Abstract
Genomic analysis of the Chlamydiaceae has revealed a multigene family encoding large, putatively autotransported polymorphic membrane proteins (Pmps) with nine members in the sexually transmitted pathogen Chlamydia trachomatis. While various pathogenesis-related functions are emerging for the Pmps, observed genotypic and phenotypic variation among several chlamydial Pmps in various Chlamydia species has led us to hypothesize that the pmp gene repertoire is the basis of a previously undetected mechanism of antigenic variation. To test this hypothesis, we chose to examine the serologic response of C. trachomatis-infected patients to each Pmp subtype. Immune serum samples were collected from four populations of patients with confirmed C. trachomatis genital infection: 40 women with pelvic inflammatory disease from Pittsburgh, PA; 27 and 34 adolescent/young females from Oakland, CA, and Little Rock, AR, respectively; and 58 adult male patients from Baltimore, MD. The Pmp-specific antibody response was obtained using immunoblot analysis against each of the nine recombinantly expressed Pmps and quantified by densitometry. Our results show that nearly all C. trachomatis-infected patients mount a strong serologic response against individual or multiple Pmp subtypes and that the antibody specificity profile varies between patients. Moreover, our analysis reveals differences in the strengths and specificities of the Pmp subtype-specific antibody reactivity relating to gender and clinical outcome. Overall, our results indicate that the Pmps elicit various serologic responses in C. trachomatis-infected patients and are consistent with the pmp gene family being the basis of a mechanism of antigenic variation.
Chlamydiae are obligate intracellular bacteria with a unique biphasic developmental cycle, consisting of the infectious extracellular form, the elementary body (EB), and the metabolically active, replicating form, the reticulate body (RB). Following host cell entry, the chlamydial developmental cycle takes place entirely within an intracellular vacuole, called an inclusion (26). Chlamydia trachomatis, a prevalent human pathogen, infects the ocular and genital mucosa. The chronic ocular disease, trachoma, affects several hundred million people in the world and is the leading cause of infectious preventable blindness (50). The urogenital infection contributes to the most common bacterial sexually transmitted diseases (STDs) in both industrialized and developing countries, with an estimated 90 million new cases each year worldwide (51). Serious sequelae of chlamydial STD include pelvic inflammatory disease (PID), infertility, and ectopic pregnancy (5, 36). In addition, chlamydial STDs are risk factors in cervical squamous cell carcinoma (1) and human immunodeficiency virus transmission (9).
Surface-exposed proteins of infectious organisms are considered major mediators in interactions between pathogens and host cells, playing significant roles in virulence. The complete sequence analysis of the C. trachomatis genome has revealed a multigene family encoding nine predicted polymorphic membrane proteins (Pmps) (41) designated PmpA to PmpI. This gene superfamily has been identified in several other Chlamydia spp., including C. pneumoniae, C. muridarum, C. caviae, C. abortus, and C. psittaci (20, 32, 33, 44). At the genome level, the pmp genes represent 13.6% and 17.5% of the Chlamydia-specific coding capacity in C. trachomatis and C. pneumoniae, respectively (35). Given the reductive evolution of the chlamydial genome, these percentages imply a critical role of this family in chlamydial biology and pathogenesis.
Pmps are heterogeneous, both in amino acid sequence and in predicted size. Their grouping as a family is based on a conserved pair of tetrapeptide motifs, GGA (with I, L, or V at the fourth position) and FXXN (where X represents any amino acid), repeated multiple times in the amino (N)-terminal half of each protein. Pmps have a sec-dependent leader sequence at the N terminus and a carboxy (C)-terminal phenylalanine residue characteristic of outer membrane proteins (18). In silico analysis suggests that Pmps are autotransported proteins, with the C terminus folding to form a beta-barrel pore that allows the translocation of the N-terminal passenger domain to the surface (19). This prediction has been supported by experimental evidence for several Pmps (22, 23, 46, 49). The surface-exposed PmpD of C. pneumoniae was shown to function as an adhesin, and both PmpB and PmpD were able to stimulate proinflammatory cytokine production through activation of the NF-κB pathway (28, 49). Moreover, a Pmp-specific monoclonal antibody was capable of inducing protective immunity against C. abortus (ovine C. psittaci) in an early study (8). Recent evidence has revealed that PmpD is a species-common, pan-neutralizing antigen of C. trachomatis (10) and that specific Pmp peptides have the capacity of eliciting protective immunity in an animal model (21). These observations imply that Pmps may be important virulence factors susceptible to antibody attack and that they hold potential as vaccine candidates.
Comparative genomics has revealed genetic variation and rearrangements among pmp gene families in different strains and isolates across Chlamydia spp., including frameshift mutations, deletions, intragenic duplications, insertion sequence-like elements, etc. (14, 16-18, 34, 39). Moreover, differential Pmp expression in C. pneumoniae has also been observed both in tissue culture and in infected animals (4, 30).
The observed differences suggest that pmp expression varies with high frequency, suggestive of a potential coupled phase and antigenic variation mechanism. Using such mechanisms, an infectious organism may alter its surface at high frequency to evade the host adaptive immune system, allowing the pathogen to develop chronic or repeated infections in mammalian hosts or to be easily transmitted (47). Similar gene families found in other pathogens, such as the opacity proteins (11, 42) of Neisseria sp., the variable major proteins of Borrelia hermsii (3), the Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein families of Mycobacterium tuberculosis (2), and the variant surface glycoproteins of Trypanosoma brucei (48), have been shown to be capable of inducing differential immune responses in infected patients or animal models (12, 45).
Previous serological analyses revealed that a Pmp-specific immune response could be elicited in C. abortus-infected animals (24, 40) and in patients with either C. pneumoniae or C. trachomatis infection (7, 15, 29, 31). In this study, we document variations in the Pmp-specific antibody response in different patient populations with confirmed C. trachomatis genital infection. Our results are consistent with the hypothesis that variable expression of the pmp family plays a role in antigenic variation.
MATERIALS AND METHODS
Chlamydia and cell culture.
