. Where mixed infections were present, only the dominant haplotype is shown and contributes to the number of isolates given. The novel genotypes VAGKAA and VAGKGS are shown in boldface.
Having identified a novel nonsynonymous mutation at codon 431 of the pfdhps gene, we looked for associations between the carriage of this mutation and geographic origin, and for evidence of associations with the other amino acid substitutions observed. Table 3 lists the country of origin reported for each of the malaria infections, grouped by pfdhps haplotype observed. Valine was present at codon 431 of pfdhps in 6 of the 15 isolates of Nigerian origin (40%) and in 1 other isolate, of unknown origin. This new mutation was therefore strongly associated with Nigerian travel (P = 0.001). The diversity of pfdhps haplotypes found among our 33 West African isolates was remarkable, but none of these carried the K540E mutation found in our index case of treatment failure; this mutation occurred only in infections from the East African countries Uganda, Kenya, and Malawi in this sample. Including our case from Mozambique, our data suggest a clear East-West split in pfdhps genotypes, and indeed, among our 46 isolates tested here, the K540E mutation was strongly associated with East African origin (P < 0.001). Also of interest is the rare occurrence together of mutations at positions 581 and 613 in five Nigerian isolates. These all carried the novel I431V substitution and so were of the quintuple mutant haplotype VSGKGS, suggesting a possible linkage of the 431, 581, and 613 codons in some Nigerian parasite populations. Two isolates carrying the pfdhps haplotype VSGKAA were also found.
TABLE 3.
pfdhps haplotypes and countries of origin for 46 P. falciparum isolates from malaria patients presenting to the HTD, London, United Kingdom, in 2006
| Haplotypea | Frequency (no. of isolates) | Country of originb |
|---|---|---|
| ISAKAA | 6 | Nigeria, Sierra Leone, Ivory Coast, Liberia, Mozambique, Guinea |
| IAAKAA | 5 | Sierra Leone, Ivory Coast, Liberia, Mali (1 ND) |
| ISGKAA | 14 | Nigeria (7), Sierra Leone (2), Ivory Coast, Ghana, Cameroon (2 ND) |
| IAGKAA | 5 | Nigeria, Ghana (4) |
| ISGEAA | 8 | Uganda (5), Kenya, Malawi (1 ND) |
| IAGKAS | 1 | Cameroon |
| VAGKAA | 2 | Nigeria (2) |
| VAGKGS | 5 | Nigeria (4) (1 ND) |
Given as one-letter amino acid codes for codons 431, 436, 437, 540, 581, and 613. ISAKAA is taken as the wild type.
Inferred from the travel history taken by the clinical staff. If more than one isolate with a given haplotype originated from a given country, the number of isolates is given in parentheses after the country name. ND, country of origin not determined.
To confirm the presence of novel pfdhps alleles in West African parasites, we selected 39 P. falciparum isolates from Nigeria and closely neighboring countries (Ghana, Sierra Leone, Togo, Cameroon, Benin) from among isolates received by the United Kingdom HPA MRL during 2007. DNA was prepared, and pfdhps sequences were amplified and sequenced, using a protocol identical to that used in the HTD analysis, but by different team members using independent reagents. The results (Table 4) verify the occurrence of the VAGKGS and VAGKAA haplotypes in Nigeria. Interestingly, two isolates carrying the K540E substitution (ISGEAA) were identified in travelers returning from Ghana. Ten of the 13 Ghanaian isolates had the ISGKAA or IAGKAA haplotype.
TABLE 4.
pfdhps haplotypes and countries of origin for 39 P. falciparum isolates of West African origin from malaria patients presenting across the United Kingdom in 2007
| Haplotypea | Frequency (no. of isolates) | Country of originb |
|---|---|---|
| ISAKAA | 2 | Nigeria (2) |
| IAAKAA | 1 | Equatorial Guinea |
| ISGKAA | 14 | Nigeria (6), Ghana (4), Sierra Leone, Cameroon, Ivory Coast, Benin |
| IAGKAA | 11 | Nigeria, Ghana (6), Sierra Leone, Ivory Coast (2), Togo |
| ISGEAA | 2 | Ghana (2) |
| IAGKAS | 4 | Nigeria (3), Ivory Coast |
| VAGKAA | 1 | Nigeria (1) |
| VAGKGS | 2 | Nigeria (2) |
| ISAKAS | 1 | Ghana |
| ISGKAS | 1 | Nigeria |
Given as one-letter amino acid codes for codons 431, 436, 437, 540, 581, and 613. ISAKAA is taken as the wild type. Novel haplotypes first described in this report are shown in boldface. Haplotypes not found among the HTD isolates presented in Table 3 are underlined.
