Abstract
The ability of Acinetobacter baumannii to adhere to and persist on surfaces as biofilms could be central to its pathogenicity. The production of pili and a biofilm-associated protein and the expression of antibiotic resistance are needed for robust biofilm formation on abiotic and biotic surfaces. This multistep process also depends on the expression of transcriptional regulatory functions, some of which could sense nutrients available to cells. This report extends previous observations by showing that although outer membrane protein A (OmpA) of A. baumannii 19606 plays a partial role in the development of robust biofilms on plastic, it is essential for bacterial attachment to Candida albicans filaments and A549 human alveolar epithelial cells. In contrast to abiotic surfaces, the interaction with biotic surfaces is independent of the CsuA/BABCDE-mediated pili. The interaction of A. baumannii 19606 with fungal and epithelial cells also results in their apoptotic death, a response that depends on the direct contact of bacteria with these two types of eukaryotic cells. Furthermore, the bacterial adhesion phenotype correlates with the ability of bacteria to invade A549 epithelial cells. Interestingly, the killing activity of cell-free culture supernatants proved to be protease and temperature sensitive, suggesting that its cytotoxic activity is due to secreted proteins, some of which are different from OmpA.
The genus Acinetobacter is a genetically diverse group of aerobic, gram-negative, nonfermenting bacteria (48, 49). Although acinetobacters are commonly described as being ubiquitous nonpathogenic bacteria, those strains belonging to the Acinetobacter baumannii-Acinetobacter calcoaceticus cluster are emerging as relevant opportunistic human pathogens due to their increases in virulence and multidrug resistance (13, 32, 34). Members of this genus play an important role in nosocomial infections and have attracted particular attention in severe cases associated with intensive care unit patients (4, 53). These infections manifest as serious diseases in compromised human hosts, particularly in cases of ventilator-acquired pneumonia, urinary tract infections, septicemia, and wound infections. More recently, A. baumannii has emerged as a serious pathogen among soldiers returning from Iraq and Afghanistan, with those suffering penetrating injuries being at the highest risk (9, 12, 13, 32, 34, 57). These are some of the reports that demonstrate the emergence of A. baumannii as a relevant pathogen that causes severe infections in civilian and military medical facilities. These infections are extremely difficult to handle because of the multiple-antibiotic-resistance phenotype of most clinical isolates.
The capacity of A. baumannii to cause disease in compromised patients and persist in the medical environment could be attributed to its resistance to major antimicrobial drugs (34) and desiccation (18), the latter of which is greater than that described for the Enterobacteriaceae and similar to that observed for Staphylococcus aureus. This remarkable resistance phenotype could be attributed to the ability of A. baumannii clinical strains to form biofilms on abiotic surfaces (27, 45, 51, 52), particularly those isolated from catheter-related urinary or bloodstream infections as well as from a case of shunt-related meningitis (36). Similarly, the ability of A. baumannii to adhere to human bronchial epithelial cells (21, 22) could explain the colonization and persistence of this bacterial pathogen in the infected human host. In the case of the interaction with abiotic surfaces, genetic and molecular analyses showed that biofilm initiation by type strain 19606 depends on pilus production via the CsuA/BABCDE usher-chaperone assembly system when cells are statically cultured in rich broth (45). However, the pili assembled via this secretion system are not required for biofilm formation on polystyrene when cells are cultured in a chemically defined medium, although these biofilms are less robust than those formed under nutrient-rich conditions (46). Mutagenesis analysis of A. baumannii strain 307-0294 showed that the production of a homologue of the staphylococcal biofilm-associated protein (Bap) is needed for the stabilization of biofilms formed on glass (27). This role together with its cell surface location suggest that this protein, which is conserved among members of a panel of 98 Acinetobacter strains, is involved in cell-to-cell interactions that support biofilm maturation. All these observations, together with the fact that a two-component regulatory system controls biofilm development (46), indicate that biofilm formation by A. baumannii on abiotic surfaces is a multistep process that involves several cellular structures and functions, some of which are regulated in response to changes in specific environmental cues, such as changes in the concentration of free extracellular cations (21, 45). Much less is known, however, about the interaction of A. baumannii with biotic surfaces. It is apparent that the adhesion of this pathogen to human bronchial epithelial cells, as well as biofilm formation on plastic, is enhanced by the presence and expression of the blaPER-1 gene (21). However, the mechanisms by which A. baumannii interacts with biotic surfaces, such as those representing human epithelia, and the regulatory processes that could control these mechanisms remain to be characterized.
To shed some light on some of the above-mentioned mechanisms, this study examines the interaction of A. baumannii 19606 with relevant biotic surfaces such as those represented by Candida albicans and A549 human alveolar epithelial cells. C. albicans has been used to study some aspects of the pathobiology of Pseudomonas aeruginosa (16). Interactions with this fungal pathogen may reflect complex bacterial-fungal processes occurring during the colonization of the human host (55). These processes have the potential of determining the outcome of infections caused by these microbial pathogens, as was described previously for P. aeruginosa burn wound infections (15) and the observation that C. albicans is the reservoir of Helicobacter pylori (37). These possibilities, which have not been explored with A. baumannii, are worthy of consideration because of the persistence of A. baumannii in medical settings, which are shared with C. albicans, and its remarkable antibiotic resistance. In a more practical sense, this experimental model could serve as a convenient tool to examine the pathobiology of A. baumannii. A549 alveolar epithelial cells may represent the host target during the respiratory infections that A. baumannii causes in humans. This work shows that major outer membrane protein A (OmpA) plays a partial role in biofilm formation on plastic but is absolutely required for attachment to fungal filaments and epithelial cells, processes that are independent of the production of the CsuA/BABCDE-mediated pili. OmpA production and direct contact of bacteria with fungal filaments and epithelial cells also play a role in their killing by apoptosis.
