Abstract
Nasopharyngeal colonization represents the initial interaction between Haemophilus influenzae and its human host. Factors that influence bacterial carriage likely affect transmission and incidence of infection. Therefore, we investigated host factors involved in limiting H. influenzae colonization in BALB/c mice, as colonization can be established in this genetic background. Unlike what is observed in the C57BL/6 background, initial colonization of BALB/c mice was mainly limited by adaptive immune components. This effect on colonization did not require either CD4- or CD8-positive T cells. Instead, initial colonization by genetically diverse strains was limited by preexisting natural antibody with a lesser contribution of complement activity and the presence of neutrophils. Natural serum immunoglobulin from mice was able to bind to the bacterial surface and exhibited complement-dependent bactericidal activity against these genetically diverse H. influenzae strains. Moreover, natural immunoglobulin G (IgG) recognizing these strains was detected at the nasopharyngeal mucosal surface. This antibody-mediated effect required exposure to the normal mouse microbial flora, since mice raised under germfree (GF) conditions showed increased levels of H. influenzae colonization that were not limited by adaptive immunity. In addition, serum IgG from GF mice exhibited less surface binding to H. influenzae, suggesting that natural antibody, induced through prior exposure to the microbial flora, mediated the observed reduction in initial colonization. The broad effect of natural IgG against genetically diverse isolates suggests the presence of conserved species-wide protective targets of antibody.
Haemophilus influenzae is a gram-negative pathogen that commonly colonizes the mucosal surface of the human nasopharynx. This species is capable of causing a wide spectrum of diseases ranging from acute infection of the respiratory tract to sepsis and meningitis. Invasive infections were most commonly associated with the encapsulated type b strain (H. influenzae type b [Hib]); however, the introduction of Hib conjugate polysaccharide vaccines has dramatically reduced the incidence of these infections. Conversely, nontypeable strains (nontypeable H. influenzae [NTHI]) do not express a polysaccharide capsule and are frequently associated with mucosal infections including otitis media, chronic bronchitis, and community-acquired pneumonia (1, 11, 26). The development of an effective vaccine to prevent NTHI infection has been hampered by the marked intra- and interstrain heterogeneity of surface antigens in this genetically diverse species.
Nasopharyngeal colonization precedes invasive disease and may exceed 50% in some populations (for a review, see reference 13). Since this organism is a strict human pathogen, this asymptomatic carriage likely represents the primary reservoir leading to horizontal spread. Therefore, factors influencing rates and density of bacterial colonization will affect transmission and incidence of infection in the population. Currently, specific host-pathogen interactions that affect H. influenzae colonization are incompletely characterized due to the lack of a convenient and genetically tractable animal model system.
Host genetic differences likely play a significant role in conferring predisposition to both colonization and infection. For example, initial colonization of artificial animal model systems could be limited by multiple factors, including the lack of an essential receptor(s), the availability of nutrients, the indigenous flora, and the innate and/or adaptive immune responses. Previous studies in our laboratory have described colonization dynamics of H. influenzae in the murine nasopharynx (29, 46), suggesting that an essential receptor or nutrient acquisition is not lacking in this artificial host. In particular, H. influenzae colonization of C57BL/6 mice is associated with recruitment of an inflammatory infiltrate comprised primarily of neutrophils to the nasal spaces, resulting in bacterial clearance mediated by multiple components of innate immunity (46). However, continued studies revealed that BALB/c mice are more susceptible to both Hib and NTHI colonization, suggesting that these innate immune factors are less effective in mice of this background.
Differing degrees of susceptibility to infection by inbred mouse strains have been characterized for numerous human bacterial pathogens including Streptococcus pneumoniae (14), Streptococcus pyogenes (34), Staphylococcus aureus (43), Mycobacterium tuberculosis (35, 38), and Salmonella enterica (4). Specifically, C57BL/6 and BALB/c mice have been shown to generate distinct immune responses against Helicobacter pylori (23, 42) and Candida albicans (41), and BALB/c mice are more susceptible to S. pneumoniae and H. influenzae infection in a model of otitis media (36). As we have previously described innate immunity-mediated clearance of H. influenzae by C57BL/6 mice and observed an increased susceptibility of BALB/c mice to various H. influenzae strains, we set forth to identify host factors that impact colonization in the BALB/c mouse background. Unlike what is observed in the C57BL/6 background, colonization of BALB/c mice was limited by adaptive immune components. Moreover, natural immunoglobulin G (IgG) antibody from conventionally reared BALB/c mice that bound to the bacterial surface was present at the nasopharyngeal mucosa, and serum Ig exhibited bactericidal activity against these genetically diverse H. influenzae strains. Our analysis of host factors affecting colonization of BALB/c mice suggests that conserved targets of IgG-mediated mucosal immunity may exist for this species.
MATERIALS AND METHODS
Mouse strains.
Female BALB/c mice (wild type [WT]) and polymeric Ig receptor (pIgR)-deficient mice (C57BL6-pIgRtm1) were purchased from Taconic Laboratories. pIgR-deficient mice contain a targeted deletion of the pIgR locus resulting in animals lacking secretory Ig (20). Germfree (GF) BALB/c WT and SCID mice were provided by J. J. Cebra (University of Pennsylvania), and maintenance of these strains has been previously described (19). C57BL/6 and B6.129-S2-Igh-6tm1Cgn/J (μMT) mice were obtained from Jackson Laboratories. μMT mice contain a targeted mutation in the heavy-chain locus of C57BL/6 IgM and do not produce mature B cells or antibody (25). Studies were conducted in compliance with the guidelines of the University of Pennsylvania, and all mice were housed in accordance with Institutional Animal Care and Use Committee protocols. Water and a standard rodent diet were provided ad libitum. Mice were inoculated at the age of 5 to 8 weeks unless otherwise specified.
Bacterial strains and culture conditions.
H. influenzae strains were grown in brain heart infusion broth (Becton Dickinson) supplemented with 2% Fildes enrichment (Remel) and 20 μg/ml β-NAD hydrate (NAD; Sigma). Strains were previously described and included H636, Eagan type b capsule (Hib); H648, a spontaneous b− mutant of Eagan lacking both copies of the cap locus (18); NTHI-1 strain TN106.P2 (33); NTHI-2 strain SR7332 (32); and NTHI-3 strain 86.028NP (31). Strains NTHI-1 and NTHI-2 were used because they had been previously shown to persist in the murine airway (9, 32). NTHI-3 was chosen because of the availability of its entire genomic sequence (16). All strains used in colonization experiments were spontaneously streptomycin-resistant mutants and were animal passaged.