Inferred from the travel history taken by the clinical staff. If more than one isolate with a given haplotype originated from a given country, the number of isolates is given in parentheses after the country name.
DISCUSSION
We report here a case of uncomplicated P. falciparum malaria contracted in Mozambique and imported to the United Kingdom in 2007. Recrudescence of P. falciparum with resumption of symptoms after treatment with quinine and SP was associated with the persistence of parasites carrying the antifolate resistance-associated haplotypes IRNI and SGEAA at the pfdhfr and pfdhps loci, respectively. Parasites with wild-type alleles at these loci were also present prior to treatment but did not persist. A survey of these loci among 46 parasite isolates from patients attending the same hospital in the 12 months prior to this case found IRNI, but not SGEAA, to be common among them. It is therefore likely that the SGEAA form of the DHPS enzyme, together with the IRN form of dihydrofolate reductase, contributed to the observed treatment failure, but we cannot rule out a contribution from other polymorphic parasite proteins that were not examined. We observed markedly different clearance times between the two episodes of parasitemia in this patient, with the primary episode (treated with quinine) clearing much faster than the second (treated with AP). While this comparison has the rare benefit of being well controlled for host factors, since the infections occurred in the same patient, our genetic analysis demonstrates differences between the two parasite populations (Table 1), so we cannot attribute the difference in clearance times entirely to the relative efficacy of the regimens employed. Nevertheless, this comparison does illustrate for the first time the utility in clinical malaria studies of qPCR coupled with the absolute quantitation now possible using the recently established international standard for P. falciparum DNA (7).
Our findings are consistent with previous work showing that, in East Africa, the pfdhps K540E mutation is closely linked to SP treatment failure, specifically by disrupting the antiparasite effects of the sulfadoxine component of the drug combination (3). This mutation is absent or rare in West Africa, where SP efficacy remains good (5). The HTD malaria patient base largely comprises infections picked up in West Africa, and by combining the results from our survey of pfdhps haplotypes among these patients with those from the 39 additional West African isolates from the MRL, the odds ratio of a malaria case imported to the United Kingdom from West Africa, rather than from East or Southern Africa, carrying the K540E mutation is 0.0041 (95% confidence interval, 0.0001 to 0.065; P < 0.0001). This is consistent with observations in the field across East, West, and Southern Africa, which have recently been summarized in highly functional graphic maps (http://www.lshtm.ac.uk/pmbu/drm/map-540.html).
The observation among Nigerian isolates of a previously undescribed amino acid substitution, valine for isoleucine at codon 431, was unexpected, and its significance remains unclear. Its association with the relatively uncommon substitutions A581G and A613S in 7 of the 10 isolates found is consistent with a recent origin of the codon 431 substitution in a parasite carrying the AGKGS haplotype. Alternatively, functional constraints on the enzyme itself may limit the ability of this mutation to combine with some other haplotypes. The role, if any, of this mutation in parasite resistance to SP needs to be determined, and further work in Nigeria and neighboring countries to examine the sensitivity of this parasite genotype to a variety of antifolate drugs is required. One intriguing possibility is that widespread use of the antibiotic combination trimethoprim-sulfamethoxazole as prophylaxis against Pneumocystis jirovecii pneumonia for people infected with human immunodeficiency virus in Africa, particularly pregnant women, may be placing additional sulfonamide selective pressure for new alleles at the pfdhps locus that are not directly related to antimalarial use.
In conclusion, the treatment of imported cases of uncomplicated P. falciparum malaria with quinine and SP is threatened by the large proportion of such infections harboring mutations in pfdhfr and pfdhps. Our data suggest that the K540E mutation in pfdhps may be particularly important, although currently this mutation is common only among cases imported from East Africa. Surveillance for further spread of this genotype into West Africa, as indicated by its occurrence in two Ghanaian isolates, is required. The antifolate sensitivity profiles of the novel pfdhps haplotypes identified here remain to be determined.
Acknowledgments
This work was supported by the United Kingdom Health Protection Agency. S.D.P. is supported by the Foundation for Innovative New Diagnostics, Geneva, Switzerland.
We thank Cally Roper for useful discussions and for suggesting the microsatellite analysis.
All the authors declare no conflict of interest.
Footnotes
Published ahead of print on 11 May 2009.