MATERIALS AND METHODS
Bacterial and fungal strains and medium conditions.
The bacterial and fungal strains used in this study are listed in Table 1. All bacterial strains were cultured in Luria-Bertani (LB) agar or broth (38) at 37°C. C. albicans strains were cultured at 30°C with yeast-peptone-dextrose (YPD) broth and agar (10 g/liter yeast extract, 20 g/liter peptone, 20 g/liter dextrose, and 15 g/liter agar when necessary).
TABLE 1.
Bacterial and fungal strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| A. baumannii | ||
| 19606 | Clinical isolate, type strain | 5 |
| 144 | csuE::EZ::TN<R6Kγori/KAN-2>; derivative of 19606 | 14 |
| 3:233 | ompA::EZ::TN<R6Kγori/KAN-2>; derivative of 19606; Kmr | This work |
| 3:233C | 3:233 harboring pMU595 | This work |
| C. albicans | ||
| SC5314 | Wild-type yeast-form strain | 16 |
| tup1 strain | Mutant filamentous-form strain | 16 |
| P. aeruginosa PAO1 | Clinical isolate | D. Frank |
| E. coli | ||
| Top10 | Used for DNA recombinant methods | Invitrogen |
| DH5α | Used for DNA recombinant methods | Gibco-BRL |
| Plasmids | ||
| pWH1266 | E. coli-A. baumannii shuttle vector; Acinetobacter lwoffii plasmid cloned into the pBR322 PvuII site; Apr Tcr | 17 |
| pMU595 | pWH1266 harboring the 19606 ompA allele; Ampr | This work |
| pRK2073 | Used as a helper in plasmid conjugation; Tpr | 26 |
Ampr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance.
DNA procedures.
Total DNA was isolated either by ultracentrifugation in CsCl density gradients (29) or by using a miniscale method (2). Plasmid DNA was isolated using commercial kits (Qiagen). DNA was sequenced with standard automated DNA sequencing methods using BigDye (Applied Biosystems) chemistry on an Applied Biosystems Prism 310 or 3100 instrument. M13 forward and reverse primers (56), primers T7 and T3 (Invitrogen), or custom-designed primers were used for this purpose. Sequences were assembled using Sequencher 4.2 (Gene Codes Corp.). Nucleotide and amino acid sequences were analyzed with DNASTAR, BLAST (http://www.ncbi.nlm.nih.gov), EMBOSS (http://liv.bmc.uu.se/emboss/), and the software available through the ExPASy Molecular Biology Server (http://www.expasy.ch).
Insertion mutagenesis, mapping, and genetic complementation.
A library of 3,000 A. baumannii 19606 insertion derivatives, generated with the EZ::TN<R6Kγori/KAN-2> Tnp transposome system as described previously (14), was screened visually for defects in biofilm formation on microtiter plates stained with crystal violet (45). The biofilm phenotype of selected derivatives was confirmed with quantitative assays using 3-ml (7.5- by 1.1-cm) polystyrene tubes and crystal violet staining as described below. Chromosomal transposon insertions affecting biofilm formation were mapped by automated DNA sequencing of EcoRI-rescued plasmid DNA (45). The ompA parental allele was PCR amplified using total 19606 DNA as a template, Pfu DNA polymerase, and primers 2913 (5′-CGGGATCCGGTAAAATCACGGCAAGC-3′) and 2914 (5′-CGCCTAGGACAAAGGTGGTATGCACG-3′), both of which included BamHI restriction sites. The amplicon was ligated into the BamHI site of A. baumannii-Escherichia coli shuttle vector pWH1266 and transformed into E. coli Top10 cells. Plasmid DNA from an ampicillin-resistant, tetracycline-sensitive transformant, named pMU595, was conjugated into A. baumannii 19606 cells using triparental mating (1) with E. coli DH5α harboring pRK2073 as a helper (26). A. baumannii 19606 transconjugants harboring complementing plasmid pMU595 were recovered by plating onto Simmons citrate agar containing kanamycin (40 μg/ml) and ampicillin (500 μg/ml). The presence and stability of pMU595 in the complemented strain were confirmed by restriction analysis and DNA sequencing of plasmid DNA isolated from cells cultured in LB broth containing 200 μg/ml ampicillin.
Biofilm assays and electron microscopy.
Biofilm formation on polystyrene was assessed by crystal violet staining of cells statically cultured in LB broth as described previously (45). Biofilms were colorimetrically quantified using ratios of the optical density at 580 nm to the optical density at 600 nm to normalize the amount of biofilm formed to the total cell content of each sample. Assays were done in duplicate at least twice by using fresh biological samples each time. For scanning electron microscopy (SEM), static cultures were grown in 50-ml conical tubes with plastic coverslips semisubmerged in 5 ml of LB broth. Coverslip samples were handled and processed as described previously (45). Samples were visualized with a Zeiss Supra Gemini 35VP field emission scanning electron microscope.