HeLa 229 cells were grown in 100-mm2 tissue culture dishes at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), gentamicin (25 μg/ml; Quality Biological, Gaithersburg, MD) and Fungizone (1.25 μg/ml; Invitrogen, Carlsbad, CA). Confluent monolayers were pretreated with DEAE-dextran (30 μg/ml), and 1 ml of inoculum containing 5 × 106 inclusion-forming units of C. trachomatis serovar E strain UW5-CX in sucrose-phosphate-glutamic acid (0.25 M sucrose, 10 mM sodium phosphate and 5 mM l-glutamic acid) was added to one dish. Infected monolayers were rocked gently for 2 h at 25°C, freshly supplemented Dulbecco's modified Eagle's medium was added, and the cultures were incubated at 37°C with 5% CO2. EBs were harvested at 48 h postinfection and purified by step gradient centrifugation using 40%, 44%, and 54% Urografin-370 (Schering AG, Germany), following standard protocols (6, 37).
Cloning and expression of rPmps.
Both full-length (Pmp-FL) and N-terminal fragments (Pmp-N) including the predicted “passenger” domain of Pmps were cloned and expressed. Briefly, DNA fragments encoding specific Pmp fragments were amplified from C. trachomatis serovar E/UW5-CX genomic DNA by PCR with the primers listed in Table 1. Primer pairs were based on the published C. trachomatis serovar D/UW-3/CX genome sequence and were designed to exclude the hydrophobic N-terminal leader sequence. Amplified products were either cloned into the PCRII TOPO vector (Invitrogen, Carlsbad, CA) for restriction mapping and then inserted into the pET30 vector (Novagen, Madison WI) or directly cloned into the pET30 vector. All clones were partially sequenced to confirm correct in-frame insertion and N-terminal fusion with the S-tag and His tag. For several Pmps, the N-terminal fragments excluding the in silico predicted beta-barrel domain were generated by digesting the plasmids carrying the full-length pmp inserts with restriction endonucleases (listed in Table 1) to remove the C-terminal sequence and then religated back into the vector. A construct expressing recombinant PmpG-FL (rPmpG-FL) could not be obtained after several attempts using different expression vectors. A construct encoding the N-terminal portion of PmpG (corresponding to amino acid residues 139 to 502) was generated and used in all subsequent experiments. All clones were transformed into Escherichia coli strain BL21 (Invitrogen, Carlsbad, CA) and overexpressed upon induction with 0.1 μM isopropyl-β-d-thiogalactopyranoside during exponential growth. Insoluble inclusion bodies, enriched with rPmps, were partially purified using Triton X-100 and ultrasonication.
TABLE 1.
Primers, vectors and restriction enzymes used for rPmp cloning
| Gene | rPmp | Primera | Primer sequence (5′ to 3′) | Vector | Restriction enzyme(s) | Amino acid residues | Molecular mass (kDa)
|
|
|---|---|---|---|---|---|---|---|---|
| Calculated | Apparent | |||||||
| pmpA | rPmpA-FL | pmpA-1 | TTTCTCTCATATTCGAAACAGC | pET30c | 48-979 | 107.8 | 105 | |
| rPmpA-N | pmpA-2 | ATAGCCTCCGCAGGATAAGCT | pET30c | PstI and BlpI | 48-602 | 65.9 | 65 | |
| pmpB | rPmpB-FL | pmpB-1 | CCCGGGTCGACCTAAAGAATTAAATTTCTCTCGCG | pET30b | 28-1732 | 188 | 190 | |
| rPmpB-N | pmpB-2 | TCACTCGAGCACCGCAGCTAGTCATTTG | pET30a | BamHI and SmaI | 28-953 | 101.6 | 135 | |
| pmpC | rPmpC-FL | pmpC-1 | CCGACTGCAATGTTAGCAAATTAGG | pET30a | 29-1692 | 191.4 | 200 | |
| pmpC-2 | ACGAGCACCGCAGTTCATCA | |||||||
| rPmpC-N | pmpC-3 | GCGGGTCCGTTACTGAGGCGAGCTCG | pET30a | 16-974 | 108.3 | 155 | ||
| pmpC-4 | CGCGTCGACCTGGCACCAACCCAATAGCAG | |||||||
| pmpD | rPmpD-FL | pmpD-1 | TGGTATATGTAGGCCCTCAAGCGG | pET30a | 45-1528 | 164.3 | 160 | |
| rPmpD-N | pmpD-2 | TCGCAATCCAGTATTCGCCTCA | pET30a | BamHI and EcoRV | 45-1079 | 142 | 140 | |
| pmpE | rPmpE-FL | pmpE-1 | GAGCAATTTCCCCATTGAGAT | pET30a | 26-962 | 112.1 | 110 | |
| rPmpE-N | pmpE-2 | CTTATGCCCAACTCAGTTCCA | pET30a | XhoI | 26-667 | 80.6 | 80 | |
| pmpF | rPmpF-FL | pmpF-1 | GCGGATATCGGATACGCTACAGTTCCGG | pET30b | 29-1032 | 116.3 | 115 | |
| rPmpF-N | pmpF-2 | GCGGAATTCAGAGCTCCTCCTGCATT | pET30b | HindIII | 29-697 | 79.3 | 80 | |
| pmpG | rPmpG-N | pmpG-1 | TCAAGGAATTTACGATGGGGAGACG | pET30a | 139-502 | 43.1 | 45 | |
| pmpG-2 | TTCCTGCACTCAAACCATAACCTCG | |||||||
| pmpH | rPmpH-FL | pmpH-1 | CCCGGGATATCCTCAAGTGTTAACACCTAATG | pET30c | 30-1009 | 110.8 | 110 | |
| rPmpH-N | pmpH-2 | CGCAAGCTTGTGCCGTTTGTCGACGACGAG | pET30c | BamHI | 30-542 | 63.6 | 65 | |
| pmpI | rPmpI-FL | pmpI-1 | TCCCCCCGGGTGAAACCGCCCTCCTCAC | pET30b | 30-879 | 97.7 | 100 | |
| pmpI-2 | CTCAAGCTTTAAAACGCAATGCTAGGC | |||||||
| rPmpI-N | pmpI-3 | TCCCCCCGGGTGAAACCGCCCTCCTCAC | pET30b | 30-488 | 55.4 | 55 | ||
| pmpI-4 | AGAAAGCTTAGGGGCGTGGATAGTTAC | |||||||
Primers designed based on the published sequence of reference strain D/UW-3/Cx (GenBank accession no. 884704, 884703, 884702, 884611, 884671, 884672, 884677, 884673, and 884675, respectively).
Patients.