Mouse model of nasopharyngeal colonization.
Mice were inoculated intranasally with 10 μl containing 107 to 108 CFU of phosphate-buffered saline (PBS)-washed, mid-log-phase H. influenzae. The animal was sacrificed at the time point specified, the trachea was cannulated, and 200 μl of PBS was instilled. Lavage fluid was collected from the nares for determination of viable counts of bacteria in serial dilutions plated on selective medium containing streptomycin (50 μg/ml) to inhibit the growth of contaminants. The lower limit of detection for bacteria in lavage fluid was 20 CFU/ml.
Depletion of CD4 and CD8 T-cell populations.
To deplete CD4-positive cells, mice were treated with 200 μg of anti-CD4 monoclonal antibody (MAb) GK1.5 intraperitoneally (i.p.) 4 days and 2 days prior to H. influenzae inoculation and boosted with 200 μg at day 1 postinoculation (p.i.). To deplete CD8-positive cells, mice were treated with 200 μg of anti-CD8 MAb 2.43 i.p. 4 days and 2 days prior to H. influenzae inoculation and boosted with 200 μg at day 1 p.i. Both antibodies, GK1.5 and 2.43, were purified from cell culture supernatants by affinity chromatography using recombinant protein G agarose (Life Technologies), as previously described (39). The efficiency of T-cell depletion was assessed by flow cytometric analysis comparing splenocytes isolated from GK1.5-treated, 2.43-treated, and untreated mice. The following antibodies were applied: rat anti-mouse CD4, to detect CD4+ T cells; rat anti-mouse CD8a, to detect CD8+ T cells; and anti-mouse CD3 molecular complex, to detect all mature T cells. All three antibodies were obtained from BD Biosciences. A total of 50,000 cells were collected for each sample, and groups were compared using FlowJo software (Tree Star).
Neutrophil and complement depletion.
MAb RB6-8C5, a rat anti-mouse IgG2b directed against Ly6G on the surface of murine myeloid (and limited subpopulations of lymphoid) lineage cells, was purified from ascites of nude mice given the RB6-8C5 hybridoma (7, 17). To deplete neutrophils, RB6-8C5 MAb was administered by i.p. injection as described previously (46). Hypocomplementemia was induced by i.p. injection of cobra venom factor (CVF; Quidel) in PBS as described previously (46). Where indicated, mice were treated with both RB6-8C5 (24 h) and CVF (18 h) prior to bacterial inoculation.
Western blot analysis.
Bacterial strains were grown to mid-logarithmic phase in supplemented brain heart infusion, pelleted, and lysed in sodium dodecyl sulfate (SDS) sample buffer. Five microliters of each lysate was loaded and separated by SDS-polyacrylamide gel electrophoresis (PAGE) on a 15% polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) transfer membranes (ThermoScientific). Membranes were then probed with the following primary antibodies at 4°C: serum or nasal lavage fluid from uninfected BALB/c mice, serum or nasal lavage fluid from uninfected SCID BALB/c mice, or serum isolated from BALB/c mice 14 days post-Hib inoculation (immune serum). Primary antibody was used at a 1:300 dilution of serum or a 1:20 dilution of nasal lavage fluid. Membranes were then incubated with a 1:10,000 dilution of alkaline phosphatase-conjugated anti-mouse IgG or IgM (whole molecule; Sigma) at room temperature. Antibody binding to H. influenzae strains was detected with 4-nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolylphosphate.
Flow cytometric analysis.
Two hundred microliters of mid-logarithmic-phase bacterial cells was pelleted and resuspended in 200 μl of Hanks’ buffer (Gibco) supplemented with 5% fetal calf serum (HyClone). A serum source was added to reaction mixtures at 1:20 or 1:100 dilutions, and all reaction mixtures were incubated at 4°C overnight. Sources of serum included samples from uninfected BALB/c, SCID, and GF BALB/c mice. Bacteria were pelleted and incubated with Hanks’ buffer plus 5% fetal calf serum containing a 1:200 dilution of goat anti-mouse IgG R-phycoerythrin conjugate (Sigma) for 1 h at 4°C. Reaction mixtures were then washed and resuspended in 200 μl of PBS containing 1% bovine serum albumin and 0.5% paraformaldehyde. A total of 10,000 cells were collected for each sample, and groups were compared using FlowJo software (Tree Star).
Bactericidal assays.
Mid-logarithmic-phase bacteria were diluted 1:10, and 20 μl was added to 140 μl Hanks’ buffer with Ca2+ and Mg2+ (Gibco). Twenty microliters of fresh serum from SCID mice was used as a complement source, and 20 μl of fresh, heat-inactivated (56°C for 30 min) BALB/c serum was added as a source of antibody. No-antibody controls contained 20 μl of fresh heat-inactivated serum from SCID mice. Reaction mixtures were incubated at 37°C for 45 min, and viable counts were determined by plating serial dilutions. Percent survival in the presence of antibody was determined by calculating the percentage of survival of each strain in the presence (heat-inactivated BALB/c serum) and absence (heat-inactivated SCID serum) of antibody.
Statistical analysis.
Statistical comparisons of colonization among groups were made by the Mann-Whitney test (GraphPad Software) unless otherwise specified.
RESULTS
Components of adaptive immunity promote mucosal clearance and limit the establishment of H. influenzae colonization.
To investigate host and bacterial factors involved in mucosal clearance of H. influenzae, we utilized a murine model of nasopharyngeal colonization. By isolating bacteria from the nasal lavage fluid following intranasal inoculation, we identified an encapsulated type b strain (Hib) and nontypeable strains (NTHI) exhibiting detectable levels of colonization of BALB/c mice. Colonization levels were examined at 3 and 14 days p.i. At 3 days, initial colonization of Hib was observed that was independent of phosphorylcholine (PC) expression (Fig. 1A), which has been shown to contribute to bacterial persistence on the mucosal surface during colonization (44). Additionally, NTHI-1 and NTHI-2 were tested because they have been previously shown to colonize BALB/c mice (9, 32), and NTHI-3 was tested because of the availability of its entire genome sequence (16). Colonization of NTHI-1 and NTHI-2 but not NTHI-3 was observed, and bacterial densities of Hib and NTHI at day 3 varied from strain to strain (Fig. 1A). Colonization of Hib and NTHI was cleared by 14 days, indicated by the detection of very few or no bacteria (Fig. 1B).
