Abstract
MurF catalyzes the last cytoplasmic step of bacterial cell wall synthesis and is essential for bacterial survival. Our previous studies used a pharmacophore model of a MurF inhibitor to identify additional inhibitors with improved properties. We now present the characterization of two such inhibitors, the diarylquinolines DQ1 and DQ2. DQ1 inhibited Escherichia coli MurF (50% inhibitory concentration, 24 μM) and had modest activity (MICs, 8 to 16 μg/ml) against lipopolysaccharide (LPS)-defective E. coli and wild-type E. coli rendered permeable with polymyxin B nonapeptide. DQ2 additionally displayed activity against gram-positive bacteria (MICs, 8 to 16 μg/ml), including methicillin (meticillin)-susceptible and -resistant Staphylococcus aureus isolates and vancomycin-susceptible and -resistant Enterococcus faecalis and Enterococcus faecium isolates. Treatment of LPS-defective E. coli cells with ≥2× MIC of DQ1 resulted in a 75-fold-greater accumulation of the MurF substrate compared to the control, a 70% decline in the amount of the MurF product, and eventual cell lysis, consistent with the inhibition of MurF within bacteria. DQ2 treatment of S. aureus resulted in similar effects on the MurF substrate and product quantities. At lower levels of DQ1 (≤1× MIC), the level of accumulation of the substrate was less pronounced (15-fold greater compared to the amount for the control). However, a 50% increase in the amount of the MurF product compared to the control was reproducibly observed, consistent with the possible upregulation of muropeptide biosynthesis upon partial inhibition of this pathway. The overexpression of cloned MurF appeared to partly alleviate the DQ1-mediated inhibition of muropeptide synthesis. The identification of MurF inhibitors such as DQ1 and DQ2 that disrupt cell wall biosynthesis suggests that MurF remains a viable target for an antibacterial agent.
Cell wall biosynthesis and the cell wall structure have long been considered useful targets for antibacterial agents, as demonstrated by antibiotics such as the β-lactams and glycopeptides (14, 29). However, of the series of steps catalyzed by the enzymes MurA through MurF that produce UDP-MurNAc-pentapeptide, a useful antibiotic, fosfomycin, has been generated only against the MurA target (6, 18), despite extensive screening efforts against all of the enzymes in this pathway (for reviews, see references 8, 11, 14, 20, 29, and 31).
MurF catalyzes the last cytoplasmic step of bacterial cell wall biosynthesis, generating UDP-MurNAc-pentapeptide from UDP-MurNAc-tripeptide and d-Ala-d-Ala (37). Previously identified inhibitors of MurF include a nonhydrolyzable ATP analog (1), phosphinate transition state analogs (25), sulfonamides (15, 22), thiazolylaminopyrimidines (4), and 8-hydroxyquinolines (5). These compounds inhibited the purified MurF enzyme but lacked antibacterial activity, presumably due to poor penetration into cells. A pharmacophore model based on the 8-hydroxyquinoline series was used to search for compounds with antibacterial activity, and this process identified several classes of compounds, including a 4-phenylpiperidine derivative (5). This inhibitor had the distinction of being the first inhibitor of the MurF enzyme which appeared to inhibit MurF within Escherichia coli cells.
Observations of conditional lethal MurF mutants of E. coli (24) and Staphylococcus aureus (33, 34) are useful for predicting the effects of a MurF inhibitor on bacteria. In E. coli, a temperature-sensitive mutant with a mutated MurF enzyme that possessed <1% of the relative activity of the wild-type enzyme exhibited (i) cell lysis at the nonpermissive temperature, (ii) the accumulation of UDP-MurNAc-tripeptide (the MurF substrate), and (iii) a decrease in the level of the UDP-MurNAc-pentapeptide product (24). In S. aureus, similar effects on the abundance of muropeptide were observed (33). These and other parameters were examined for two new MurF inhibitors, the diarylquinolines DQ1 and DQ2.
MATERIALS AND METHODS
MurF enzymatic assay.
The 50% inhibitory concentrations (IC50s) for E. coli MurF were determined as described previously (5).
Microbiology studies.
All bacterial strains were from the strain collection of Johnson & Johnson Pharmaceutical Research & Development, L.L.C. MICs were determined by CLSI broth microdilution assays (9). The checkerboard methodology was used for determination of the MICs of compound DQ2 in combination with vancomycin (10). For growth curve generation, CFU quantitation, and muropeptide analysis, 125-ml cultures of E. coli OC2530 or S. aureus ATCC 29213 were grown to an A600 of 0.3. The compounds (500× stock solutions of cycloserine in distilled H2O, DQ1 or DQ2 in dimethyl sulfoxide [DMSO]) were added at the indicated multiple of the MIC, and incubation was continued; DMSO was added to the control culture. After 30 min, the muropeptides were extracted and quantified as described previously (5). Upon addition of compound, aliquots (100 μl each) of the cultures were transferred to replicate wells of a Bioscreen C microplate reader (Growth Curves USA, Piscataway, NJ) to generate growth curves on the basis of the changes in the optical density (OD). After 30 min and 3 h, aliquots of the cultures were removed from microplate wells for CFU quantitation (5).
Microscopy.
E. coli OC2530 was grown as described above for growth curve generation, and either DMSO or 0.5× MIC DQ1 was added. For light microscopy, a 20-μl aliquot of cells was placed onto a glass slide and fixed with SHUR/Mount (Triangle Biomedical Sciences, Inc., Durham, NC), followed by placement of a coverslip. The bacteria were observed at a ×100/×1.25 oil immersion magnification on a Nikon Eclipse E800 microscope (Melville, NY). Representative pictures were taken for both sets of cultures. For transmission electron microscopy (TEM), aliquots (0.5 ml) of the cultures were treated with glutaraldehyde at a final concentration of 1% 5 h after addition of DMSO or DQ1. The cells were incubated at room temperature for 1 h and centrifuged (10,000 × g), and the supernatants were discarded. The cell pellets were suspended in 0.5 ml phosphate-buffered saline; the cells were then negatively stained with 1% ammonium molybdate and photographed under the transmission electron microscope at a magnification of ×15,000 (16).