Four different patient populations were recruited in this study. The first sample population consisted of 40 women with mild to moderate chlamydial PID from Pittsburgh, PA, selected from the PID Evaluation and Clinical Health (PEACH) study of 831 women with clinically suspected PID, defined by pelvic pain of less than 30 days duration, pelvic organ tenderness, leukorrhea, and mucopurulent cervicitis or untreated cervicitis (27). The second population included 13- to 20-year-old female patients attending adolescent clinics in Oakland, CA, for routine sexually transmitted infection testing. Thirty-four C. trachomatis-positive serum samples and two serum samples from adolescent healthy females were collected from this group. The third population was a similar group of adolescent female patients attending Arkansas Children's Hospital Research Institute. Twenty-seven serum samples from patients with either current or previously reported infection were included. The two adolescent female populations were recruited for this study, and their demographics are summarized in Table S1 in the supplemental material. C. trachomatis infection was confirmed using the Amplicor PCR test (Roche Diagnostics) or positive serology against purified EB proteins (Little Rock, AR) and Amplicor or ompA genotyping of the sample (Oakland, CA). The last population was a group of 18- to 35-year-old male patients from Baltimore STD clinics (13). Most of these patients were symptomatic, while others were called in due to their contact with infected female patients. Fifty-eight serum samples from confirmed C. trachomatis-positive (GenProbe Aptima Combo 2 assay; GenProbe Inc., San Diego, CA) patients were collected. The four populations are designated (i) PEACH PID women, (ii) Oakland adolescent females, (iii) Arkansas adolescent females, and (iv) Baltimore male adults. Two serum samples from healthy adults, one from each gender, were also included. All patients whose sera were used in this study are a part of independent studies at each site and have been independently approved by the appropriate institutional review boards (or human subject protection committees), and informed consent for the provision of the serum samples used in this study has been obtained.
SDS-PAGE and immunoblot analyses.
Purified EBs or partially purified rPmp polypeptides (rPmpA-FL, rPmpB-N, rPmpC-N, rPmpD-FL, rPmpE-FL, rPmpF-FL, rPmpG-N, rPmpH-FL, and rPmpI-N) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5% precast criterion or 15-well gels and transferred to polyvinylidene difluoride membranes (Amersham Biosciences, NJ). Amounts of rPmps in each lane were standardized by immunoblot analysis with anti-His tag antibody (1:1,500; GE Healthcare) so that each lysate contained densitometrically equivalent amounts of full-size rPmp antigen. Serum samples were used at a dilution of 1:500 when incubated with immunoblots of purified EB lysates. For immunoblots of rPmps, all serum samples were preadsorbed 30 min prior to use with inclusion bodies of E. coli expressing an irrelevant antigen (recombinant β-galactosidase prepared by a method similar to that used for rPmps) to remove any nonspecific reactivity. Membranes were incubated with the preadsorbed serum samples at 1:100 dilution, and bound antibody was detected with alkaline phosphatase-conjugated anti-human immunoglobulin G (1:5,000; KPL, MD). The blots were visualized with the chemifluorescent alkaline phosphatase substrate ECF (Amersham Biosciences, NJ), using a Molecular Dynamics Typhoon 9400 imager (Amersham Biosciences, NJ). Duplicate experiments were performed for 65 of the 159 serum samples tested without significant variation by visual assessment or densitometric quantitation.
Quantification and statistical analysis of anti-Pmp antibody reactivity of C. trachomatis-infected patient sera.
The antibody reactivity against the highest-molecular-mass band in each lane (corresponding to full-size rPmp) was analyzed using ImageQuant 5.2 image analysis software (Molecular Dynamics, Sunnyvale, CA). The serum response against each rPmp was quantified by measuring the volume of the band (band intensity × band area). The background level of each membrane was subtracted from the band intensity, and the data were normalized against His-tag-specific antibody reactivity. Because the distribution of the Pmp-specific antibody responses was highly skewed, we examined the median and interquartile ranges of the reactivity. The results are visualized using a graded color system based on the thresholds of the 25% percentile (Q1), 50% percentile (median), and 75% percentile (Q3) values. The data from the Oakland adolescent female and Arkansas adolescent female groups were pooled for the purpose of statistical analysis. Relative frequencies of the antibody response against individual rPmps in the three populations were analyzed using the chi-square test. The Mann-Whitney and Kruskal-Wallis tests were conducted to compare the relative reactivities between any two populations and those between all three populations, respectively.
RESULTS
Analysis of the antibody response to purified EBs during infection with C. trachomatis.
A subset of serum samples from the four populations of patients with C. trachomatis genital infection was randomly selected for immunoblot analysis against C. trachomatis purified EBs (Fig. 1). While antibody reactivity was undetectable for healthy (C. trachomatis-negative) control sera, all confirmed C. trachomatis-positive patient serum samples tested displayed antibody reactivities toward specific EB protein bands. This corroborates screening results from the four collaborating institutions that provided the serum samples. Moreover, it demonstrates that immunoblot analysis is a suitable method for the detection of EB-specific antibodies in infected patients. Interestingly, different patients display differential antibody responses against a wide range of polypeptides of the purified EBs, especially high-molecular-mass bands, which may correspond to Pmps.
FIG. 1.
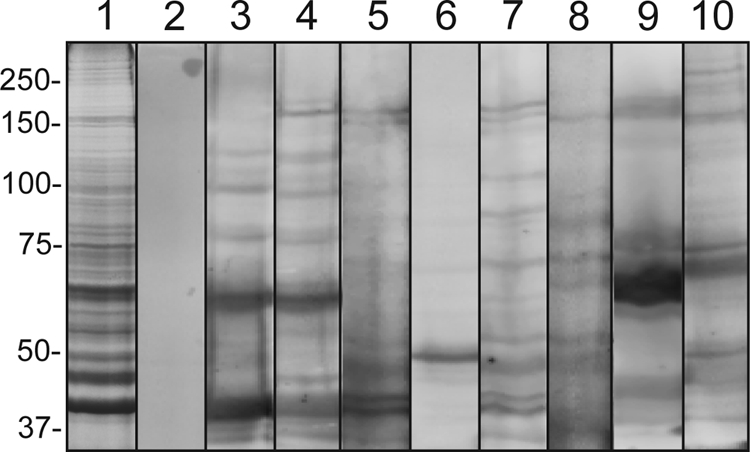
Serum reactivity of C. trachomatis-infected patients against purified EB lysates. Proteins from urographin-purified EBs were separated by SDS-PAGE and analyzed by immunoblot analysis. Lane 1, Coomassie blue-stained EB proteins; lane 2, control serum from C. trachomatis-negative patient; lanes 3 to 10, serum samples from C. trachomatis-infected patients (lanes 3 and 4, PEACH PID group; lanes 5 and 6, Oakland adolescent female group; lanes 7 and 8, Arkansas adolescent female group; lanes 9 and 10, Baltimore male adult group). Molecular mass markers (kDa) are indicated in each row.