FIG. 1.
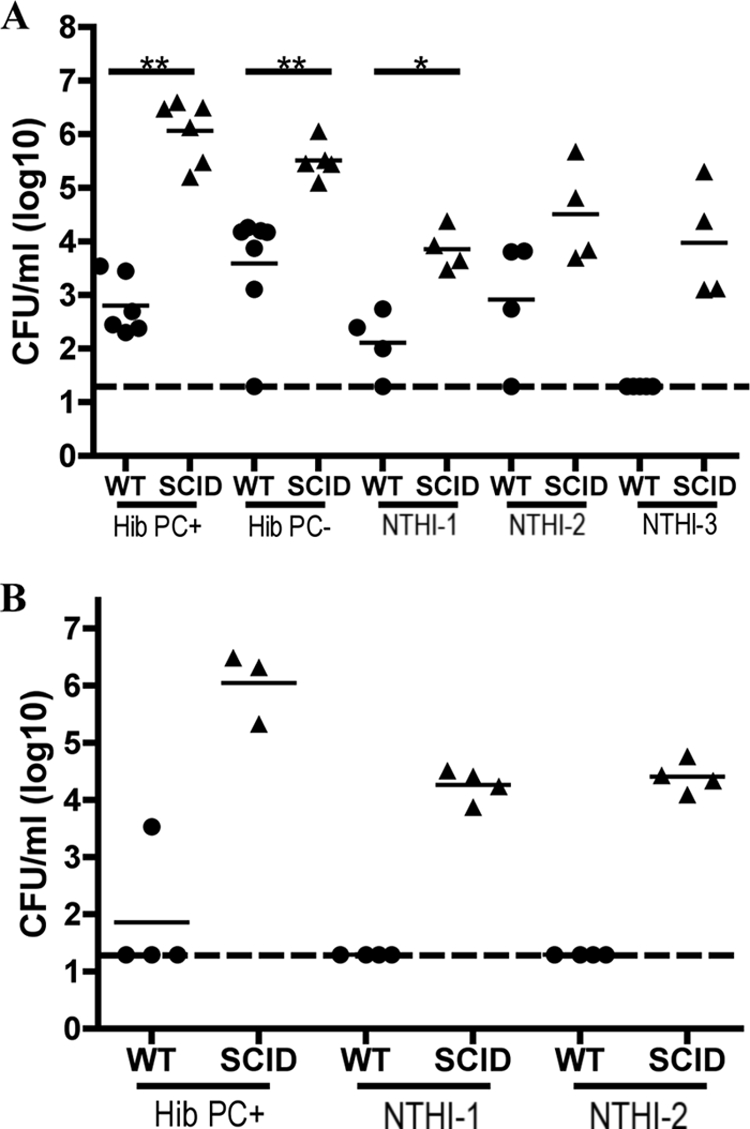
Adaptive immunity limits H. influenzae colonization. The density of the H. influenzae strain indicated in the upper respiratory tract lavage fluid of BALB/c WT (circles) and SCID (triangles) mice was determined at 3 days (A) and 14 days (B) post-intranasal inoculation. Bacterial strains included Hib with or without PC expression, NTHI-1 strain TN106.P2, NTHI-2 strain SR7332, and NTHI-3 strain 86.028NP. The log CFU/ml of each H. influenzae strain in the lavage fluid is indicated for each mouse, and horizontal bars denote mean values. The dashed line indicates the limit of detection. Statistical differences were determined using the Mann-Whitney test (*, P < 0.05; **, P < 0.01).
The role of adaptive immune responses in limiting and clearing H. influenzae colonization was investigated by inoculating BALB/c SCID mice with each of these strains and determining colonization levels at 3 and 14 days p.i. As expected, SCID mice exhibited attenuated clearance of Hib and the two NTHI strains tested at 14 days p.i., suggesting that the onset of adaptive immune responses is essential to promote clearance of H. influenzae (Fig. 1B). Interestingly, in comparison to immunocompetent WT mice, SCID mice exhibited significantly impaired early clearance of Hib and NTHI-1 at 3 days p.i. (Fig. 1A). Moreover, NTHI-3, which is cleared by day 3 from immunocompetent mice, colonized SCID mice (Fig. 1A). Cumulatively, these data suggest that, in addition to promoting the clearance of H. influenzae after the predicted onset of specific adaptive immune responses, components of adaptive immunity also limit the establishment of H. influenzae colonization.
CD4- and CD8-positive T cells do not limit the establishment of H. influenzae colonization.
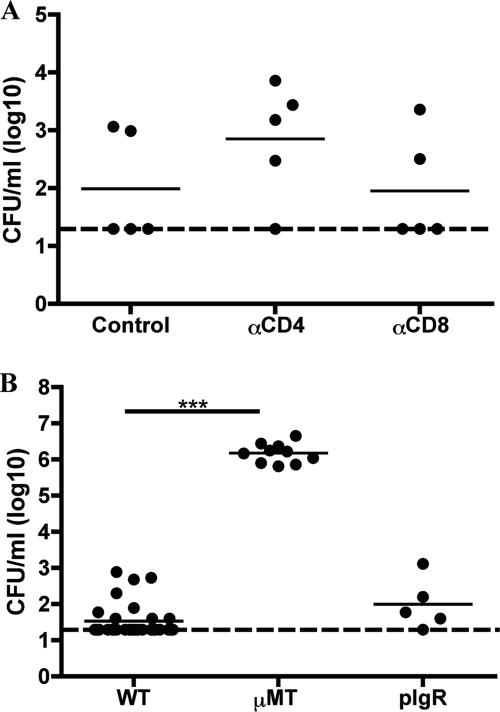
To assess whether the establishment of H. influenzae colonization is CD4+ T cell dependent, mice were treated with GK1.5, an IgG2b subclass MAb recognizing L3T4 on CD4+ T cells (10). We used the Hib PC+ strain, as it exhibited the most consistent levels of colonization (Fig. 1A and data not shown). Fluorescence-activated cell sorting analysis confirmed the depletion of >99% of CD4+ cells from the splenocyte population of these anti-CD4-treated mice (data not shown). Mice treated with anti-CD4 exhibited colonization levels of Hib that were similar to those of no-antibody-treated controls (P > 0.05; Fig. 2A). To determine if CD8+ T cells attenuate levels of H. influenzae colonization, mice were treated with MAb 2.43 recognizing Lyt-2 expressed by this cell population, resulting in the depletion of >95% of CD8+ T cells from the splenocyte population of these anti-CD8-treated mice (data not shown). Similar to the results for the anti-CD4-treated mice, treatment with anti-CD8 did not alter bacterial colonization levels (P > 0.05; Fig. 2A). Cumulatively, these data suggest that neither CD4- nor CD8-positive T cells individually represent the component(s) of adaptive immunity that is limiting the initial colonization of H. influenzae.