Overexpression of MurF in E. coli.
The MurF-coding region was excised with NdeI-BamHI from a construct similar to that described previously (4), except that it lacked a histidine tag, and was ligated into the XbaI-BamHI site of the expression vector pASK-IBA3C (IBA, St. Louis, MO) (32) under the control of the tetA promoter by the use of double-stranded DNA oligomers (5′-CTAGATAACGAGGGCAAAA-3′ and 3′-TATTGCTCCCGTTTTAT-5′) to re-create the vector sequences upstream of the translational start codon. The resultant plasmid, pMurF, was electroporated into E. coli OC2530; 3 μg/ml chloramphenicol was sufficient to prevent the growth of nontransformed strain OC2530 and was used throughout the study. The expression of cloned MurF mRNA upon induction with anhydrotetracycline (AHT; IBA) was confirmed by reverse transcription-PCR with a LightCycler instrument (Roche, Indianapolis, IN) and primers MurFor (5′-CAACACGCTTTATACGGCAGGCAA-3′) and MurRev (5′-CTGATGGTTCGCGCCAAGTTCAAT-3′), which specifically detected mRNA from the cloned (but not the chromosomal) gene. The expression of the cloned MurF protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Invitrogen, Carlsbad, CA) of cell lysates prepared with BugBuster lysis solution (EMD Biosciences). For the experiment described in Table 3, E. coli OC2530 cells harboring pASK (empty vector) or pMurF were grown with aeration in 125 ml Mueller-Hinton broth to an A600 of 0.3, followed by addition of AHT (100 ng/ml) and continued incubation. After 1 h, the compounds were added as described above, and the cells were processed for muropeptide quantitation.
TABLE 3.
Relative amounts of UDP-MurNAc-tripeptide and -pentapeptide in E. coli OC2530 overexpressing MurFa
| Strain and compoundb | Amtc
|
UDP-MurNAc-tripeptide/UDP-MurNAc-pentapeptide ratio | |
|---|---|---|---|
| UDP-MurNAc-tripeptide | UDP-MurNAc-pentapeptide | ||
| E. coli OC2530/pASK | |||
| None (control) | 63 ± 4 | 880 ± 17 | 0.07 |
| DQ1 | 1,196 ± 71 | 213 ± 18 | 5.6 |
| Cycloserine | 2,255 ± 750 | 146 ± 3 | 15 |
| E. coli OC2530/pMurF | |||
| None (control) | 220 ± 65 | 2,674 ± 696 | 0.08 |
| DQ1 | 933 ± 115 | 351 ± 0 | 2.7 |
| Cycloserine | 5,087 ± 494 | 181 ± 4 | 28 |
The results are representative of those from two independent experiments.
Each of the compounds was used at 2× MIC.
Amounts are mAU of the high-pressure liquid chromatography peak area.
RESULTS
Identification of E. coli MurF inhibitor DQ1.
A pharmacophore model was used to screen a library of compounds to select candidate compounds to be tested in E. coli MurF binding and enzyme inhibition assays (5). The diarylquinoline compound DQ1 (Fig. 1) bound to MurF and inhibited its enzymatic activity (IC50, 24 ± 4 μM). DQ1 also demonstrated activity against lipopolysaccharide (LPS)-defective (permeable) E. coli OC2530 (MIC, 8 μg/ml) (Table 1) but had no measurable activity at the concentration tested against wild-type (non-LPS-defective) E. coli, Enterococcus faecalis, Enterococcus faecium, and S. aureus (both methicillin [meticillin]-susceptible and -resistant strains) (MICs, >32 μg/ml). Cycloserine, which inhibits the production of d-Ala-d-Ala via the inhibition of both the d-alanine racemase and ligase enzymes (27), was used as a comparator throughout these experiments. Cycloserine had an MIC of 64 μg/ml for E. coli OC2530 (Table 1), eightfold less active than DQ1. In contrast, cycloserine was more potent than DQ1 against wild-type E. coli (MICs, 16 to 32 μg/ml) and displayed activity (MICs, 32 to 128 μg/ml) against the strains of gram-positive bacteria tested.
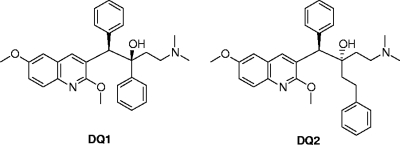
FIG. 1.
Structures of the MurF inhibitors DQ1 and DQ2.
TABLE 1.
Susceptibilities of bacterial strains to MurF inhibitors DQ1 and DQ2
| Strain | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| DQ1 | DQ1 plus 2 μg/ml PMBN | PMBN only | DQ2 | Cycloserine | |
| E. coli OC2530 (LPS defective) | 8 | 2 | 16 | 8 | 64 |
| E. coli OC2605 | >32 | 4 | 32 | >32 | 16 |
| E. coli OC9040 | >32 | 8 | 64 | >32 | 32 |
| E. coli ATCC 25922 | >32 | 8 | 64 | >32 | 32 |
| E. faecalis ATCC 29212 | >32 | >32 | >64 | 8 | 128 |
| E. faecium OC3312 | >32 | >32 | >64 | 8 | 64 |
| S. aureus ATCC 29213 | >32 | >32 | >64 | 8 | 32 |
| Methicillin-resistant S. aureus OC3726 | >32 | >32 | >64 | 8 | 32 |
| Methicillin-resistant S. aureus OC2878 | >32 | >32 | >64 | 8 | 64 |
| S. aureus OC4172 | >32 | >32 | >64 | 8 | 32 |
The hydrophobic nature of DQ1 suggested that poor permeability might be responsible for the lack of measureable MICs for wild-type E. coli. In support of this hypothesis, wild-type E. coli strains rendered permeable with a sub-MIC level of polymyxin B nonapeptide (PMBN) (36) were susceptible to DQ1, which exhibited MICs of 4 to 8 μg/ml (Table 1), fourfold lower than the MICs of cycloserine. The MICs of cycloserine, which uses d-Ala and Gly transport systems for entry into bacteria (38), were not affected by PMBN (5).