Generation of rPmps.
To determine if C. trachomatis Pmps are antigenic and antibody responses can be elicited to Pmps in naturally infected humans, rPmps, including rPmp-FL and rPmp-N, were cloned and expressed in E. coli strain BL21. The observed molecular masses of the rPmps were consistent with those predicted from the sequence as revealed by immunoblotting with anti-His-tag antibody (Fig. 2b), except for rPmpB-N and rPmpC-N, whose apparent molecular masses were higher (Table 1). This discrepancy may be due to artifactual folding of high-molecular-mass polypeptides in the SDS-PAGE gel. Confirmation of the Pmp specificity of the recombinant polypeptides was obtained independently of this study; monospecific antibodies obtained using the recombinant Pmps as immunogens specifically recognized surface C. trachomatis antigens within inclusions by immunofluorescence or on purified EBs by immunoelectron microscopy, and polypeptides of purified EBs consistent in molecular mass with Pmp polypeptides detected by proteomic analysis (38) (C. Tan and P. M. Bavoil, unpublished data).
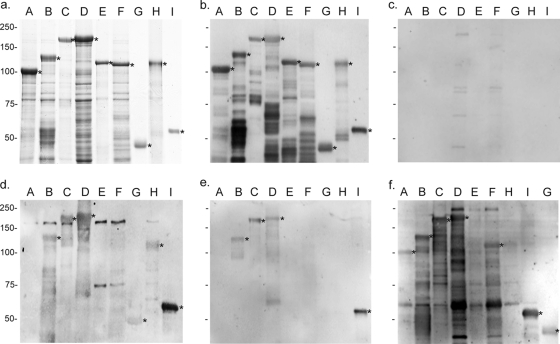
FIG. 2.
Serum reactivity of C. trachomatis-infected patients against the Pmp family. Partially purified proteins rPmpA to -I (subtypes are indicated above each lane) were fractionated by SDS-PAGE and Coomassie blue-stained (a) or immunoblotted with anti-His tag antibody (1:1,500) (b), control serum from a healthy individual (1:100) (c), or serum samples (1:100) from patients with C. trachomatis genital infection. Representatives of the PEACH PID group (serum no. 19) (d), Oakland adolescent females (serum no. 19) (e), and Baltimore male adults (serum no. 21) (f) are shown. Stars indicate specific reactivity against full-size proteins rPmpA to -I. Patient serum samples reacting with bands that do not include full-size rPmp were recorded as negative. Molecular mass markers (kDa) are indicated in each row.
Differential Pmp-specific antibody responses in patients with C. trachomatis genital infection.
The partially purified proteins rPmpA to -I were used as target antigens to analyze antibodies in serum samples from the following four patient populations with confirmed C. trachomatis genital infection by immunoblot analysis: PEACH PID women, Oakland adolescent females, Arkansas adolescent females, and Baltimore male adults. In total, we screened 159 C. trachomatis patient serum samples and four healthy, C. trachomatis-negative control serum samples in a blinded fashion. Our results indicate that all negative control serum samples from healthy individuals were nonimmunoreactive, while all but three serum samples from C. trachomatis-positive patients in the Oakland adolescent female group exhibited anti-Pmp antibody responses. Of the three Pmp-negative serum samples from infected patients, one was tested against purified EBs (Fig. 1, lane 6). Even though this patient showed anti-EB reactivity, antibodies against high-molecular-mass bands were not detected, consistent with a negative anti-Pmp response. In each population, each rPmp was recognized at least once, demonstrating the antigenicity of the recombinant polypeptides. Each Pmp-reactive serum sample displayed a unique antibody profile against individual or multiple rPmps. Representative immunoblot images are shown in Fig. 2d to f. Because lower-molecular-mass bands cannot be distinguished as Pmp specific from E. coli contaminant bands, only the reactivity against the highest-molecular-mass band corresponding to the full-size rPmp served as a representative gauge of antibody reactivity to a given Pmp in a given serum sample.
The four populations included in this study represent groups of patients that are demographically and clinically distinct, including differences in age, gender, geographical region, and clinical findings. However, because of the similarity in clinical symptoms and demographics (gender and age) the Oakland adolescent female and Arkansas adolescent female groups were pooled for all subsequent statistical analyses. We compared the frequencies of the antibody response against individual Pmps in the three populations by using chi-square analysis (Table 2). Results showed that the relative frequencies of the anti-PmpA, -E, -F, and -H responses were not statistically different, while those of the anti-PmpB, -C, -D, -G, and -I responses were statistically different among the three groups. The pooled adolescent female population displayed a statistically significant lower frequency of the antibody response across all rPmps. Despite differences in frequency of the specific responses, PmpB, -C, -D, and -I were the major Pmps recognized in all populations. For female patients, PmpB was recognized most frequently (82% of serum samples), while for male patients, PmpD reactivity was most frequent (91% of serum samples). In contrast, PmpA and -E were recognized with the lowest frequencies in all patient populations.
TABLE 2.
Frequency of serum antibody reactivity against individual rPmps
| Patient source | No. of patients | Frequency (no. of patients [%]) of antibody response against Pmp subtypea:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | ||
| PEACH PID females | 40 | 5 (13) | 37 (93) | 32 (80) | 30 (75) | 9 (23) | 17 (43) | 23 (58) | 14 (35) | 30 (75) |
| Pooled adolescent females | 61 | 10 (16) | 46 (75) | 31 (51) | 33 (54) | 6 (10) | 18 (30) | 14 (23) | 10 (16) | 27 (44) |
| Baltimore adult males | 58 | 17 (29) | 41 (81) | 48 (83) | 53 (91) | 5 (9) | 20 (34) | 15 (26) | 14 (24) | 43 (74) |
| Total | 159 | 32 (20) | 124 (78) | 111 (70) | 116 (73) | 20 (13) | 55 (35) | 52 (33) | 38 (24) | 100 (63) |
Differences in anti-Pmp antibody frequency are not statistically significant (P > 0.05) for PmpA, -E, -F, and -H, and they are statistically significant for PmpB and -G (P < 0.05) and for PmpC, -D, and -I (P < 0.001).