Protein analysis.
Total membrane fractions were isolated from sonically disrupted bacterial cells by high-speed centrifugation (1). Proteins were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12.5% polyacrylamide gels and either stained with Coomassie blue or electrotransferred onto nitrocellulose filters (1). The protein blots were probed with anti-OmpA serum, and the immunocomplexes were detected by chemiluminescence using protein A labeled with horseradish peroxidase (1). Polyclonal anti-OmpA serum was raised in rabbits injected with OmpA electroeluted from polyacrylamide gels after total membrane proteins were size fractionated by SDS-PAGE and stained with Coomassie blue (1). Protein electroelution was done with an IBI electroeluter under the conditions recommended by the instrument manufacturer. Specific antibodies were immunopurified with nitrocellulose strips containing the OmpA protein band (30).
Bacterial-fungal attachment and killing assays.
C. albicans SC5314 and the tup1 isogenic constitutive filamentous derivative (7) were grown in YPD broth at 30°C for 48 h prior to being washed in preconditioned medium (PCM). PCM, which described previously (16), is defined as the filter-sterilized supernatant of an early-stationary-phase-grown bacterial culture. Bacterial cultures were grown overnight in LB broth at 37°C with constant shaking. After washing in their respective PCM by centrifugation, approximately 2 × 106 bacterial cells (in 0.5 ml PCM) were added to 0.5 ml PCM containing either 1 × 106 SC5314 cells or 1 × 105 tup1 strain cells. One-milliliter cocultures were incubated without shaking at 37°C for 24 h to 72 h prior visualization using fluorescence or electron microcopy. For electron microscopy, coincubation samples grown for 24 h were fixed, dehydrated, critical-point dried, gold coated, and visualized with a Zeiss Supra 85V Gemini series scanning electron microscope as described previously (45). Micrograph observations were confirmed by counting the number of visible bacteria attached to 50 μm of fungal filaments in 10 different fields of each sample. For fluorescence microscopy, 50-μl aliquots were taken from each sample after 24 h of coincubation, washed with phosphate-buffered saline (PBS), and stained using the Live/Dead Backlight viability stain (Molecular Probes). Samples were examined with an Olympus FV500 laser scanning confocal microscope. C. albicans cells incubated in sterile LB broth and bacterial cells either treated with 0.5% formaldehyde or heated at 65°C for 30 min prior to addition to the fungal cells were used as controls. The secretion of potential bacterium-killing factors was examined by incubating fungal cells with culture supernatants grown overnight that were obtained from each bacterial strain and which were filtered through a 0.22-mm filter. The thermostability and protease sensitivity of killing factors secreted by bacterial cells were tested by preincubating cell-free culture supernatants at 65°C for 30 min or with 50 μg/ml proteinase K for 30 min at 37°C followed by the addition of phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 100 μg/ml. P. aeruginosa PAO1 cells and cognate culture supernatants were used as positive controls. Fungal killing results obtained with microscopy were confirmed by viable plate counts. For this purpose, bacterial cell-fungal cell cocultures were established as described above, and at 24-h intervals, 10 μl of each sample was taken, diluted in YPD broth, and plated onto YPD agar containing 60 μg/ml tetracycline, 30 μg/ml chloramphenicol, and 30 μg/ml gentamicin. CFU were determined after plates were incubated at 30°C for 48 h. These experiments were repeated three times using fresh samples each time.
Evaluation of apoptosis using the TUNEL assay.
Fungal cell death due to apoptosis was tested with the DeadEnd terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (Promega) as previously described (35), with some modifications. Briefly, fungal filaments were grown and used in attachment assays as described above. After 24 h of coincubation, a 100-μl sample was taken and washed vigorously with PBS solution three times. Next, the fungal samples were fixed with 4% formaldehyde in PBS at room temperature for 2 h, and the samples were then suspended in 100 μl of fresh 4% formaldehyde in PBS and placed onto a poly-l-lysine-coated glass coverslip for 2 h. Afterward, the cells were washed three times with PBS, protoplasted for 15 min as previously described (35), and then permeabilized with 0.2% Triton X-100 in PBS for 5 min. The coverslips were then washed twice with PBS and equilibrated with reaction buffer for 10 min, and DNA fragments were labeled via a deoxynucleotidyltransferase reaction at 37°C for 1 h. The coverslips were immersed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) solution provided with the kit for 15 min to stop the end-labeling reaction. After washing twice with PBS, the coverslips were mounted and examined with an Olympus AX-70 wide-field multimode microscope.
A549 adhesion and invasion assays.
To determine bacterial adhesion, A549 human alveolar epithelial cells were grown in 5% CO2 at 37°C in Ham's medium supplemented with 10% heat-inactivated fetal bovine serum to 70% confluence in six-well plates or on tissue culture-treated plastic coverslips placed in six-well plates. Confluent monolayers were washed three times with prewarmed PBS solution before infection with 2 × 106 bacterial cells per well and subsequent incubation for 60 min at 37°C. For quantitative purposes, the infected monolayers were washed three times with PBS and then lysed in 1 ml of sterile deionized water. Dilutions of the lysates were plated onto LB agar and incubated at 37°C for 24 h. The following day, the colonies were counted to determine the number of bacteria that attached to A549 cells. For microscopy purposes, the coverslips were washed five times with prewarmed PBS and fixed with 4% formaldehyde in PBS for 2 h. For SEM analysis, coverslip samples were secondarily fixed with 1.5% osmium tetroxide for 1 h before dehydration with progressively increasing concentrations of ethanol. The coverslips were then subjected to critical-point drying before being mounted and coated with 20 nm of gold. Samples were viewed with a Zeiss Supra Gemini 35VP field emission scanning electron microscope. For TUNEL assays, samples were processed as described above and examined with an Olympus AX-70 wide-field multimode microscope. Experiments using cell-free culture supernatants were done as described above for the analysis of bacterium-fungal filament interactions.