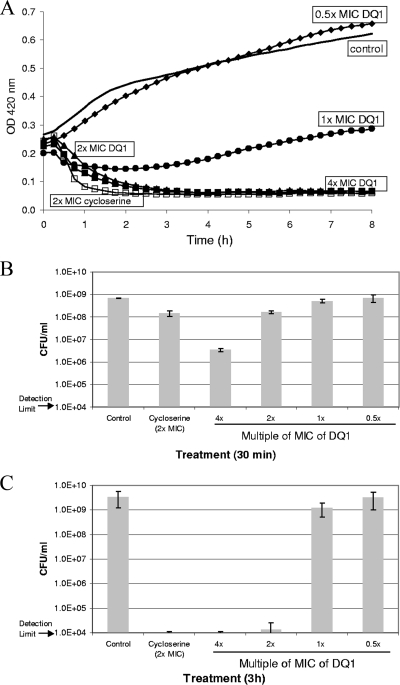
The growth of E. coli OC2530 in the presence of various amounts (0.5× to 4× MIC) of DQ1 was examined (Fig. 2A). At 2× and 4× MICs, DQ1 caused cell lysis, as suggested by a decline in the OD and as confirmed by microscopic examination of the cultures, similar to the behavior of cycloserine at 2× MIC. At 1× MIC, the DQ1-treated culture exhibited partial growth inhibition compared to the growth of the control culture. At 0.5× MIC, the growth curves of the DQ1-treated and the control cultures were similar, except that at approximately 4 h after compound addition, the DQ1-treated culture displayed a reproducible crossover in the OD and had slightly higher OD values than the control culture, which is discussed below.
FIG. 2.
(A) Growth curves of E. coli OC2530 treated with DQ1. DQ1 was added to mid-logarithmic-phase cultures at 0.5× MIC (diamonds), 1× MIC (circles), 2× MIC (triangles), or 4× MIC (solid squares); and the OD was monitored. The growth curves of a control (to which DMSO was added; solid line) and a cycloserine-treated culture (2× MIC; open squares) are also shown. Aliquots of the cultures were removed for quantitation of CFU at 30 min (B) and 3 h (C).
The CFU in the control, cycloserine-treated, and DQ1-treated cultures were quantified at 30 min and 3 h (Fig. 2B and C). At 30 min after compound addition at 2× MIC, both the cycloserine- and the DQ1-treated cultures displayed an approximately 1-log10-unit drop in the numbers of CFU relative to the number for the control. At 4× MIC, the DQ1-treated culture displayed an approximately 2-log10-unit decrease in the number of CFU relative to the control. At 1× and 0.5× MIC of DQ1, very little change in the number of CFU relative to the control was observed.
At 3 h after compound addition, the decline in the numbers of CFU at 2× MIC of cycloserine and at ≥2× MIC of DQ1 was at least 5 log10 units relative to the control (Fig. 2C). Thus, DQ1, like cycloserine, exhibited a bactericidal mode of action. At lower concentrations of DQ1 (≤1× MIC), the numbers of CFU at 3 h were similar to the untreated control. The number of CFU achieved with 1× MIC of DQ1 is consistent with the OD value of the corresponding growth curve (Fig. 2A): the threefold lower OD at 1× MIC of DQ1 compared to the control culture at 3 h should yield a decrease in the number of CFU of 0.3 log10 unit, a value that is within the error bars for these two samples.
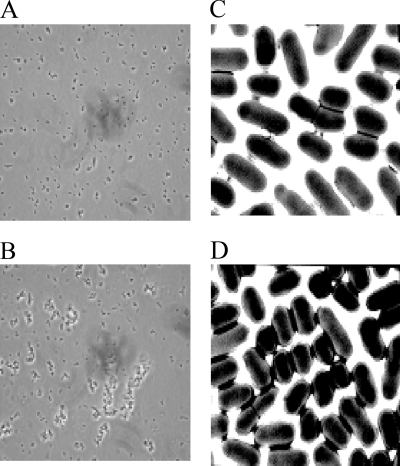
The crossover detected when the OD values of the growth curve of the control culture were compared with those of the culture treated with 0.5× MIC of DQ1 was reproducibly observed (Fig. 2A). These cultures were examined by light microscopy (Fig. 3A and B). The control culture contained dispersed, rod-shaped E. coli cells (Fig. 3A). In contrast, for the culture treated with 0.5× MIC of DQ1, aggregation of the cells was observed (Fig. 3B), which may be the cause of the increase in the OD value compared to the control. Cycloserine at 0.5× MIC also caused cell aggregation and the OD crossover (data not shown). Cultures containing larger amounts of DQ1 (≥1× MIC) showed dose-dependent cell lysis and the presence of significant amounts of cell debris.
FIG. 3.
Microscopy of E. coli OC2530 incubated for 5 h without DQ1 (A and C) and with 0.5× MIC of DQ1 (B and D). (A and B) light microscopy; (C and D) TEM.
The appearance of control cells and cells treated with 0.5× MIC of DQ1 was examined in greater detail by TEM (Fig. 3C and D). The control culture generally contained individual, rod-shaped cells, and there was evidence of pairs of dividing cells (Fig. 3C). In contrast, the DQ1-treated cells were more rounded and less rod-like and contained fewer dividing cells (Fig. 3D). Most striking in the DQ1-treated culture was the presence of aggregates of dozens of cells. These aggregates were possibly caused by the production of colanic acid, a capsular exopolysaccharide substance whose synthesis is induced by sub-MICs of β-lactams but not by inhibitors of protein synthesis or DNA replication (19, 28).