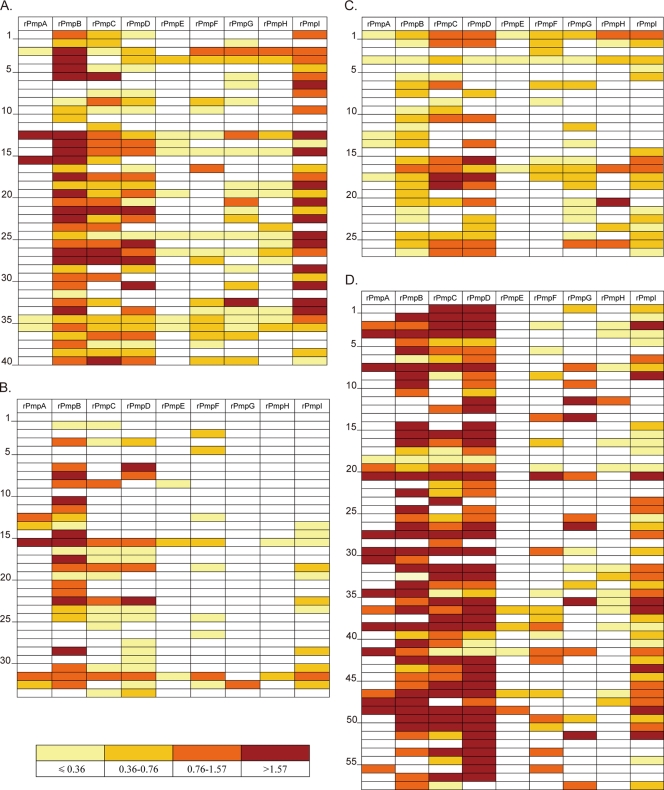
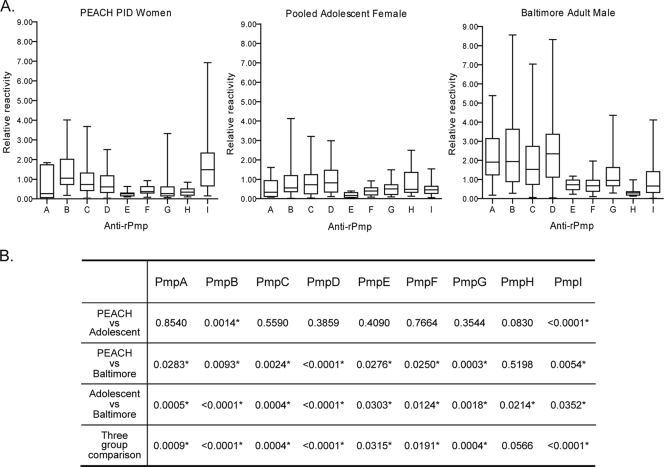
The Pmp subtype-specific antibody reactivity was quantified and normalized against the anti-His tag reactivity in order to compare the responses against individual Pmps and between different patients. We used an interquartile-value-based color grading system to display patient-specific Pmp reactivity for all populations (Fig. 3), namely, from light yellow to dark red; the antibody responses change from the lowest 25% to the highest 25% of the pooled values. Figure 3 graphically illustrates patient-to-patient variation of the anti-Pmp humoral immune response and indicates that the major antigens being recognized in all four populations were PmpB, -C, -D, and -I. The interquartile distribution of Pmp subtype-specific reactivity is further illustrated for each population in Fig. 4A. Most of the antibody reactivities against individual rPmps display a wide range between the minimum and the maximum reactivities, with a distribution highly skewed toward the lower end. Because of the skewed distribution, nonparametric tests were performed to compare the relative antibody reactivities. The Kruskal-Wallis test was used to compare Pmp-specific responses between all populations, and then, for rigor, the Mann-Whitney test was done for comparisons between each of the three paired combinations (Fig. 4B). The two female populations (PEACH PID and pooled adolescent females) showed similar reactivity against all Pmps except PmpB and PmpI, for which the PEACH PID patients had a stronger antibody response. Moreover, for each rPmp, the antibody reactivity of the Baltimore male adult group was stronger than that of either female population, except for the anti-PmpH reactivities, which were similar between the PEACH PID and Baltimore male adult groups.
FIG. 3.
Profiling of the Pmp-specific antibody responses in C. trachomatis-infected patients. The antibody reactivity against rPmps was quantified and color-coded for each patient as described in Materials and Methods. Reactivity lower than Q1 is coded as light yellow, that between Q1 and the median as gold, that between the median and Q3 as orange, and that above Q3 as dark red. (A) PEACH PID women; (B) Oakland adolescent females; (C) Arkansas adolescent females; and (D) Baltimore male adults. The patient serum numbers are indicated along the side of each panel.
FIG. 4.
Quartile distribution and statistical significance of Pmp-specific relative antibody reactivity in the three patient populations. (A) Antibody reactivity against each rPmp was quantified using ImageQuant 5.2, and the distributions of immunoreactivity were analyzed and compared in the following three patient populations: PEACH PID women, pooled adolescent females, and Baltimore male adults. The box corresponds to reactivity between Q1 and Q3 of the total distribution. The horizontal line inside the box denotes the median, and the upper and lower horizontal lines correspond to the maximum and minimum reactivities of the total distribution, respectively. (B) Statistical analysis was performed to compare the relative antibody reactivities against individual rPmps between different patient populations as described in Materials and Methods. The P values are listed for each comparison, and the asterisks indicate statistically significant difference in two or three group comparisons.
DISCUSSION
Previous studies have shown that Pmps of Chlamydia spp. are immunogenic; however, relatively little is known regarding the specificity of the immune response against these antigens in the case of C. trachomatis. The availability of a complete panel of rPmps and serum samples from C. trachomatis-infected patients allowed for the examination of Pmp-specific antibody responses at the Pmp subtype level in four different patient groups. Immunoblot analysis revealed that 156 out of 159 patients exhibited serum antibody reactivity to at least one rPmp. Reactivity to rPmp also correlated with the reactivity to high-molecular-mass protein bands ranging from 75 to 150 kDa in purified EBs of C. trachomatis (Fig. 1). This strongly suggests that the observed antibody reactivity to rPmps is representative of the serum immune response to native chlamydial Pmps. Conversely, this result suggested that immunoblot analysis using denatured rPmp antigen would be a suitable assay of antibody reactivity against native Pmps in infected patients. Notwithstanding, it is likely that patient antibodies also recognize Pmp conformational epitopes that are not represented in denatured rPmps and thus would not be detected by our method. Hence, although the immunoblot method developed in this study provides a semiquantitative assay representative of the patient antibody response to each Pmp, which is suitable for comparative analysis, it falls short of providing a true antibody titer to each specific Pmp subtype.