To determine bacterial invasion, A549 monolayers were grown in six-well plates, infected, and washed as described above for the adhesion assays. The monolayers were then treated with 2 ml of gentamicin (10 mg/ml in PBS) for 30 min, the shortest time point that resulted in the killing of all bacteria added to the monolayers. Afterward, the cells were washed three times with PBS, and the cells were trypsinized and resuspended in PBS. Epithelial cells were collected by centrifugation and lysed in 1 ml of sterile deionized water. Dilutions of the lysates were plated onto LB agar and incubated at 37°C for 24 h. The following day, the colonies were counted to determine the number of bacteria that had invaded the eukaryotic cells. The adhesion and invasion assays were repeated three times using fresh biological samples each time.
RESULTS
Role of an OmpA homologue in biofilm formation on plastic.
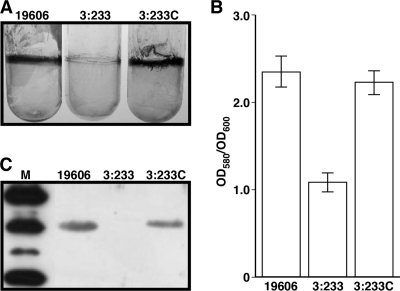
Screening of 3,000 A. baumannii EZ::TN<R6Kγori/KAN-2> insertion mutants using microtiter plate biofilm assays resulted in the identification of several derivatives that were affected in biofilm formation as reported previously (14, 45). Among them were derivatives in which biofilm formation was diminished but not abolished when cells were statically cultured overnight in LB broth. The biofilm-deficient phenotype of one of these derivatives, A. baumannii 19606 mutant strain 3:233, was confirmed using 3-ml polystyrene tubes, where it formed significantly reduced biofilms compared with those formed by the parental strain under the same experimental conditions (Fig. 1A). Quantitative biofilm assays proved that there was a significant difference between the biofilms formed by the 19606 parental strain and the 3:233 isogenic insertion derivative (Fig. 1B). Rescue cloning and nucleotide sequence analysis of the 3:233 derivative mapped the transposon insertion within a gene coding for a translation product that is highly related (E values higher than 1e−5) to major outer membrane protein A described for a wide range of bacteria, including A. baumannii (6, 11, 50), which plays a role in several important cellular functions (42). This coding region represents a putative monocistronic operon located between two divergently transcribed predicted genes coding for a conserved hypothetical protein and a FimT-related protein. The role of ompA in biofilm formation was confirmed by the observation that the conjugation of pMU595, which carries only the ompA wild-type allele cloned into Escherichia coli-A. baumannii shuttle vector pWH1266, into 3:233 was enough to restore the parental biofilm phenotype in this isogenic derivative (Fig. 1A and B). Taken together, these observations indicate that the biofilm defect of the 3:233 derivative is not due to polar effects or multiple insertions of the transposon used to generate the insertion bank screened in this work. It is important to note that the parental strain, the 3:233 OmpA mutant, and the 3:233C OmpA-complemented derivative did not display significant differences in growth rates when cultured in shaken or static LB broth (data not shown). Furthermore, restriction analysis and DNA sequencing proved that pMU595 was stably maintained as an independent replicon without detectable rearrangements in the complemented derivative 3:233C (data not shown).
FIG. 1.
Biofilm assays and detection of OmpA produced by A. baumannii 19606 derivatives. (A) Crystal violet staining of biofilms formed on 3-ml plastic tubes by parental strain 19606, the OmpA-deficient derivative 3:233, and ompA-complemented strain 3:233C. (B) Quantification of the biofilms shown in A. Error bars, 1 standard deviation (SD). OD580, optical density at 580 nm. (C) Western blotting of total membrane proteins (7 μg per lane) isolated from cells of the strains shown in A. Lane M, molecular mass markers. The top, middle, and bottom most-intense bands represent the mobilities of the 50-kDa, 37-kDa, and 25-kDa markers, respectively. After SDS-PAGE, proteins were blotted onto nitrocellulose and probed with anti-OmpA antiserum.
Immunoblot analysis with anti-OmpA serum showed that the expression of ompA and the formation of biofilms on plastic are associated with the production of a 38-kDa protein that was detected in the total membrane fraction of 19606 cells (Fig. 1C). Such a protein band was also detected in the total membrane fraction of 3:233 mutant cells only when they harbored the pMU595 ompA complementing plasmid.
Effect of ompA inactivation on biofilm architecture.