FIG. 2.
Nonsecretory antibody, but not CD4- or CD8-positive T cells, limits Hib colonization. (A) Individual depletion of either CD4- or CD8-positive cells does not affect colonization. The density of Hib PC+ in the upper respiratory tract lavage fluid of BALB/c mice treated with no antibody (control), MAb GK1.5 (anti-CD4), or MAb 2.43 (anti-CD8) to deplete CD4- or CD8-expressing cells, respectively, at 3 days p.i. (B) Nonsecretory antibody limits Hib PC+ colonization. The density of Hib in the upper respiratory tract lavage fluid of C57BL/6 (WT), antibody-deficient (μMT), and secretory antibody-deficient (pIgR) mice at 3 days post-Hib inoculation. The log CFU/ml of each H. influenzae strain in the lavage fluid is indicated for each mouse, and horizontal bars denote mean values. The dashed line indicates the limit of detection. Statistical differences were determined using the Mann-Whitney test (***; P < 0.001).
Natural antibody limits H. influenzae colonization.
Due to the lack of availability of knockout animals in the BALB/c background, we initially investigated the role of antibody in limiting H. influenzae colonization by using antibody-deficient (μMT) C57BL/6 mice. At day 3 p.i., μMT mice had a greatly increased density of Hib colonization compared to that of the WT (P < 0.0001; Fig. 2B). Moreover, at day 3 p.i., immunocompetent C57BL/6 mice cleared NTHI-1 (zero/five mice exhibited detectable colonization levels) compared to μMT mice challenged with NTHI-1, which remained colonized (five/five mice; range, 102 to 104 CFU/ml). The increased clearance of Hib and NTHI is likely caused by the presence of preexisting natural antibody in immunocompetent mice, as it occurs prior to the predicted onset of adaptive immune responses leading to the production of specific antibody. To specifically investigate the role of secretory antibody (secretory IgA and secretory IgM), we had previously tested pIgR-deficient mice (46). Interestingly, these mice, which do not transport antibody to the mucosal surface, exhibited Hib colonization levels similar to those of the WT mice (Fig. 2B). Together these data suggest that naturally acquired, nonsecretory antibody limits initial H. influenzae colonization at the mucosal surface.
The presence of neutrophils and complement components is associated with decreased levels of H. influenzae colonization.
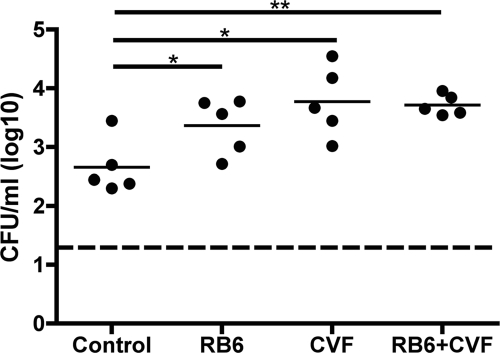
Since our previous work showed that H. influenzae colonization promotes an acute inflammatory response associated with the influx of neutrophils (46), we investigated the roles of both neutrophils and complement in colonization of BALB/c mice by Hib. To assess whether colonization of H. influenzae is limited by neutrophils, mice were treated with RB6-8C5, a rat MAb recognizing murine Ly6G, prior to intranasal challenge. This treatment has been shown to deplete neutrophils from peripheral blood (17) and prevent their recruitment to the nasopharynx in colonized mice (29). Mice treated with RB6-85C had an increased density of Hib colonization at day 3 p.i. compared to that of WT mice (P = 0.03; Fig. 3). To determine the role of complement in colonization, mice were treated with CVF to induce hypocomplementemia prior to bacterial challenge. Colonization levels of Hib were increased in treated mice compared to WT mice (P = 0.016). Moreover, there was not an additive effect on colonization levels when mice were treated with both RB6-85C and CVF prior to bacterial inoculation (Fig. 3). This suggests that complement and neutrophils function in the same pathway to decrease H. influenzae colonization. This effect of complement and neutrophils, however, is less dramatic than a lack of antibody as observed in μMT and SCID mice. Therefore, it is possible that the role for natural antibody in limiting H. influenzae colonization extends beyond promotion of phagocytic and complement-mediated killing.
FIG. 3.
Neutrophils and complement components limit Hib colonization. The density of Hib PC+ at 3 days p.i. in the nasal lavage fluid of BALB/c mice treated with rat IgG (control), RB6-85C (i.p. antibody treatment to deplete neutrophil-like cells), CVF (i.p. CVF pretreatment to deplete complement), or RB6-85C and CVF. The log CFU/ml of Hib in the lavage fluid is indicated for each mouse, and horizontal bars denote mean values. The dashed line indicates the limit of detection. Statistical differences were determined using the Mann-Whitney test (*, P < 0.05; **, P < 0.01).
GF mice do not exhibit limited colonization.
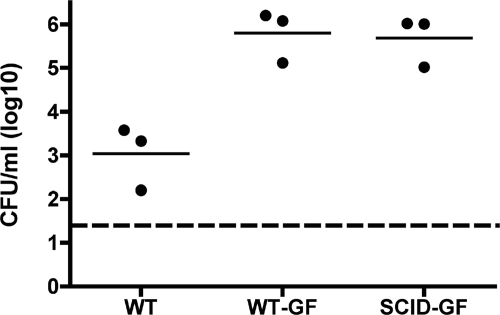
At 3 days p.i., adaptive immune components of conventionally reared mice limit H. influenzae colonization (Fig. 1A). Furthermore, the data in the C57BL/6 background suggest that natural antibody is essential to prevent effective bacterial colonization. Therefore, we wanted to determine whether the antibodies limiting colonization are generated by exposure to other microbial species by use of BALB/c mice reared under GF conditions. At 7 days p.i., conventional WT mice exhibit a decrease in colonization density compared to levels in WT-GF and SCID-GF mice (Fig. 4). Therefore, in mice lacking a normal microbial flora, adaptive immunity is no longer associated with a decrease in H. influenzae colonization levels. Cumulatively, these data support the involvement of preexisting antibody that is generated through prior microbial exposure in limiting initial colonization.