Quantitation of muropeptides in DQ1-treated E. coli.
Previous experiments with conditional MurF deletion mutants have demonstrated that lowering the amount of functional MurF enzyme increases the amount of the UDP-MurNAc-tripeptide substrate and decreases the amount of the UDP-MurNAc-pentapeptide product (24). Inhibition of the MurF enzyme would be expected to have similar effects on muropeptide levels.
To determine whether MurF was inhibited in E. coli cells treated with DQ1, muropeptides were extracted and quantified (Table 2). In the untreated control culture, very little of the MurF substrate UDP-MurNAc-tripeptide was present, in contrast to UDP-MurNAc-pentapeptide, which was about 100-fold more abundant than the tripeptide, in agreement with the findings of previous studies (24). For the positive control, treatment of cells with cycloserine caused UDP-MurNAc-tripeptide to accumulate (30-fold relative to the amount in the untreated culture), consistent with cycloserine's role in preventing the formation of d-Ala-d-Ala (38) and, thus, the formation of UDP-MurNAc-pentapeptide.
TABLE 2.
Relative amounts of UDP-MurNAc-tripeptide and -pentapeptide at different concentrations of MurF inhibitors DQ1 and DQ2
| Test set, strain, compound, and concn | Amta
|
UDP-MurNAc-tripeptide/UDP-MurNAc-pentapeptide ratiob | |
|---|---|---|---|
| UDP-MurNAc-tripeptide | UDP-MurNAc-pentapeptide | ||
| Set 1,cE. coli OC2530 | |||
| None (control) | 1 | 1.00 | 0.01 |
| DQ1 | |||
| 0.5× MIC | 6 | 1.62 | 0.04 |
| 1× MIC | 27 | 1.48 | 0.22 |
| 2× MIC | 89 | 0.33 | 3.3 |
| 4× MIC | 63 (98)d | 0.15 (0.23) | 5.0 |
| Cycloserine, 2× MIC | 30 (63) | 0.12 (0.25) | 3.0 |
| Set 2,eE. coli OC2530 | |||
| None (control) | 1 | 1.00 | 0.02 |
| DQ2, 2× MIC | 17 | 0.19 | 1.9 |
| Set 3,eS. aureus ATCC 29213 | |||
| None (control) | 1f | 1.00 | <0.03f |
| DQ2 | |||
| 1× MIC | >10 | 0.60 | 0.11 |
| 2× MIC | >38 | 0.34 | 0.71 |
| 4× MIC | >67 | 0.21 | 2.0 |
| Cycloserine, 2× MIC | ≫100 | 0.33 | ≫63 |
Relative value of the high-pressure liquid chromatography (HPLC) peak area normalized to the control value. Control values for UDP-MurNAc-tripeptide and -pentapeptide were as follows: for set 1, 17 and 1,400 milli-absorbance units (mAU), respectively; for set 2, 50 and 2,300 mAU, respectively; and for set 3, <12 and 460 mAU, respectively.
Ratio of HPLC peak areas.
Average of four independent experiments.
Values in parentheses are corrected for the amount of muropeptide lost to the supernatant due to cell lysis (∼35% for 4× MIC of DQ1, ∼50% for cycloserine).
The results are representative of those from two independent experiments.
The abundance of UDP-MurNAc-tripeptide was below the limit of detection by HPLC (12 mAU).
A dose-dependent increase in the amount of the MurF substrate UDP-MurNAc-tripeptide was observed at 0.5× to 2× MIC of DQ1 (Table 2). At 4× MIC of DQ1 at 30 min, it was evident from visual and microscopic inspection of the culture that the cells were starting to lyse. Although the relative amounts of UDP-MurNAc-tripeptide and UDP-MurNAc-pentapeptide should be accurate in this sample (Table 2), the absolute amounts were less than those that would be found if all cells were intact, due to the loss of muropeptides into the culture supernatant. From microscopy, we estimated that intact cells and, by extension, the observed amounts of UDP-MurNAc-tripeptide and -pentapeptide were approximately 35% lower than expected. Similarly, for cycloserine, we estimated a loss of about 50% of total muropeptides from cell lysis. The corrected amounts of muropeptide for the samples treated with cycloserine and 4× MIC of DQ1 are shown in Table 2.
Concomitant with the DQ1 dose-dependent increase in the amount of the MurF UDP-MurNAc-tripeptide substrate, a corresponding decrease in the amount of the MurF UDP-MurNAc-pentapeptide product was observed (Table 2). The increase in the substrate level and the decrease in the product level led to a shift in the substrate/product ratio from 0.01 (when no DQ1 was present) to 5.0 (when 4× MIC of DQ1 was present) (Table 2). These results are in good agreement with the accumulation of UDP-MurNAc-tripeptide and the decline in the amount of UDP-MurNAc-pentapeptide seen in the temperature-sensitive MurF deletion mutant of E. coli, which exhibited substrate/product ratios of 0.1 at the permissive temperature (when functional MurF enzyme was present) and 4.6 at the nonpermissive temperature (when nonfunctional MurF enzyme was present) (24).
Unexpectedly, a reproducible increase in the absolute amount of UDP-MurNAc-pentapeptide was observed in the presence of low levels of DQ1 (≤1× MIC; Table 2). To determine if murF mRNA levels were altered as a result of DQ1 treatment, reverse transcription-PCR was performed with the RNA from E. coli OC2530, without and with DQ1 treatment (0.5× to 4× MIC). No difference in murF mRNA levels was observed between samples treated with DQ1 and the control sample (data not shown), indicating that MurF gene expression was unchanged.
Identification of MurF inhibitor DQ2 with activity against gram-positive bacteria.