We quantified the antibody reactivity to each rPmp subtype for each patient. Since it was not experimentally feasible to distinguish reactivity to degraded Pmp-specific immunoreactive peptides from that to E. coli contaminating proteins, only the reactivity against the intact full-size protein was used for quantification. Our results reveal that Pmps are capable of eliciting quantitatively and qualitatively different antibody responses in patients with C. trachomatis genital infection and that individual patients mount antibody responses to different subsets of Pmp proteins. A possible cause for the observed differential Pmp immunoreactivity resides in the variable in vivo expression of the pmp genes, which may be highly regulated in the context of a genital infection. Regulated pmp expression (7, 15, 29, 31) may allow Chlamydia to alter its antigenic coat to evade host immunity, thus facilitating the development of chronic infection. A graphic summary of the anti-Pmp response in all patient serum samples is shown in Fig. 3.
Although the sequence and content of the pmp loci are highly conserved between different strains and serovars of C. trachomatis (14), differential transcriptional regulation or posttranslational modification in different strains cannot be ruled out. Moreover, intrinsic geographical, behavioral (e.g., increased frequency of repeat infection with age), or gender-specific factors are also not accounted for in our study. For instance, our comparison of C. trachomatis-infected males and females from different locations does not account for potential distinct pathogenic or serovar-specific properties of the infecting strain(s) or possible genetic differences or differences in antigen presentation in the infected human populations. A logical extension of our study will be a similar comparison of Pmp-specific antibody profiles in C. trachomatis-infected males and females from the same geographical location.
Table 2 summarizes the relative frequencies of the antibody response to each rPmp subtype in the three different patient populations. PmpB, -C, -D, and -I were more frequently recognized than were the other Pmps in all groups. This indicates that some Pmps, including PmpB, -C, -D, and -I, may be more abundantly expressed or specifically exposed at the chlamydial surface to elicit a relatively stronger antibody response. Alternatively, the antibody response to each Pmp subtype may be differentially regulated. For instance, the host immune system may preferentially recognize protein structures that are present in some but not all Pmps. Swanson et al. have recently shown that the native form of PmpD of C. trachomatis exists as an oligomer with a flower-like structure (43). They also found out that PmpD is a target of broadly cross-reactive neutralizing antibodies (10). The relatively high prevalence of PmpD-specific antibodies (73% of patients) supports the concept that PmpD may be important in the development of a multicomponent Chlamydia vaccine in the future. Gomes et al. (15) and Nunes et al. (29) have investigated the serum antibody response to recombinant forms of PmpC, -D, and -F in a group of female adolescent patients from Oakland, CA, by dot blot analysis. Results from their studies indicate that patients infected with specific strains of C. trachomatis make antibodies reactive to PmpC and PmpD, but not to PmpF, which is consistent with our observations.
Cross-sectional comparisons of anti-Pmp antibody reactivities in different patient populations are shown in Fig. 4. The two adolescent female populations were pooled for the purpose of statistical analysis since both groups include patients of similar age and from similar urban areas. Although differences in ethnicity between the two urban sites may impact disease development, the focus of our analysis is the Pmp-specific antibody profile in a heterogeneous population, irrespective of the infecting strain or the infection status. Indeed, several infecting strains are usually found in urban sites with potential additional variation within strains (25). When comparing the frequencies of the anti-Pmp response between populations, the least frequently recognized rPmps (rPmpA, -E, -F, and -H) tend to also be similarly less immunoreactive in all populations. In contrast, rPmps that are recognized more strongly and more frequently (rPmpB, -C, -D, and -I) reveal some differences in antibody response between populations. The major differences in relative reactivity of Pmp-specific antibody occur between the male population and either of the two adolescent female populations. Male patients tend to display overall stronger anti-Pmp reactivity, suggesting a possible gender specificity of the antibody response. The PID and adolescent female patients displayed similar antibody responses, except for PmpB and PmpI, for which the PID group showed stronger reactivity than did the adolescent females. This suggests that during chronic or repeated infection, infecting chlamydiae stimulate the host immune response against Pmps, further suggesting that the Pmp-specific response contributes to the development of chronic inflammation leading to PID and infertility. Furthermore, the development of strong antibody reactivity to specific Pmp subtypes (PmpB and PmpI) in PID patients implies that these patients may be more susceptible to develop inflammatory outcomes. The possibility that PmpB and PmpI antibody titers could be used as predictable markers of the onset of PID and other long-term sequelae should be investigated.
Figure 3 shows that the majority of the patient serum samples recognize multiple rPmps. Pmps are similar in sequence; hence, the possibility that some of the observed reactivity to multiple Pmps may be due to cross-reactivity cannot be excluded. Although serum samples from all four C. trachomatis-negative control samples were nonreactive against any rPmp, respiratory infection was not assessed in these patients, so the possibility that potentially cross-reactive Pmps of C. pneumoniae contributed to the observed antibody responses cannot be excluded. Antigenic cross-reactivity between PmpB and PmpC of C. trachomatis is nevertheless possible since these Pmps belong to the same subfamily and share 44% sequence identity. However, our experimental design using a complete panel of rPmps allowed heterologous rPmps to serve as internal negative controls for one another. Moreover, the human immune system most likely generates polyclonal antibodies that recognize mutually exclusive epitopes on individual Pmps. In an independent part of this project, guinea pig polyclonal antibodies to each Pmp subtype were obtained. All antibodies recognized rPmps by immunoblot analysis and specifically labeled C. trachomatis inclusions by immunofluorescence, strongly supporting the antigenic and immunogenic specificity of individual anti-Pmp antibody responses in vivo (Tan and Bavoil, unpublished).
Our study reveals varied anti-Pmp antibody profiles in patients from four geographically distinct C. trachomatis-infected populations. In addition, Pmp subtype-specific and gender-specific antibody responses are demonstrated. These observations imply variable expression of the pmp gene family during infection, suggesting that the C. trachomatis pmp gene family is the basis of a mechanism of antigenic variation for the purpose of immune evasion. Ongoing efforts include in vitro investigations of the prediction that pmp expression varies in a highly regulated manner.