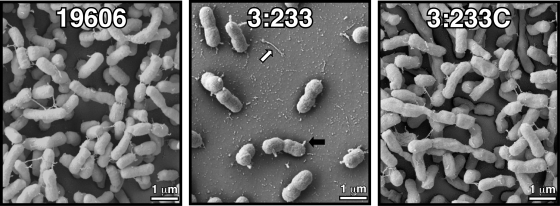
SEM analysis of biofilms formed on plastic showed that, as expected from results reported in previous work (45), parental strain 19606 formed cell aggregates at the liquid-air interface with an architecture compatible with that of bacterial biofilms (Fig. 2). In contrast, the 3:233 OmpA mutant formed significantly simpler and fewer cell aggregates. The complementation of strain 3:233 with a plasmid copy of the parental ompA allele resulted in the formation of biofilms with a structure similar to that displayed by parental strain 19606 (Fig. 2). These observations are in agreement with the results obtained with crystal violet biofilm assays (Fig. 1).
FIG. 2.
SEM analysis of bacterial biofilms formed on plastic surfaces at the liquid-air interface. The black and white arrows indicate cell protrusions and pili on the plastic surface, respectively.
It is interesting to note that unlike the filaments that connect the 19606 biofilm cells, those produced by the 3:233 OmpA mutant were instead found to be detached from the cell clusters on the plastic surface (Fig. 2). Furthermore, a large proportion of the 3:233 mutant cells showed the presence of short and round projections (Fig. 2) not seen on the 19606 biofilm cells. These short round projections may represent vesicle-like structures protruding from the cell surface rather than truncated pili, since almost no cell appendages could be observed on the surface of the 3:233 mutant cells. These cell structure alterations may reflect those observed previously during transmission electron microscopy analysis of the A. baumannii 19606 KS37 ompA mutant (11).
Attachment to eukaryotic cells.
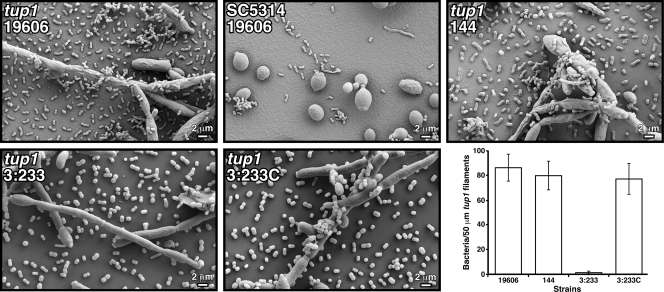
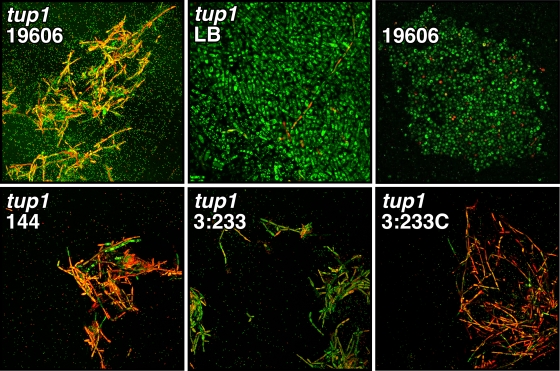
Most of the work reported so far has examined the interaction of A. baumannii with abiotic surfaces, and little is known about the interaction of this pathogen with biotic surfaces that could be relevant to the pathobiology of this bacterium. To address this issue, we adapted the C. albicans model used to study P. aeruginosa virulence (16). SEM analysis of fungal-bacterial cocultures showed that A. baumannii 19606 cells attached to C. albicans tup1 strain filaments, where they formed aggregates in which bacterial cells were stacked on top of each other on the surface of the fungal filaments (Fig. 3). Such structures were not detected when this fungal strain was incubated in sterile LB broth (data not shown). In contrast, the bacterial cells did not attach to SC5314 cells, which showed the expected yeast cell morphology (Fig. 3). Although not shown, P. aeruginosa PAO1 cells, which were used as a control, displayed a similar behavior. This observation is in agreement with the results previously reported for this experimental model (16). Interestingly, the A. baumannii 144 derivative, which does not attach to and form biofilms on plastic due to the inactivation of csuE and the consequential lack of CsuA/BABCDE-mediated pilus production (45), also attached to the fungal filaments and formed structures similar to those seen with parental strain 19606 (Fig. 3). In contrast to the results obtained with the parental strain and the 144 derivative, the interruption of the ompA gene resulted in a drastic defect in fungal cell-bacterial cell interactions, with almost no bacterial cells being attached to tup1 strain filaments after coincubation for 24 h (Fig. 3). However, the attachment phenotype of this 19606 isogenic insertion derivative was restored to the level of the parental strain when it was genetically complemented with pMU595 (Fig. 3). Figure 3 (bottom right) shows that micrograph observations are supported by the quantification of visible bacteria attached to tup1 strain filaments.
FIG. 3.
SEM analysis of bacterial attachment to C. albicans cells. SC5314 yeast cells and tup1 strain filaments were coincubated with bacterial cells from parental strain 19606 or cells from the isogenic derivative 144, 3:233, or 3:233C. All micrographs were taken at a ×2,000 magnification. The bottom right represents the quantification of visible bacteria attached to 50 μm of tup1 strain filaments. Error bars, 1 SD.