REFERENCES
- 1.Anderson, T. J., X. Z. Su, M. Bockarie, M. Lagog, and K. P. Day. 1999. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 119:113-125. [DOI] [PubMed] [Google Scholar]
- 2.Djimdé, A., O. K. Doumbo, R. W. Steketee, and C. V. Plowe. 2001. Application of a molecular marker for surveillance of chloroquine-resistant falciparum malaria. Lancet 358:890-891. [DOI] [PubMed] [Google Scholar]
- 3.Dorsey, G., C. Dokomajilar, M. Kiggundu, S. G. Staedke, M. R. Kamya, and P. J. Rosenthal. 2004. Principal role of dihydropteroate synthase mutations in mediating resistance to sulfadoxine-pyrimethamine in single-drug and combination therapy of uncomplicated malaria in Uganda. Am. J. Trop. Med. Hyg. 71:758-763. [PubMed] [Google Scholar]
- 4.Dunyo, S., R. Ord, R. Hallett, M. Jawara, G. Walraven, E. Mesa, R. Coleman, M. Sowe, N. Alexander, G. A. Targett, M. Pinder, and C. J. Sutherland. 2006. Randomised trial of chloroquine/sulphadoxine-pyrimethamine in Gambian children with malaria: impact against multidrug-resistant P. falciparum. PLoS Clin. Trials 1:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hallett, R. L., S. Dunyo, R. Ord, M. Jawara, M. Pinder, A. Randall, A. Alloueche, G. Walraven, G. A. Targett, N. Alexander, and C. J. Sutherland. 2006. Chloroquine/sulphadoxine-pyrimethamine for Gambian children with malaria: transmission to mosquitoes of multidrug-resistant Plasmodium falciparum. PLoS Clin. Trials 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legrand, E., B. Volney, J. B. Meynard, O. Mercereau-Puijalon, and P. Esterre. 2008. In vitro monitoring of Plasmodium falciparum drug resistance in French Guiana: a synopsis of continuous assessment from 1994 to 2005. Antimicrob. Agents Chemother. 52:288-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Padley, D. J., A. B. Heath, C. Sutherland, P. L. Chiodini, and S. A. Baylis. 2008. Establishment of the 1st World Health Organization International Standard for Plasmodium falciparum DNA for nucleic acid amplification technique (NAT)-based assays. Malar. J. 7:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce, R. J., C. Drakeley, D. Chandramohan, F. Mosha, and C. Roper. 2003. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of northern Tanzania. Antimicrob. Agents Chemother. 47:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodger, A., G. S. Cooke, R. Ord, C. J. Sutherland, and G. Pasvol. 2008. Synchronised cluster of falciparum malaria cases in an airport departure lounge. Emerg. Infect. Dis. 14:1284-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schellenberg, D., C. Menendez, J. J. Aponte, et al. 2005. Intermittent preventive antimalarial treatment for Tanzanian infants: follow-up to age 2 years of a randomised, placebo-controlled trial. Lancet 365:1481-1483. [DOI] [PubMed] [Google Scholar]
- 11.Sharp, S. 2006. Ph.D. thesis. University of London, London, United Kingdom.
- 12.Smith, A. D., D. J. Bradley, V. Smith, M. Blaze, R. H. Behrens, P. L. Chiodini, and C. J. Whitty. 2008. Imported malaria and high risk groups: observational study using UK surveillance data 1987-2006. BMJ 337:a120. doi: 10.1136/bmj.a120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutherland, C. J., T. Haustein, N. Gadalla, M. Armstrong, J. F. Doherty, and P. L. Chiodini. 2007. Chloroquine-resistant Plasmodium falciparum infections among UK travellers returning with malaria after chloroquine prophylaxis. J. Antimicrob. Chemother. 59:1197-1199. [DOI] [PubMed] [Google Scholar]
- 14.Sutherland, C. J., M. Laundy, N. Price, M. Burke, Q. L. Fivelman, G. Pasvol, J. L. Klein, and P. L. Chiodini. 2008. Mutations in the Plasmodium falciparum cytochrome b gene are associated with delayed parasite recrudescence in malaria patients treated with atovaquone-proguanil. Malar. J. 7:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutherland, C. J., C. J. Drakeley, and D. Schellenberg. 2007. How is childhood development of immunity to Plasmodium falciparum enhanced by certain antimalarial interventions? Malar. J. 6:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wootton, D. G., H. Opara, G. A. Biagini, M. K. Kanjala, S. Duparc, et al. 2008. Open-label comparative clinical study of chlorproguanil-dapsone fixed dose combination (Lapdap) alone or with three different doses of artesunate for uncomplicated Plasmodium falciparum malaria. PLoS ONE 3:e1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization. 2005. Susceptibility of Plasmodium falciparum to antimalarial drugs: report on global monitoring 1996-2004, p. 35-39. World Health Organization, Geneva, Switzerland.