FIG. 4.
Adaptive immune components from conventionally reared but not GF mice limit Hib colonization. The density of Hib PC+ in the nasal lavage fluid of conventionally reared BALB/c mice (WT), GF BALB/c mice (WT-GF), and GF SCID mice (SCID-GF) at 7 days p.i. The log CFU/ml of Hib in the lavage fluid is indicated for each mouse, and horizontal bars denote mean values. The dashed line indicates the limit of detection.
Targets of preexisting antibody appear to be conserved among H. influenzae strains.
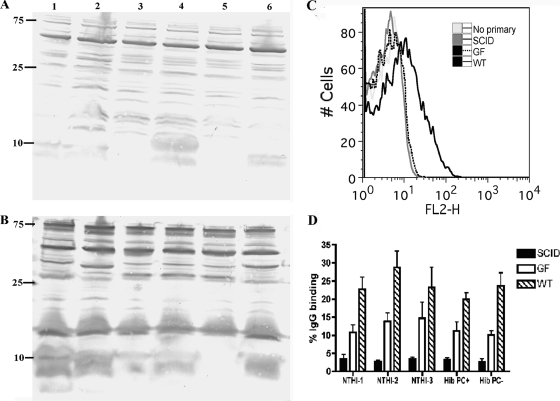
To determine if serum from uninfected, conventional mice contains IgG antibodies that recognize H. influenzae strains, whole-cell lysates were probed by Western blot analysis with either serum from uninfected BALB/c mice or serum that was isolated from mice 17 days after Hib intranasal inoculation (immune serum). Serum from uninfected mice contained IgG antibodies that recognized multiple components of H. influenzae lysates, and many of these targets appear similar across the panel of strains (Fig. 5A). Immune sera exhibited similar banding patterns at an increased intensity, implying that sera isolated from mice boosted by Hib colonization generated specific antibodies to targets that appear to be conserved among H. influenzae strains (Fig. 5B).
FIG. 5.
Natural antibody from conventionally reared mice recognizes targets expressed by all H. influenzae strains. (A and B) IgG antibodies from uninfected conventionally reared BALB/c mice recognize conserved H. influenzae targets by Western blot analysis. Whole-cell lysates of Hib with or without PC and without capsule (Cap−), NTHI-1, NTHI-2, and NTHI-3 were separated by SDS-PAGE, transferred to PVDF membranes, and probed with sera from uninfected BALB/c mice (A) or immune sera (B) followed by alkaline phosphatase-conjugated anti-mouse IgG. Strains tested included NTHI-1 (lane 1), NTHI-2 (lane 2), Hib PC− (lane 3), Hib PC+ (lane 4), Hib Cap− (lane 5), and NTHI-3 (lane 6). Molecular mass markers are in kDa. (C and D) Natural antibodies from conventionally reared BALB/c mice bind to the surface of all H. influenzae strains tested. Flow cytometric analysis was conducted on each H. influenzae strain incubated with no serum (“No primary”) or uninfected sera from conventionally reared uninfected BALB/c mice (WT), GF uninfected mice (GF), or conventionally reared SCID mice (SCID) followed by phycoerythrin-labeled anti-mouse whole IgG. (C) Fluorescence intensity is represented on the x axis, and binding of the antibody to the surface of Hib (PC+) is indicated by a shift of the curve to the right. The image is representative of three independent experiments. Each serum source was added to reaction mixtures at a 1:20 dilution. (D) The percent IgG binding was determined by calculating the percentage of 10,000 events with an increase in mean fluorescence intensity compared to that of no-serum controls. Each H. influenzae strain was incubated with sera from WT (hatched bars), GF (open bars), or SCID (solid bars) BALB/c mice at a 1:100 dilution. Values are derived from three independent experiments and are shown ± standard deviations.
To investigate if serum antibodies from conventional and GF mice are reactive with H. influenzae surface components, we incubated Hib and NTHI strains with uninfected sera from conventional WT, conventional SCID (SCID), and GF WT (GF) mice. Samples were then incubated with a phycoerythrin-labeled anti-mouse IgG, and antibody binding was determined by flow cytometry. Binding of serum antibody to whole Hib was observed with normal mouse sera and absent when Hib was incubated with sera from either SCID or GF mice (Fig. 5C). This increase in IgG binding when H. influenzae was incubated with WT sera compared to the binding when it was incubated with GF sera was observed for all strains (Fig. 5D). Cumulatively, these data indicate that natural serum IgG antibodies from conventional mice recognize surface targets expressed by a diverse array of H. influenzae strains. Moreover, fewer H. influenzae-specific antibodies recognizing H. influenzae surface components are present in GF mouse sera. This observation further supported our hypothesis that prior exposure to the microbial flora is the source of preexisting antibodies that limit colonization.
The presence of natural antibody is associated with increased bactericidal killing of all H. influenzae strains.
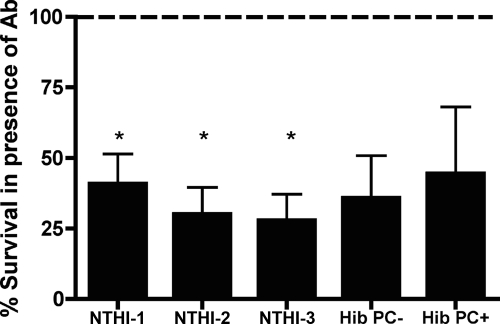
To determine if natural serum antibodies from uninfected conventionally reared mice promote bactericidal killing of H. influenzae, each strain was incubated with conventional SCID serum as a complement source and either heat-inactivated conventional WT serum that contains antibody or heat-inactivated conventional SCID serum (antibody-negative control). The percent survival in the presence of antibody was determined by calculating the CFU/ml of each strain in the presence (heat-inactivated BALB/c serum) divided by the CFU/ml of each strain in the absence (heat-inactivated SCID serum) of antibody. Lower percent survival of all H. influenzae strains was observed in the presence of antibody (Fig. 6). This demonstrates that natural antibody from conventionally reared mice exhibits bactericidal activity.