Supplementary Material
Acknowledgments
This work was supported by NIH grant AI51417 to P.M.B., A.M.S. and D.D. A.M.S. and D.D. wish to acknowledge the Arkansas Biosciences Institute and the Arkansas Children's Hospital Research Institute and NIH grant AI039499 for related support. C.L.H. and R.B.N. acknowledge support from grant HS08358-05 from the Agency for Healthcare Research and Quality Development.
C.T. and P.M.B. are grateful to Kelley Hovis for editing of the manuscript; Nisha George, Udit Pandya, and Roxanne Howell for their laboratory assistance; and the members of the Oram lab for useful comments.
Editor: A. Camilli
Footnotes
Published ahead of print on 1 June 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anttila, T., P. Saikku, P. Koskela, A. Bloigu, J. Dillner, I. Ikaheimo, E. Jellum, M. Lehtinen, P. Lenner, T. Hakulinen, A. Narvanen, E. Pukkala, S. Thoresen, L. Youngman, and J. Paavonen. 2001. Serotypes of Chlamydia trachomatis and risk for development of cervical squamous cell carcinoma. JAMA 28547-51. [DOI] [PubMed] [Google Scholar]
- 2.Banu, S., N. Honore, B. Saint-Joanis, D. Philpott, M. C. Prevost, and S. T. Cole. 2002. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 449-19. [DOI] [PubMed] [Google Scholar]
- 3.Barbour, A. G. 1990. Antigenic variation of a relapsing fever Borrelia species. Annu. Rev. Microbiol. 44155-171. [DOI] [PubMed] [Google Scholar]
- 4.Birkelund, S., K. Knudsen, A. S. Madsen, E. Falk, P. Mygind, and G. Christiansen. 1998. Differential expression of Chlamydia pneumoniae Omp4 and Omp5 after infection of C57-Black mice? p. 275-282. In R. S. Stephens et al. (ed.), Chlamydial infections. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, Napa, CA.
- 5.Brunham, R. C., B. Binns, F. Guijon, D. Danforth, M. L. Kosseim, F. Rand, J. McDowell, and E. Rayner. 1988. Etiology and outcome of acute pelvic inflammatory disease. J. Infect. Dis. 158510-517. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 311161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, L. A., C.-C. Kuo, S.-P. Wang, and J. T. Grayston. 1990. Serological response to Chlamydia pneumoniae infection. J. Clin. Microbiol. 281261-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cevenini, R., M. Donati, E. Brocchi, F. De Simone, and M. La Placa. 1991. Partial characterization of an 89-kDa highly immunoreactive protein from Chlamydia psittaci A/22 causing ovine abortion. FEMS Microbiol. Lett. 81111-116. [DOI] [PubMed] [Google Scholar]
- 9.Chesson, H. W., and S. D. Pinkerton. 2000. Sexually transmitted diseases and the increased risk for HIV transmission: implications for cost-effectiveness analyses of sexually transmitted disease prevention interventions. J. Acquir. Immune Defic. Syndr. 2448-56. [DOI] [PubMed] [Google Scholar]
- 10.Crane, D. D., J. H. Carlson, E. R. Fischer, P. Bavoil, R. C. Hsia, C. Tan, C. C. Kuo, and H. D. Caldwell. 2006. Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc. Natl. Acad. Sci. USA 1031894-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dehio, C., S. D. Gray-Owen, and T. F. Meyer. 1998. The role of neisserial Opa proteins in interactions with host cells. Trends Microbiol. 6489-495. [DOI] [PubMed] [Google Scholar]
- 12.Delogu, G., and M. J. Brennan. 2001. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect. Immun. 695606-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaydos, C., N. E. Maldeis, A. Hardick, J. Hardick, and T. Quinn. 2009. Mycoplasma genitalium compared to Chlamydia, Gonorrhea, and Trichomonas as an etiologic agent of urethritis in men attending STD clinics. Sex. Transm. Infect., in press. [DOI] [PMC free article] [PubMed]
- 14.Gomes, J. P., W. J. Bruno, M. J. Borrego, and D. Dean. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 1864295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes, J. P., R. C. Hsia, S. Mead, M. J. Borrego, and D. Dean. 2005. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microb. Infect. 7410-420. [DOI] [PubMed] [Google Scholar]
- 16.Gomes, J. P., A. Nunes, W. J. Bruno, M. J. Borrego, C. Florindo, and D. Dean. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 188275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimwood, J., L. Olinger, and R. S. Stephens. 2001. Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect. Immun. 692383-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimwood, J., and R. S. Stephens. 1999. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics 4187-201. [DOI] [PubMed] [Google Scholar]
- 19.Henderson, I. R., and A. C. Lam. 2001. Polymorphic proteins of Chlamydia spp.—autotransporters beyond the proteobacteria. Trends Microbiol. 9573-578. [DOI] [PubMed] [Google Scholar]
- 20.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21385-389. [DOI] [PubMed] [Google Scholar]
- 21.Karunakaran, K. P., J. Rey-Ladino, N. Stoynov, K. Berg, C. Shen, X. Jiang, B. R. Gabel, H. Yu, L. J. Foster, and R. C. Brunham. 2008. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J. Immunol. 1802459-2465. [DOI] [PubMed] [Google Scholar]
- 22.Kiselev, A. O., W. E. Stamm, J. R. Yates, and M. F. Lampe. 2007. Expression, processing, and localization of PmpD of Chlamydia trachomatis serovar L2 during the chlamydial developmental cycle. PLoS ONE 2e568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longbottom, D., J. Findlay, E. Vretou, and S. M. Dunbar. 1998. Immunoelectron microscopic localisation of the OMP90 family on the outer membrane surface of Chlamydia psittaci. FEMS Microbiol. Lett. 164111-117. [DOI] [PubMed] [Google Scholar]
- 24.Longbottom, D., M. Russell, G. E. Jones, F. A. Lainson, and A. J. Herring. 1996. Identification of a multigene family coding for the 90 kDa proteins of the ovine abortion subtype of Chlamydia psittaci. FEMS Microbiol. Lett. 142277-281. [DOI] [PubMed] [Google Scholar]