Analogs of DQ1 were examined for their ability to inhibit the MurF enzyme and for their antibacterial activities. Compound DQ2 (Fig. 1), which is a chain-extended analog of DQ1 with a relative stereochemistry opposite that of DQ1 at the position β to the quinoline ring, inhibited MurF with an IC50 of 29 ± 3 μM, comparable to that of DQ1. Similar to DQ1, DQ2 displayed activity against LPS-defective E. coli OC2530 (MIC, 8 μg/ml) and was not active against wild-type E. coli at the concentration tested (MIC, >32 μg/ml) (Table 1); the inclusion of PMBN lowered the MICs of DQ2, similar to the case for DQ1, for wild-type E. coli (Table 1 and data not shown). However, in contrast to DQ1, DQ2 exhibited an MIC of 8 μg/ml for the tested strains of E. faecalis, E. faecium, and S. aureus (both methicillin susceptible and resistant), thus extending the activity of the series to gram-positive bacteria. The DQ2 MICs for LPS-defective E. coli and the gram-positive bacteria were 4- to 16-fold lower than the cycloserine MICs observed (Table 1). For eight additional strains of enterococci, both vancomycin susceptible and resistant, DQ2 displayed MICs of 16 μg/ml (E. faecalis) and 8 μg/ml (E. faecium) both in the absence and in the presence of vancomycin (data not shown).
Against E. coli OC2530, DQ2 exhibited behavior similar to that seen for DQ1, including its effects on the growth curves, reductions in the numbers of CFU, and cell aggregation (at <1× MIC). DQ2 treatment of E. coli OC2530 also altered the muropeptide profile in a fashion similar to that of DQ1, with the accumulation of UDP-MurNAc-tripeptide and a reduction in the amount of UDP-MurNAc-pentapeptide (Table 2).
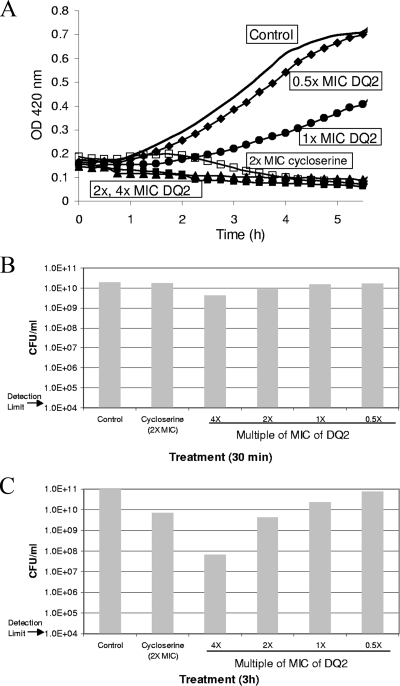
The treatment of S. aureus ATCC 29213 with DQ2 resulted in the dose-dependent inhibition of cell growth (Fig. 4A); however, in contrast to E. coli OC2530, S. aureus ATCC 29213 did not appear to be lysed either by cycloserine or by DQ2, as determined by macroscopic and microscopic examination of the cultures. Assessment of the numbers of CFU (at 30 min and 3 h after exposure to the compound; Fig. 4B and C) suggested that both cycloserine and DQ2 at 2× MIC were bacteriostatic against S. aureus ATCC 29213, in contrast to the bactericidal mode of action observed against E. coli OC2530 (Fig. 2C). DQ2 did not cause a change in the morphology of S. aureus cells, even upon prolonged incubation (>12 h), and cell aggregation was not observed either from determination of the OD or by microscopy (data not shown).
FIG. 4.
(A) Growth curves of S. aureus ATCC 29213 treated with DQ2. DQ2 was added to mid-logarithmic-phase cultures at 0.5× MIC (diamonds), 1× MIC (circles), 2× MIC (triangles), or 4× MIC (solid squares); and the OD was monitored. The growth curves of a control (to which DMSO was added; solid line) and a cycloserine-treated culture (2× MIC; open squares) are also shown. Aliquots of the cultures were removed for quantitation of CFU at 30 min (B) and 3 h (C).
The muropeptide profile of S. aureus ATCC 29213 treated with DQ2 (at 1×, 2×, and 4× MIC) was examined (Table 2). In contrast to the control culture of E. coli OC2530, in which UDP-MurNAc-tripeptide was detectable (albeit only at 1% of the level of abundance of UDP-MurNAc-pentapeptide; Table 2), the amount of UDP-MurNAc-tripeptide present was below the limit of detection (12 milli-absorbance units [mAU], or ∼2 μM UDP-MurNAc [4]) in the control culture of S. aureus ATCC 29213. DQ2 treatment of S. aureus resulted in the dose-dependent accumulation of the MurF substrate UDP-MurNAc-tripeptide, with a concomitant reduction in the amount of the MurF product UDP-MurNAc-pentapeptide (Table 2). With cycloserine treatment (2× MIC), the decrease in the abundance of UDP-MurNAc-pentapeptide was similar to that observed with DQ2; however, the level of accumulation of UDP-MurNAc-tripeptide was much greater for cycloserine than for DQ2. Sobral et al. likewise observed a more dramatic effect from cycloserine than from mutant MurF in S. aureus, with cycloserine (but not mutant MurF) eliminating the production of UDP-MurNAc-pentapeptide (34).
Overexpression of MurF in E. coli.
To determine the effect of increased amounts of MurF enzyme on muropeptide biosynthesis in E. coli, plasmid-borne MurF, under the control of the AHT-inducible tetA promoter, was introduced into strain OC2530, and the muropeptides were quantified (Table 3). Compared to the level of the MurF product UDP-MurNAc-pentapeptide in the control strain (harboring empty vector pASK), the MurF-overexpressing strain (harboring pMurF) showed an approximately threefold increase in the level of the MurF product UDP-MurNAc-pentapeptide (2,674 and 880, respectively). The amount of the MurF substrate UDP-MurNAc-tripeptide also increased by about threefold (220 and 63, respectively), suggesting that the muropeptide biosynthesis enzymes upstream of MurF may be upregulated in response to MurF overexpression.