FIG. 6.
Natural antibody contributes to bactericidal killing of genetically diverse H. influenzae. The H. influenzae strain indicated was incubated with a complement source (SCID serum) and either a source of antibody (heat-inactivated BALB/c serum) or a no-antibody control (heat-inactivated SCID serum) at 37°C for 45 min, and viable counts were determined. Percent survival in the presence of antibody (y axis) was determined by calculating the ratio of bacterial survival in the presence of WT serum (presence of antibody) to bacterial survival in the presence of SCID serum (absence of antibody). Values are derived from three independent experiments and are shown ± standard deviations. Statistical differences were determined using the one-sample t test (*, P < 0.05).
The murine nasopharyngeal mucosa contains natural IgG antibody recognizing H. influenzae components.
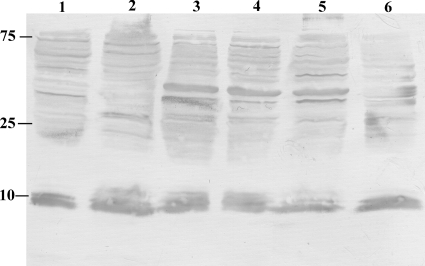
Although the serum of BALB/c mice contains natural IgG antibodies that recognize H. influenzae strains, we wanted to determine if anti-H. influenzae antibody (IgM or IgG) is also present at the primary site of colonization, the nasopharyngeal mucosa. Whole-cell lysates of each H. influenzae strain were separated by SDS-PAGE, transferred to PVDF membranes, and incubated with nasal lavage fluids from uninfected BALB/c mice. No IgM binding was detected in nasal lavage fluids (data not shown). In contrast, nasal lavage fluids contained IgG antibodies that recognized multiple components for all H. influenzae lysates (Fig. 7), confirming the presence of anti-H. influenzae IgG at the mucosal surface.
FIG. 7.
Natural IgG antibody recognizing H. influenzae strains is present on the mucosal surface. Western analysis showing whole-cell lysates of Hib with or without PC and without capsule (Cap−), NTHI-1, NTHI-2, and NTHI-3 separated by SDS-PAGE and transferred to PVDF membranes. Immunoblotting was carried out with nasal lavage fluid obtained from uninfected BALB/c mice followed by alkaline phosphatase-conjugated anti-mouse IgG to detect mucosal IgG recognizing H. influenzae targets. Strains tested included NTHI-1 (lane 1), NTHI-2 (lane 2), Hib PC− (lane 3), Hib PC+ (lane 4), Hib Cap− (lane 5), and NTHI-3 (lane 6). Molecular mass markers are in kDa.
DISCUSSION
In this study we were able to take advantage of the intrinsic differences associated with heterologous host species and genetic differences within a host species to further define immune components essential to limiting H. influenzae colonization. Innate immune factors in C57BL/6 mice promote the rapid clearance of H. influenzae (46). Conversely, comparatively higher levels of colonization of BALB/c mice by encapsulated and nontypeable (NTHI) strains may indicate a less effective innate immune response, rendering this murine strain more susceptible to H. influenzae infection. Using the BALB/c mouse model, we determined that adaptive immunity is the major factor that limits initial H. influenzae colonization and that this effect is not associated with the presence of either CD4- or CD8-positive T cells. Further studies showed that the main component of adaptive immunity limiting levels of initial colonization is likely preexisting or natural IgG. Natural serum antibodies from BALB/c mice are able to bind to the surface of and exhibit bactericidal activity against genetically diverse H. influenzae strains, and natural IgG recognizing these strains is present at the nasopharyngeal mucosal surface.
The generation of this antibody-mediated protective effect requires prior exposure to normal mouse microbial flora since mice raised as GF exhibited higher levels of Hib colonization that were not limited by adaptive immune responses. Furthermore, serum IgG from GF mice exhibited less binding to genetically diverse H. influenzae strains. Previous studies using GF animals have focused on the role of the gut microbiota in the development and maturation of local immune responses (for reviews, see references 8 and 21). However, it has also been shown that GF mice have decreased serum Ig levels (5). The specific impact of these immunological defects on the nasopharyngeal mucosa of GF mice has not been characterized. One previous study determined that nasal colonization levels of Klebsiella pneumoniae were similar in GF and the corresponding WT mice (27), suggesting that the general immune deficiencies associated with these mice do not universally impact clearance mechanisms of the nasopharyngeal mucosa. In our system, where initial colonization is not limited by adaptive immunity in GF mice, it is possible that the normal murine flora promotes expression of cross-reactive antibodies recognizing broadly reactive targets that limit H. influenzae colonization.
The production of germ line-encoded natural antibodies by naïve animals has been previously described and is most commonly of the IgM subclass (2). These antibodies have been associated with control of viral and bacterial infection, and the protective effects mediated by these antibodies are similar for WT and GF mice (37). Here we describe a different mechanism in which the presence of IgG antibody produced by naïve animals develops in response to the normal mouse microbiota. This antibody may promote complement-dependent bactericidal activity or phagocytic killing. Our findings suggest that natural antibody may additionally limit colonization by complement- and neutrophil-independent mechanisms. For example, antibody may have neutralizing effects by, for example, blocking adherence to host cells or promoting bacterial agglutination that leads to clearance.
Since natural antibody recognizing PC has been previously described for both mice (28) and humans (15), and GF BALB/c mice do not express anti-PC antibodies until they are conventionalized (28), we investigated the role of anti-PC antibody in our system. PC is an immunodominant structure incorporated into the lipopolysaccharide (LPS) of H. influenzae in a phase-variable manner (45), and anti-PC antibodies are protective against infection with another respiratory pathogen, S. pneumoniae (6). Therefore, we wanted to determine if H. influenzae lacking PC expression would exhibit higher colonization levels by thwarting the effects of natural anti-PC antibody. PC expression, however, did not affect the establishment of colonization. Moreover, reactivity to LPS constituents, which could represent a target conserved among H. influenzae strains, was not observed in the Western analysis of H. influenzae lysates probed with BALB/c serum. Cumulatively, these data suggest that preexisting antibody to PC-decorated LPS is not limiting H. influenzae colonization. The specific bacterial target(s) of the natural antibody is currently being investigated.