- 25.Millman, K., C. M. Black, R. E. Johnson, W. E. Stamm, R. B. Jones, E. W. Hook, D. H. Martin, G. Bolan, S. Tavare, and D. Dean. 2004. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 1862457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ness, R. B., D. E. Soper, H. E. Richter, H. Randall, J. F. Peipert, D. B. Nelson, D. Schubeck, S. G. McNeeley, W. Trout, D. C. Bass, K. Hutchison, K. Kip, and R. C. Brunham. 2008. Chlamydia antibodies, Chlamydia heat shock protein, and adverse sequelae after pelvic inflammatory disease: the PID Evaluation and Clinical Health (PEACH) study. Sex. Transm. Dis. 35129-135. [DOI] [PubMed] [Google Scholar]
- 28.Niessner, A., C. Kaun, G. Zorn, W. Speidl, Z. Turel, G. Christiansen, A. S. Pedersen, S. Birkelund, S. Simon, A. Georgopoulos, W. Graninger, R. de Martin, J. Lipp, B. R. Binder, G. Maurer, K. Huber, and J. Wojta. 2003. Polymorphic membrane protein (PMP) 20 and PMP 21 of Chlamydia pneumoniae induce proinflammatory mediators in human endothelial cells in vitro by activation of the nuclear factor-kappaB pathway. J. Infect. Dis. 188108-113. [DOI] [PubMed] [Google Scholar]
- 29.Nunes, A., J. P. Gomes, S. Mead, C. Florindo, H. Correia, M. J. Borrego, and D. Dean. 2007. Comparative expression profiling of the Chlamydia trachomatis pmp gene family for clinical and reference strains. PLoS ONE 2e878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen, A. S., G. Christiansen, and S. Birkelund. 2001. Differential expression of Pmp10 in cell culture infected with Chlamydia pneumoniae CWL029. FEMS Microbiol. Lett. 203153-159. [DOI] [PubMed] [Google Scholar]
- 31.Puolakkainen, M., C. C. Kuo, A. Shor, S. P. Wang, J. T. Grayston, and L. A. Campbell. 1993. Serological response to Chlamydia pneumoniae in adults with coronary arterial fatty streaks and fibrolipid plaques. J. Clin. Microbiol. 312212-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 281397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 312134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocha, E. P., O. Pradillon, H. Bui, C. Sayada, and E. Denamur. 2002. A new family of highly variable proteins in the Chlamydophila pneumoniae genome. Nucleic Acids Res. 304351-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rockey, D. D., J. Lenart, and R. S. Stephens. 2000. Genome sequencing and our understanding of chlamydiae. Infect. Immun. 685473-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schachter, J. 1978. Chlamydial infections. N. Engl. J. Med. 298428-435. [DOI] [PubMed] [Google Scholar]
- 37.Schachter, J., and P. B. Wyrick. 1994. Culture and isolation of Chlamydia trachomatis, p. 377-390. In V. L. Clark and P. M. Bavoil (ed.), Methods in enzymology, vol. 236. Bacterial pathogenesis, part B. Interaction of pathogenic bacteria with host cells. Academic Press, Inc., San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, A. C., K. Gevaert, H. Demol, B. Hoorelbeke, J. Vandekerckhove, M. R. Larsen, P. Roepstorff, A. Holm, G. Christiansen, and S. Birkelund. 2002. Comparative proteome analysis of Chlamydia trachomatis serovar A, D and L2. Proteomics 2164-186. [DOI] [PubMed] [Google Scholar]
- 39.Shirai, M., H. Hirakawa, K. Ouchi, M. Tabuchi, F. Kishi, M. Kimoto, H. Takeuchi, J. Nishida, K. Shibata, R. Fujinaga, H. Yoneda, H. Matsushima, C. Tanaka, S. Furukawa, K. Miura, A. Nakazawa, K. Ishii, T. Shiba, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of outer membrane protein genes omp and pmp in the whole genome sequences of Chlamydia pneumoniae isolates from Japan and the United States. J. Infect. Dis. 181S524-S527. [DOI] [PubMed] [Google Scholar]
- 40.Souriau, A., J. Salinas, C. De Sa, K. Layachi, and A. Rodolakis. 1994. Identification of subspecies- and serotype 1-specific epitopes on the 80- to 90-kilodalton protein region of Chlamydia psittaci that may be useful for diagnosis of chlamydial induced abortion. Am. J. Vet. Res. 55510-514. [PubMed] [Google Scholar]
- 41.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282754-759. [DOI] [PubMed] [Google Scholar]
- 42.Stern, A., M. Brown, P. Nickel, and T. F. Meyer. 1986. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell 4761-71. [DOI] [PubMed] [Google Scholar]
- 43.Swanson, K. A., L. D. Taylor, S. D. Frank, G. L. Sturdevant, E. R. Fischer, J. H. Carlson, W. M. Whitmire, and H. D. Caldwell. 2009. Chlamydia trachomatis polymorphic membrane protein D is an oligomeric autotransporter with a higher-order structure. Infect. Immun. 77508-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson, N. R., C. Yeats, K. Bell, M. T. Holden, S. D. Bentley, M. Livingstone, A. M. Cerdeno-Tarraga, B. Harris, J. Doggett, D. Ormond, K. Mungall, K. Clarke, T. Feltwell, Z. Hance, M. Sanders, M. A. Quail, C. Price, B. G. Barrell, J. Parkhill, and D. Longbottom. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tundup, S., N. Pathak, M. Ramanadham, S. Mukhopadhyay, K. J. Murthy, N. Z. Ehtesham, and S. E. Hasnain. 2008. The co-operonic PE25/PPE41 protein complex of Mycobacterium tuberculosis elicits increased humoral and cell mediated immune response. PLoS ONE 3e3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandahl, B. B., A. S. Pedersen, K. Gevaert, A. Holm, J. Vandekerckhove, G. Christiansen, and S. Birkelund. 2002. The expression, processing and localization of polymorphic membrane proteins in Chlamydia pneumoniae strain CWL029. BMC Microbiol. 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Woude, M. W., and A. J. Baumler. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17581-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanhamme, L., E. Pays, R. McCulloch, and J. D. Barry. 2001. An update on antigenic variation in African trypanosomes. Trends Parasitol. 17338-343. [DOI] [PubMed] [Google Scholar]
- 49.Wehrl, W., V. Brinkmann, P. R. Jungblut, T. F. Meyer, and A. J. Szczepek. 2004. From the inside out—processing of the chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51319-334. [DOI] [PubMed] [Google Scholar]
- 50.Whitcher, J. P., M. Srinivasan, and M. P. Upadhyay. 2001. Corneal blindness: a global perspective. Bull. World Health Org. 79214-221. [PMC free article] [PubMed] [Google Scholar]
- 51.World Health Organization. 2001. p. 1-43. In Global prevalence and incidence of selected curable sexually transmitted infections: overview and estimates. World Health Organization, Geneva, Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.