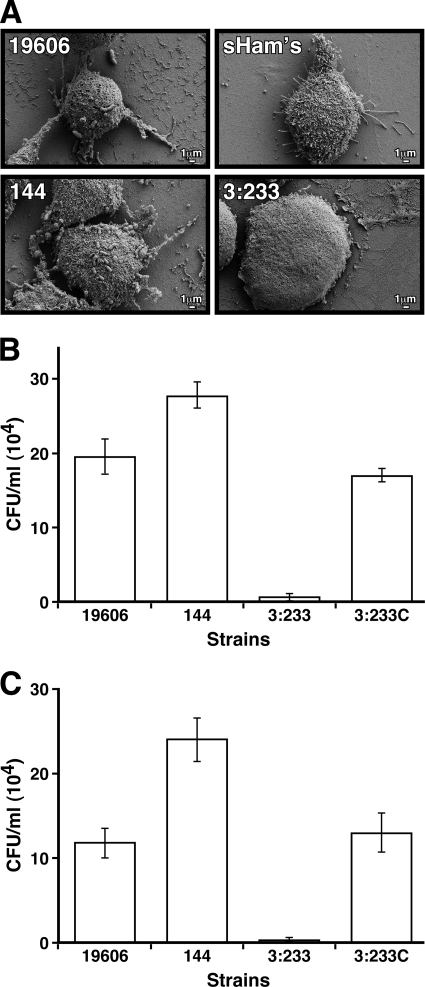
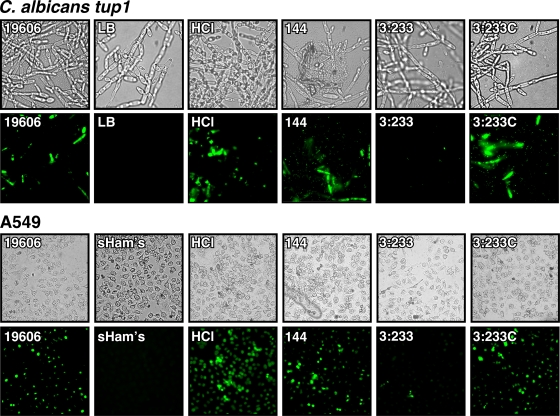
We also examined the interaction of A. baumannii 19606 with A549 human alveolar epithelial cells, which could be the host target during the respiratory infections that this pathogen causes in humans. SEM showed that 19606 cells attached to the surface of A549 cells, causing cell rounding, a loss of cell projections, as well as their detachment from the plates after a 30-min infection, a response which was also detected when monolayers were infected with 144 bacterial cells (Fig. 4A). Interestingly, microscopy observations and attachment assays showed that more 144 bacterial cells adhered to the surface of the epithelial cells than parental 19606 cells (Fig. 4A and B). In contrast to the results obtained with the parental strain and CsuE mutant strain 144, infection of A549 monolayers with 3:233 OmpA-deficient cells resulted in almost no bacterial attachment, although their presence in the tissue culture medium caused an almost total loss of cell projections and a remarkable flattening of the eukaryotic cells (Fig. 4A and B). Results obtained with invasion assays paralleled the electron microscopy and attachment data showing that in contrast to 3:233 OmpA mutant cells, whose invasion ability was drastically impaired, cells of parental strain 19606 and complemented strain 3:233C were able to invade A549 cells to comparable levels, while CsuE mutant strain 144 displays an enhanced invasion phenotype (Fig. 4C).
FIG. 4.
Bacterial adhesion and invasion of A549 alveolar epithelial cells. (A) SEM analysis of A549 cells incubated in sterile supplemented Ham's tissue culture medium (sHam's) or in the presence of cells from parental strain 19606 or cells from the isogenic derivative 144, 3:233, or 3:233C. (B) Bacterial counts obtained after A549 cells were infected and lysed. (C) Bacterial counts obtained after A549 cells were infected, treated with gentamicin, and then lysed. Error bars, 1 SD.
Killing of eukaryotic cells.
The coincubation of A. baumannii 19606 cells with C. albicans tup1 strain filaments resulted in, in addition to bacterial attachment, significant killing of fungal filaments as determined by laser scanning confocal microscopy (LSCM) of Live/Dead-stained samples (Fig. 5). In contrast, very few tup1 strain filaments and SC5314 yeast cells were dead when they were incubated in sterile LB broth or in the presence of 19606 cells, respectively (Fig. 5). Figure 5 also shows that the presence of A. baumannii 144 cells also resulted in bacterial attachment and significant killing of tup1 strain filaments. However, the killing of the fungal filaments was significantly reduced but not abolished when they were coincubated with A. baumannii 19606 mutant 3:233 OmpA-deficient cells. The killing-deficient phenotype of the 19606 OmpA mutant was reversed to that of the parental strain or the 144 derivative when it was genetically complemented with pMU595. It is important to note that although the magnitude of the effect produced by the presence of 3:233 cells is lower than that observed with the parental strain, it was significantly higher than that observed after the incubation of tup1 strain cells in sterile LB broth (Fig. 5).
FIG. 5.
LSCM of Live/Dead-stained tup1 strain filaments or SC5314 yeast cells incubated in sterile LB broth or in the presence of 19606 parental cells or cells from the isogenic derivative 144, 3:233, or 3:233C. Micrographs were taken at a ×400 magnification.