The MICs of DQ1 for E. coli OC2530 harboring pMurF or pASK (in the presence of 3 μg/ml chloramphenicol to maintain the plasmid and 100 ng/ml AHT to induce expression) were 16 μg/ml for each strain; the cycloserine MIC for each strain was 64 μg/ml. However, the overexpression of MurF did have a modest but reproducible effect of increasing the amount of the MurF product UDP-MurNAc-pentapeptide in the presence of the MurF inhibitor DQ1 by about twofold (351 and 213, respectively; which decreased the substrate/product ratio from 5.6 to 2.7; Table 3). In contrast, cycloserine treatment of the MurF-overexpressing E. coli strain had the opposite effect, increasing the substrate/product ratio from 15 to 28.
DISCUSSION
In the current report, we describe new inhibitors of E. coli MurF, compounds DQ1 and DQ2, whose effects on E. coli are consistent with the inhibition of bacterial cell wall synthesis: (i) cell lysis (at >1× MIC), (ii) a decrease in the amount of the MurF product UDP-MurNAc-pentapeptide, and (iii) an increase in the amount of the MurF substrate UDP-MurNAc-tripeptide. DQ1 treatment of LPS-defective E. coli resulted in dose-dependent cell lysis and a >5-log10-unit reduction in the number of CFU (when DQ1 was used at 2× MIC and the cells were treated for 3 h), indicating bactericidal activity. These data are consistent with the behavior of a conditional MurF deletion mutant of E. coli that exhibited cell lysis upon a shift to the nonpermissive temperature (24).
An additional, unexpected finding was that the treatment of E. coli with low levels of DQ1 (≤1× MIC) appeared to increase the amount of the MurF product UDP-MurNAc-pentapeptide by 50 to 60% compared to that for the untreated control, leading to the question of whether MurF activity is upregulated by low levels of DQ1. No change in murF mRNA levels was observed, indicating that putative upregulation was not due to an increase in the level of MurF gene expression. It is possible that the inhibition of the MurF enzyme by a sub-MIC level of DQ1 could serve as a signal to increase UDP-MurNAc-pentapeptide production, possibly by the stimulation of MurF or of enzymes upstream in the pathway. An example of the modulation of peptidoglycan biosynthesis is provided by MurA, which appears to be negatively regulated by bound UDP-MurNAc (26). The release of bound UDP-MurNAc would be expected to increase MurA activity and, possibly, the output of the downstream steps in this pathway, such as MurB through MurF. Alternatively, a sub-MIC level of DQ1 may decrease UDP-MurNAc-pentapeptide turnover, possibly by the downregulation of enzymes downstream of MurF in the muropeptide biosynthetic pathway, such as MraY (37).
Muropeptide quantitation experiments, which demonstrated an increase in the amount of MurF substrate UDP-MurNAc-tripeptide and a concomitant decrease in the amount of UDP-MurNAc-pentapeptide upon DQ1 treatment, indicated that MurF was indeed inhibited within bacterial cells. However, this does not prove that MurF inhibition was responsible for cell death and does not exclude the possibility that DQ1 has multiple bacterial targets. Our efforts to obtain E. coli mutants resistant to DQ1 were unsuccessful, possibly due to the trivial reason that it may be more likely to obtain less permeable variants of OC2530 (the only strain of E. coli susceptible to DQ1 in the absence of a permeabilizing agent such as PMBN), as putative mutants were cross-resistant to a variety of unrelated antibiotics (data not shown). Alternatively, if DQ1 acts on multiple targets, the isolation of resistant mutants would be unlikely.
Although the overexpression of MurF in E. coli transformants did not lead to an increase in the MIC of DQ1, a modest twofold increase in the ability of the transformed strain to accumulate the MurF product was observed in the presence of DQ1 and may be consistent with MurF as the target of DQ1. As discussed above, DQ1 may act on multiple targets. In particular, the muropeptide synthetases MurC, MurD, and MurE share some structural and functional homology with MurF (7, 12); and we cannot exclude the possibility that DQ1 inhibits these or other E. coli targets. However, the inhibition of MurC, MurD, and MurE would block the production of UDP-MurNAc-monopeptide, -dipeptide, and -tripeptide, respectively, thereby preventing the DQ1- or DQ2-mediated accumulation of the MurF substrate UDP-MurNAc-tripeptide, which is also the MurE product. Since UDP-MurNAc-tripeptide did accumulate upon DQ1 or DQ2 treatment of the bacteria, it can be postulated that the inhibition of MurF by these compounds is more extensive than the possible inhibition of MurC, MurD, and MurE. It has been suggested that the simultaneous inhibition of multiple targets of cell wall biosynthesis by a single compound is a viable strategy for antibiotic discovery (29, 30).
The observation that compounds DQ1 and DQ2 were not able to inhibit the growth of wild-type E. coli at the concentrations tested, presumably due to poor permeation into cells, is a significant limitation to their potential as agents against gram-negative bacteria. Although the utility of DQ1 and DQ2 against gram-negative bacteria appeared to be limited to strains made permeable either by mutation (LPS deficiency) or by addition of a permeabilizing agent (e.g., PMBN), DQ2 extends the range of antibacterial activity of the series to gram-positive bacteria, including S. aureus (both methicillin susceptible and resistant) and enterococci (both vancomycin susceptible and resistant). The phenethyl group of DQ2, in comparison to the phenyl group of DQ1, is expected to improve the flexibility of the lateral chain, which may favor DQ2's interaction with the S. aureus MurF target.