Although natural antibody limited H. influenzae colonization of BALB/c mice, these strains effectively colonize humans; therefore, it is interesting to speculate about the difference between these two hosts. Ig concentrations within nasal secretions of healthy humans consist primarily of secretory IgA but also contain comparably lower levels of IgG and secretory IgM (24). It is possible that the proportions of particular antibody types differ in mouse and in human nasal secretions or that these murine antibodies recognizing broadly reactive H. influenzae targets are lacking in the human host. Alternatively, it is possible that H. influenzae is able to thwart the effects of this antibody in its natural host. An example of such a mechanism is the production of IgA protease by H. influenzae and other pathogens inhabiting the human nasopharynx (22, 30, 40). Production of this enzyme, which is highly specific for IgA of humans and other great apes, likely interferes with the effectiveness of protective IgA1 antibodies. Perhaps H. influenzae is also capable of utilizing a method of interfering with IgG-mediated protection in the human host.
Previous studies have demonstrated that children colonized with H. influenzae exhibit a predominately IgA-mediated mucosal antibody response and develop strain-specific bactericidal antibodies (3, 12). Moreover, children with a greater mucosal antibody response rapidly eliminate their H. influenzae strain compared to children exhibiting prolonged colonization or colonization with multiple strains (12). Therefore, in the human host, antibodies mediate strain-specific protection from H. influenzae colonization; however, the genetic diversity associated with this organism permits subsequent colonization of new strains. Therefore, identifying the protective targets expressed by genetically diverse H. influenzae strains that are recognized by natural IgG antibody from naïve BALB/c mice may reveal a species-wide vaccine candidate.
Acknowledgments
We thank Bruce Green and Robert Munson for providing strains. We additionally acknowledge Zhe Zhang for her technical expertise and assistance.
This work was supported by U.S. Public Health Service grant numbers AI44231 and AI38446 to J.N.W.
Editor: A. Casadevall
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Apisarnthanarak, A., and L. M. Mundy. 2005. Etiology of community-acquired pneumonia. Clin. Chest Med. 2647-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avrameas, S. 1991. Natural autoantibodies: from ‘horror autotoxicus’ to ‘gnothi seauton.’ Immunol. Today 12154-159. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, J. M., H. S. Faden, B. G. Loos, T. F. Murphy, and P. L. Ogra. 1992. Recurrent otitis media with non-typable Haemophilus influenzae: the role of serum bactericidal antibody. Int. J. Pediatr. Otorhinolaryngol. 231-13. [DOI] [PubMed] [Google Scholar]
- 4.Borrego, A., L. C. Peters, J. R. Jensen, O. G. Ribeiro, W. H. Koury Cabrera, N. Starobinas, M. Seman, O. M. Ibañez, and M. De Franco. 2006. Genetic determinants of acute inflammation regulate Salmonella infection and modulate Slc11a1 gene (formerly Nramp1) effects in selected mouse lines. Microbes Infect. 82766-2771. [DOI] [PubMed] [Google Scholar]
- 5.Bos, N. A., H. Kimura, C. G. Meeuwsen, H. De Visser, M. P. Hazenberg, B. S. Wostmann, J. R. Pleasants, R. Benner, and D. M. Marcus. 1989. Serum immunoglobulin levels and naturally occurring antibodies against carbohydrate antigens in germ-free BALB/c mice fed chemically defined ultrafiltered diet. Eur. J. Immunol. 192335-2339. [DOI] [PubMed] [Google Scholar]
- 6.Briles, D. E., M. Nahm, K. Schroer, J. Davie, P. Baker, J. Kearney, and R. Barletta. 1981. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J. Exp. Med. 153694-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryder, D., Y. Sasaki, O. J. Borge, and S. E. Jacobsen. 2004. Deceptive multilineage reconstitution analysis of mice transplanted with hemopoietic stem cells, and implications for assessment of stem cell numbers and lineage potentials. J. Immunol. 1721548-1552. [DOI] [PubMed] [Google Scholar]
- 8.Cebra, J. J. 1999. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 691046S-1051S. [DOI] [PubMed] [Google Scholar]
- 9.Cutter, D., K. W. Mason, A. P. Howell, D. L. Fink, B. A. Green, and J. W. St. Geme III. 2002. Immunization with Haemophilus influenzae Hap adhesin protects against nasopharyngeal colonization in experimental mice. J. Infect. Dis. 1861115-1121. [DOI] [PubMed] [Google Scholar]
- 10.Dialynas, D. P., Z. S. Quan, K. A. Wall, A. Pierres, J. Quintans, M. R. Loken, M. Pierres, and F. W. Fitch. 1983. Characterization of the murine T cell surface molecule, designated L3T4, identified by monoclonal antibody GK1.5: similarity of L3T4 to the human Leu-3/T4 molecule. J. Immunol. 1312445-2451. [PubMed] [Google Scholar]
- 11.Eldika, N., and S. Sethi. 2006. Role of nontypeable Haemophilus influenzae in exacerbations and progression of chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 12118-124. [DOI] [PubMed] [Google Scholar]
- 12.Faden, H., L. Duffy, A. Williams, D. A. Krystofik, and J. Wolf. 1995. Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first 2 years of life. J. Infect. Dis. 172132-135. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Rodriguez, J. A., and M. J. Fresnadillo Martinez. 2002. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J. Antimicrob. Chemother. 50(Suppl. S2)59-73. [DOI] [PubMed] [Google Scholar]
- 14.Gingles, N. A., J. E. Alexander, A. Kadioglu, P. W. Andrew, A. Kerr, T. J. Mitchell, E. Hopes, P. Denny, S. Brown, H. B. Jones, S. Little, G. C. Booth, and W. L. McPheat. 2001. Role of genetic resistance in invasive pneumococcal infection: identification and study of susceptibility and resistance in inbred mouse strains. Infect. Immun. 69426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg, H. B., T. L. McCool, and J. N. Weiser. 2004. Cross-reactivity of human immunoglobulin G2 recognizing phosphorylcholine and evidence for protection against major bacterial pathogens of the human respiratory tract. J. Infect. Dis. 1901254-1263. [DOI] [PubMed] [Google Scholar]