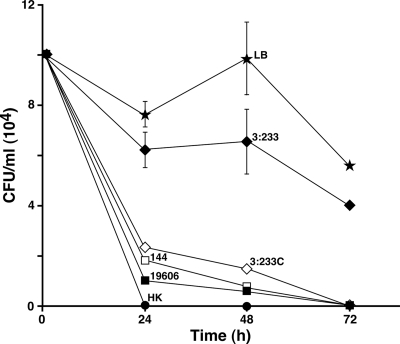
The microscopy results described above were confirmed with viable plate count assays, which showed that the presence of 19606, 144, and 3:233C cells results in a drastic reduction in the number of viable tup1 strain filaments after 24 h of coincubation, with almost complete fungal death by 72 h compared with the CFU values obtained by incubating the fungal filaments with sterile LB broth (Fig. 6). In contrast, the coincubation of C. albicans tup1 strain cells with A. baumannii 19606 mutant strain 3:233 cells resulted in a rather milder, although significant, reduction of the viability of the fungal filaments throughout the course of the 72-h incubation period.
FIG. 6.
C. albicans viability assays. tup1 strain filaments were incubated in sterile LB broth or in the presence of bacterial cells from parental strain 19606 or the isogenic derivative 144, 3:233, or 3:233C. Heat-killed (HK) tup1 strain filaments were used as a negative control. The results represent three independent experiments performed under the same experimental conditions. Error bars, 1 SD.
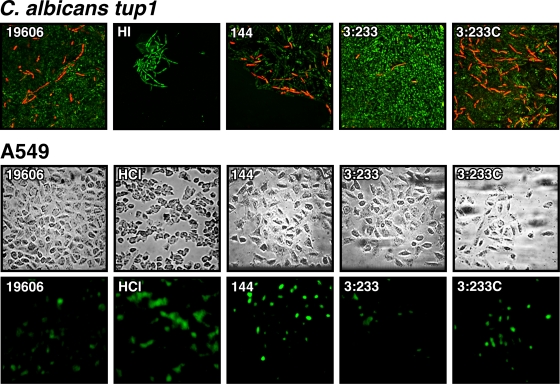
The possibility that A. baumannii 19606 kills tup1 strain filaments by apoptosis, as was previously reported for human epithelial cells (10, 11, 22, 23), was tested using a TUNEL assays as described previously for testing the role of Ras in C. albicans programmed cell death (35). Figure 7 (top) shows that the incubation of fungal filaments in the presence of 19606 cells resulted in positive apoptosis signals that were also detected when the filaments were incubated with HCl, which was used as a positive control (43; K. Del Rio-Tsonis, Miami University, personal communication), but not when tup1 strain filaments were incubated with sterile LB broth, which was used as a negative control. The presence of 144 and complemented 3:233C cells also induced apoptosis, while almost no apoptotic signals were observed when the fungal filaments were coincubated with 3:233 OmpA-deficient cells. Infection of A549 monolayers with 19606, 144, and 3:233C cells also caused detectable apoptosis, as was observed when the alveolar cells were incubated in supplemented Ham's medium containing 2 M HCl. Such a response was either absent or almost abolished when the monolayers were incubated with sterile Ham's medium or infected with 3:233 OmpA-deficient bacteria, respectively (Fig. 7, bottom).
FIG. 7.
TUNEL assays of C. albicans tup1 strain filaments and A549 monolayers cultured in sterile media (LB and supplemented Ham's medium [sHam's]) in the absence or in the presence of 2 M HCl (HCl), conditions which were used as negative and positive controls, respectively. The eukaryotic cells were also coincubated with bacterial cells from parental strain 19606 or with cells from the isogenic derivatives 144, 3:233, and 3:233C. Fungal filaments and A549 cells were imaged at ×600 and ×400 magnifications, respectively.
Effect of bacterial culture supernatants on eukaryotic cell viability.
Whether the ability of A. baumannii to kill eukaryotic cells depended on the direct interaction of bacterial cells with fungal filaments and alveolar epithelial cells was tested by repeating the killing assays described above using cell-free supernatants of bacterial cultures grown overnight. Figure 8 (top) shows that the incubation of tup1 strain filaments with A. baumannii 19606 and 144 culture supernatants for 24 h results in readily detectable fungal death. This phenomenon was not observed when the 19606 cell-free culture supernatant was heat inactivated at 65°C for 30 min (Fig. 8) or treated with proteinase K and PMSF (Fig. 9) prior to addition to the fungal filaments. A much more reduced killing effect was detected when the tup1 strain filaments were incubated with the culture supernatants of OmpA-deficient strain 3:233, although this defect was corrected when culture supernatants of ompA-complemented strain 3:233C were used in these assays (Fig. 8). Testing of A549 monolayers using a similar approach showed that the presence of cell-free supernatants from 19606, 144, and 3:233C cultures or the presence of 2 M HCl caused similar killing effects, while the presence of the OmpA mutant strain 3:233 culture supernatant resulted in an apparent reduction in the apoptotic signal (Fig. 8). As observed with the tup1 strain filaments, the heat inactivation of cell-free culture supernatants abolished their capacity to induce apoptosis in A549 cells (data not shown).
FIG. 8.