The mechanism of action of DQ2 in S. aureus strain ATCC 29213 appears to be similar to that in LPS-defective E. coli strain OC2530, on the basis of the comparable shift in the UDP-linked muropeptide precursor pool, with the accumulation of UDP-MurNAc-tripeptide at the expense of UDP-MurNAc-pentapeptide; however, some important differences were observed. In E. coli, DQ1 or DQ2 treatment results in lysis and cell death; in contrast, in S. aureus, DQ2 treatment does not cause lysis and has a static effect. These effects are in agreement with the phenotypes of the corresponding MurF conditional mutants of these organisms (33, 34), providing further support that the mechanism of action of the DQs involves the inhibition of MurF. In addition, at ≤1× MIC of the DQs, an increased amount of UDP-MurNAc-pentapeptide was observed in E. coli but not in S. aureus. These findings are consistent with the differential effects of penicillin treatment on E. coli (23) and S. aureus (35): UDP-MurNAc-pentapeptide accumulates in S. aureus, indicating that feedback regulation does not occur (35). In contrast, in E. coli, UDP-MurNAc-pentapeptide is feedback regulated and does not accumulate (23).
In enterococci, the mechanism of vancomycin resistance involves the vancomycin-induced production of d-Ala-d-Lac, which is incorporated into peptidoglycan via MurF (3). d-Ala-d-Lac, in comparison to d-Ala-d-Ala, does not efficiently sequester vancomycin, thereby providing resistance to the antibiotic. Thus, in the MIC determinations with vancomycin in combination with DQ2, it appears that the activity of DQ2 was not affected by the presence of d-Ala-d-Lac in place of d-Ala-d-Ala.
We note that the MurF inhibitors DQ1 and DQ2 belong to the same chemical class as the diarylquinoline ATP synthase inhibitor TMC207 (R207910), an anti-tuberculosis compound (2, 21); however, the DQ compounds in E. coli and TMC207 in mycobacteria appear to exhibit distinctly different mechanisms of action. In particular, the mycobacterial ATP synthase enzyme appears to be essential, but the E. coli homolog can be deleted without the loss of bacterial viability (17) and so is unlikely to be the killing target of the DQ compounds. Also, the treatment of E. coli with the known ATP synthase inhibitor dicyclohexylcarbodiimide (13) did not cause an increase in UDP-MurNAc-tripeptide levels (S. Crespo-Carbone, unpublished data). Furthermore, DQ1 did not affect ATP levels in E. coli OC2530 (Anil Koul, personal communication).
In conclusion, we have described new MurF inhibitors that demonstrate the feasibility of inhibiting this target in both gram-negative and gram-positive organisms. The identification of additional broad-spectrum compounds with improved properties, especially with an enhanced ability to permeate gram-negative bacteria, is warranted.
Acknowledgments
We thank Anil Koul, Koen Andries, and Simon Lynch for useful comments on the manuscript. We thank Luc Vranckx for measurement of ATP levels and Ellyn Wira for assistance with microbiology studies.
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Anderson, M. S., S. S. Eveland, H. R. Onishi, and D. L. Pompliano. 1996. Kinetic mechanism of the Escherichia coli UDPMurNAc-tripeptide d-alanyl-d-alanine-adding enzyme: use of a glutathione S-transferase fusion. Biochemistry 35:16264-16269. [DOI] [PubMed] [Google Scholar]
- 2.Andries, K., P. Verhasselt, J. Guillemont, H. W. H. Goehlmann, J.-M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambau, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223-227. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., and P. Courvalin. 1993. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37:1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baum, E. Z., S. M. Crespo-Carbone, D. Abbanat, B. Foleno, A. Maden, R. Goldschmidt, and K. Bush. 2006. Utility of muropeptide ligase for identification of inhibitors of the cell wall biosynthesis enzyme MurF. Antimicrob. Agents Chemother. 50:230-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum, E. Z., S. M. Crespo-Carbone, A. Klinger, B. D. Foleno, I. Turchi, M. Macielag, and K. Bush. 2007. A MurF inhibitor that disrupts cell wall biosynthesis in Escherichia coli. Antimicrob. Agents Chemother. 51:4420-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergogne-Berezin, E. 2005. Fosfomycin and derivatives, p. 972-982. In A. Bryskier (ed.) Antimicrobial agents: antibacterials and antifungals. ASM Press, Washington, DC.
- 7.Bouhss, A., D. Mengin-Lecreulx, D. Blanot, J. van Heijenoort, and C. Parquet. 1997. Invariant amino acids in the Mur peptide synthetases of bacterial peptidoglycan synthesis and their modification by site-directed mutagenesis in the UDP-MurNAc:l-alanine ligase from Escherichia coli. Biochemistry 36:11556-11563. [DOI] [PubMed] [Google Scholar]
- 8.Bryskier, A., and C. Dini. 2005. Peptidoglycan synthesis inhibitors, p. 377-400. In A. Bryskier (ed.) Antimicrobial agents: antibacterials and antifungals. ASM Press, Washington, DC.
- 9.CLSI. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. CLSI, Wayne, PA.
- 10.Eliopoulos, G., and R. Moellering. 1996. Antimicrobials combinations, p. 330-396. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, MD.
- 11.El Zoeiby, A., F. Sanschagrin, and R. C. Levesque. 2003. Structure and function of the Mur enzymes: development of novel inhibitors. Mol. Microbiol. 47:1-12. [DOI] [PubMed] [Google Scholar]
- 12.Eveland, S. S., D. L. Pompliano, and M. S. Anderson. 1997. Conditionally lethal Escherichia coli murein mutants contain point defects that map to regions conserved among murein and folyl poly-γ-glutamate ligases: identification of a ligase superfamily. Biochemistry 36:6223-6229. [DOI] [PubMed] [Google Scholar]
- 13.Fillingame, R. H. 1975. Identification of the dicyclohexylcarbodiimide-reactive protein component of the adenosine 5′-triphosphate energy-transducing system of Escherichia coli. J. Bacteriol. 124:870-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, D. W. 2002. The bacterial cell wall as a source of antibacterial targets. Exp. Opin. Ther. Targets 6:1-19. [DOI] [PubMed] [Google Scholar]
- 15.Gu, Y. G., A. S. Florjancic, R. F. Clark, T. Zhang, C. S. Cooper, D. D. Anderson, C. G. Lerner, J. O. McCall, Y. Cai, C. L. Black-Schaefer, G. F. Stamper, P. J. Hajduk, and B. A. Beutel. 2004. Structure-activity relationships of novel potent MurF inhibitors. Bioorg. Med. Chem. Lett. 14:267-270. [DOI] [PubMed] [Google Scholar]
- 16.Hayat, M. A., and S. E. Miller. 1990. Negative staining. McGraw-Hill Publishing Co., New York, NY.