- 16.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 1874627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hestdal, K., F. W. Ruscetti, J. N. Ihle, S. E. Jacobsen, C. M. Dubois, W. C. Kopp, D. L. Longo, and J. R. Keller. 1991. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J. Immunol. 14722-28. [PubMed] [Google Scholar]
- 18.Hoiseth, S. K., C. J. Connelly, and E. R. Moxon. 1985. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect. Immun. 49389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, H.-Q., N. A. Bos, and J. J. Cebra. 2001. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect. Immun. 693611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen, F. E., M. Pekna, I. N. Norderhaug, B. Haneberg, M. A. Hietala, P. Krajci, C. Betsholtz, and P. Brandtzaeg. 1999. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med. 190915-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, D., T. King, and R. Aminov. 2007. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat. Res. 62258-69. [DOI] [PubMed] [Google Scholar]
- 22.Kilian, M., J. Mestecky, and R. E. Schrohenloher. 1979. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect. Immun. 26143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 2001. Importance of the host genetic background on immune responses to Helicobacter pylori infection and therapeutic vaccine efficacy. FEMS Immunol. Med. Microbiol. 3141-46. [DOI] [PubMed] [Google Scholar]
- 24.Kirkeby, L., T. T. Rasmussen, J. Reinholdt, and M. Kilian. 2000. Immunoglobulins in nasal secretions of healthy humans: structural integrity of secretory immunoglobulin A1 (IgA1) and occurrence of neutralizing antibodies to IgA1 proteases of nasal bacteria. Clin. Diagn. Lab. Immunol. 731-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitamura, D., J. Roes, R. Kühn, and K. Rajewsky. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350423-426. [DOI] [PubMed] [Google Scholar]
- 26.Klein, J. O. 1994. Otitis media. Clin. Infect. Dis. 19823-833. [DOI] [PubMed] [Google Scholar]
- 27.Lau, H. Y., G. B. Huffnagle, and T. A. Moore. 2008. Host and microbiota factors that control Klebsiella pneumoniae mucosal colonization in mice. Microbes Infect. 101283-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberman, R., M. Potter, E. B. Mushinski, W. Humphrey, and S. Rudikoff. 1974. Genetics of a new IgVH (T15 idiotype) marker in the mouse regulating natural antibody to phosphorylcholine. J. Exp. Med. 139983-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lysenko, E. S., A. J. Ratner, A. L. Nelson, and J. N. Weiser. 2005. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 1e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Male, C. J. 1979. Immunoglobulin A1 protease production by Haemophilus influenzae and Streptococcus pneumoniae. Infect. Immun. 26254-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason, K. M., R. S. Munson, Jr., and L. O. Bakaletz. 2003. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 713454-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason, K. W., D. Zhu, C. A. Scheuer, J. C. McMichael, G. W. Zlotnick, and B. A. Green. 2004. Reduction of nasal colonization of nontypeable Haemophilus influenzae following intranasal immunization with rLP4/rLP6/UspA2 proteins combined with aqueous formulation of RC529. Vaccine 223449-3456. [DOI] [PubMed] [Google Scholar]
- 33.McGehee, J. L., J. D. Radolf, G. B. Toews, and E. J. Hansen. 1989. Effect of primary immunization on pulmonary clearance of nontypable Haemophilus influenzae. Am. J. Respir. Cell Mol. Biol. 1201-210. [DOI] [PubMed] [Google Scholar]
- 34.Medina, E., O. Goldmann, M. Rohde, A. Lengeling, and G. S. Chhatwals. 2001. Genetic control of susceptibility to group A streptococcal infection in mice. J. Infect. Dis. 184846-852. [DOI] [PubMed] [Google Scholar]
- 35.Medina, N. 1998. Resistance ranking of some common inbred mouse strains to Mycobacterium tuberculosis and relationship to major histocompatibility complex haplotype and Nramp1 genotype. Immunology 93270-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melhus, A., and A. F. Ryan. 2003. A mouse model for acute otitis media. APMIS 111989-994. [DOI] [PubMed] [Google Scholar]
- 37.Ochsenbein, A. F., T. Fehr, C. Lutz, M. Suter, F. Brombacher, H. Hengartner, and R. M. Zinkernagel. 1999. Control of early viral and bacterial distribution and disease by natural antibodies. Science 2862156-2159. [DOI] [PubMed] [Google Scholar]
- 38.Pan, H., B.-S. Yan, M. Rojas, Y. V. Shebzukhov, H. Zhou, L. Kobzik, D. E. Higgins, M. J. Daly, B. R. Bloom, and I. Kramnik. 2005. Ipr1 gene mediates innate immunity to tuberculosis. Nature 434767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan, Z. K., G. Ikonomidis, A. Lazenby, D. Pardoll, and Y. Paterson. 1995. A recombinant Listeria monocytogenes vaccine expressing a model tumour antigen protects mice against lethal tumour cell challenge and causes regression of established tumours. Nat. Med. 1471-477. [DOI] [PubMed] [Google Scholar]
- 40.Plaut, A. G., J. V. Gilbert, M. S. Artenstein, and J. D. Capra. 1975. Neisseria gonorrhoeae and Neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science 1901103-1105. [DOI] [PubMed] [Google Scholar]
- 41.Schofield, D. A., C. Westwater, and E. Balish. 2005. Divergent chemokine, cytokine and beta-defensin responses to gastric candidiasis in immunocompetent C57BL/6 and BALB/c mice. J. Med. Microbiol. 5487-92. [DOI] [PubMed] [Google Scholar]
- 42.van Doorn, N. E., F. Namavar, M. Sparrius, J. Stoof, E. P. van Rees, L.-J. van Doorn, and C. M. Vandenbroucke-Grauls. 1999. Helicobacter pylori-associated gastritis in mice is host and strain specific. Infect. Immun. 673040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Kockritz-Blickwede, M., M. Rohde, S. Oehmcke, L. S. Miller, A. L. Cheung, H. Herwald, S. Foster, and E. Medina. 2008. Immunological mechanisms underlying the genetic predisposition to severe Staphylococcus aureus infection in the mouse model. Am. J. Pathol. 1731657-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiser, J. N., N. Pan, K. L. McGowan, D. Musher, A. Martin, and J. Richards. 1998. Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zola, T. A., E. S. Lysenko, and J. N. Weiser. 2008. Mucosal clearance of capsule-expressing bacteria requires both TLR and nucleotide-binding oligomerization domain 1 signaling. J. Immunol. 1817909-7916. [DOI] [PubMed] [Google Scholar]