- 17.Jensen, P. R., and O. Michelsen. 1992. Carbon and energy metabolism of atp mutants of Escherichia coli. J. Bacteriol. 174:7635-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahan, F. M., J. S. Kahan, P. J. Cassidy, and H. Kropp. 1974. Mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235:364-386. [DOI] [PubMed] [Google Scholar]
- 19.Kaldalu, N., R. Mei, and K. Lewis. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob. Agents Chemother. 48:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotnik, M., P. S. Anderluh, and A. Prezelj. 2007. Development of novel inhibitors targeting intracellular steps of peptidoglycan biosynthesis. Curr. Pharm. Des. 13:2283-2309. [DOI] [PubMed] [Google Scholar]
- 21.Koul, A., N. Dendouga, K. Vergauwen, B. Molenberghs, L. Vranckx, R. Willebrords, Z. Ristic, H. Lill, I. Dorange, J. Guillemont, D. Bald, and K. Andries. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 3:323-324. [DOI] [PubMed] [Google Scholar]
- 22.Longenecker, K. L., G. F. Stamper, P. J. Hajduk, E. H. Fry, C. G. Jakob, J. E. Harlan, R. Edalji, D. M. Bartley, K. A. Walter, L. R. Solomon, T. F. Holzman, Y. G. Gu, C. G. Lerner, B. A. Beutel, and V. S. Stoll. 2005. Structure of MurF from Streptococcus pneumoniae crystallized with a small molecule inhibitor exhibits interdomain closure. Protein Sci. 14:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lugtenberg, E. J., L. De Haas-Menger, and W. H. Ruyters. 1972. Murein synthesis and identification of cell wall precursors of temperature-sensitive lysis mutants of Escherichia coli. J. Bacteriol. 109:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lugtenberg, E. J. J., and A. Van Schijndel-Van Dam. 1972. Temperature-sensitive mutants of Escherichia coli K-12 with low activities of the l-alanine adding enzyme and the d-alanyl-d-alanine adding enzyme. J. Bacteriol. 110:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, D. J., S. M. Hammond, D. Anderluzzi, and T. D. H. Bugg. 1998. Aminoalkylphosphinate inhibitors of d-Ala-d-Ala adding enzyme. J. Chem. Soc. Perkins Trans. 1:131-142. [Google Scholar]
- 26.Mizyed, S., A. Oddone, B. Byczynski, D. W. Hughes, and P. J. Berti. 2005. UDP-N-acetylmuramic acid (UDP-MurNAc) is a potent inhibitor of MurA (enolpyruvyl-UDP-GlcNAc synthase). Biochemistry 44:4011-4017. [DOI] [PubMed] [Google Scholar]
- 27.Reitz, R. H., H. D. Slade, and F. C. Neuhaus. 1967. Biochemical mechanisms of resistance by streptococci to the antibiotics d-cycloserine and O-carbamyl-d-serine. Biochemistry 6:2561-2570. [DOI] [PubMed] [Google Scholar]
- 28.Sailer, F. C., B. M. Meberg, and K. D. Young. 2003. β-Lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol. Lett. 226:245-249. [DOI] [PubMed] [Google Scholar]
- 29.Silver, L. L. 2006. Does the cell wall of bacteria remain a viable source of targets for novel antibiotics? Biochem. Pharmacol. 71:996-1005. [DOI] [PubMed] [Google Scholar]
- 30.Silver, L. L. 2007. Multi-targeting by monotherapeutic antibacterials. Nat. Rev. Drug Discov. 6:41-55. [DOI] [PubMed] [Google Scholar]
- 31.Silver, L. L. 2003. Novel inhibitors of bacterial cell wall synthesis. Curr. Opin. Microbiol. 6:431-438. [DOI] [PubMed] [Google Scholar]
- 32.Skerra, A. 1994. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151:131-135. [DOI] [PubMed] [Google Scholar]
- 33.Sobral, R. G., A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2006. Role of murF in cell wall biosynthesis: isolation and characterization of a murF conditional mutant of Staphylococcus aureus. J. Bacteriol. 188:2543-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobral, R. G., A. M. Ludovice, S. Gardete, K. Tabei, H. De Lencastre, and A. Tomasz. 2003. Normally functioning murF is essential for the optimal expression of methicillin resistance in Staphylococcus aureus. Microb. Drug Resist. 9:231-241. [DOI] [PubMed] [Google Scholar]
- 35.Strominger, J. L., J. T. Park, and R. E. Thompson. 1959. Composition of the cell wall of Staphylococcus aureus: its relation to the mechanism of action of penicillin. J. Biol. Chem. 234:3263-3268. [PubMed] [Google Scholar]
- 36.Tsubery, H., I. Ofek, S. Cohen, and M. Fridkin. 2000. Structure-function studies of polymyxin B nonapeptide: implications to sensitization of gram-negative bacteria. J. Med. Chem. 43:3085-3092. [DOI] [PubMed] [Google Scholar]
- 37.van Heijenoort, J. 2001. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18:503-519. [DOI] [PubMed] [Google Scholar]
- 38.Wargel, R. J., C. A. Shadur, and F. C. Neuhaus. 1971. Mechanism of d-cycloserine action: transport mutants for d-alanine, d-cycloserine, and glycine. J. Bacteriol. 105:1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]