Killing of tup1 strain filaments and A549 cells by cell-free bacterial culture supernatants. (Top) LSCM of Live/Dead-stained tup1 strain filaments incubated with cell-free supernatants of cultures of strains 19606, 144, 3:233, and 3:233C grown overnight. HI, the 19606 culture supernatant was heat inactivated prior to addition to fungal filaments. Micrographs were taken at a ×400 magnification. (Bottom) TUNEL assays of A549 monolayers cultured in supplemented Ham's (sHam's) medium containing 2 M HCl (HCl) or cell-free supernatants of cultures of strains 19606, 144, 3:233, and 3:233C grown overnight. Fungal filaments and A549 cells were imaged at ×600 and ×400 magnifications, respectively.
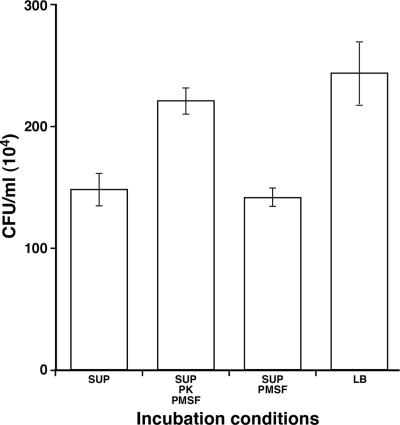
FIG. 9.
Protease sensitivity of the cytotoxic factor(s) secreted by A. baumannii 19606. Fungal filaments were coincubated for 24 h with stationary-phase untreated 19606 cell-free supernatants (SUP) or cell-free supernatants that were treated with either proteinase K (PK) and PMSF or PMSF prior coincubation with tup1 strain filaments. Coincubation with sterile LB broth was used as a control. Error bars, 1 SD.
DISCUSSION
Screening of an A. baumannii 19606 bank of insertion derivatives for mutants in which biofilm formation was altered but not abolished resulted in the identification of strain 3:233. This isogenic derivative harbors a transposon insertion within a gene coding for a product that has the same predicted amino acid sequence (GenBank accession number AY485227) reported for A. baumannii 19606 outer membrane protein 38 (11). Although that report was the first to provide evidence for the apoptotic role of this protein, that work and subsequent publications from the same research group (10, 19, 24, 25) did not provide experimental evidence supporting the role of this outer membrane protein in the ability of A. baumannii to interact with abiotic and biotic surfaces. Thus, the data presented in this report indicate that although not essential, as it is in the case of CsuA/BABCDE pili (45), OmpA is needed for the formation of robust biofilms on polystyrene. Whether OmpA plays a role at the initiation or maturation step of biofilm development is not clear at present. It is also unclear whether OmpA plays a direct or indirect role in bacterial attachment and biofilm formation. Our electron microscopy data indicate that OmpA inactivation results in apparent alterations of the bacterial cell wall, probably due to the destabilization of the outer membrane, as evidenced by the appearance of vesicle-like structures on the cells. Such a possibility is supported by the importance of OmpA in stabilizing the outer membrane of E. coli K1 (54) and the observation that only the inner membrane could be detected in cells of the A. baumannii 19606 OmpA isogenic-deficient derivative KS37 (11). Based on these considerations, it is tempting to speculate that the biofilm defect of the 3:233 insertion derivative is due to the lack of or poor anchoring of the CsuA/BABCDE pili on the cell surface, which are needed for the interaction with plastic surfaces (45). Although this is an interesting hypothesis, the possibility that OmpA also has a direct function in the interaction of cells with abiotic surfaces cannot be discarded, considering the role of OmpA in biofilm formation by E. coli. OmpA is upregulated in clinical and laboratory isolates of E. coli during biofilm formation in hydrogels (31) and plays a role in the interaction of cells with abiotic surfaces (28), and its absence significantly reduces biofilm mass when E. coli is cultured under nutrient-rich conditions in flow cell devices (3). Unfortunately, none of these reports indicated whether changes in or an abolishment of OmpA production is associated with the type of alterations in cell appearance that we describe in this report.
In contrast to the results obtained with abiotic surfaces, OmpA proved to be essential for the ability of A. baumannii 19606 to attach to biotic surfaces such as those represented by C. albicans and human alveolar epithelial cells. Equally contrasting is the fact that the interaction of A. baumannii with these two types of eukaryotic cells is independent of the CsuA/BABCDE-mediated pili. Taken together, these observations indicate that different A. baumannii cell products and structures, most of which remain to be identified, are involved in the interaction of this pathogen with abiotic and biotic surfaces. Furthermore, the differential attachment response that we obtained with C. albicans tup1 strain filaments and SC5314 yeast cells, which mimics the results obtained with P. aeruginosa when tested with this experimental model (16), indicates that the composition of the eukaryotic cell surface is also an important factor in eukaryote-pathogen interactions. It is interesting that not only is the A. baumannii-A549 interaction pilus independent, but the absence of these cell appendages also favors bacterial attachment and invasion of epithelial cells. This could be due to a better exposure of other unknown bacterial adhesion and biofilm factors when these pili are absent. The adhesion of A. baumannii to human alveolar epithelial cells is in agreement with data from previous reports describing the interaction of this bacterial pathogen with other types of human respiratory cells such as human bronchial epithelial cells (21, 22). This indicates that this pathogen interacts with different components of the human respiratory tract. However, our observation that OmpA is required for attachment to epithelial cells contradicts data reported previously by Choi et al. (11). This disagreement could be due to either differences in experimental conditions or the utilization of different cell lines. In spite of these discrepancies, our observations regarding the role of this major outer membrane protein in host-pathogen interactions are supported by reports showing that OmpA is involved either in bacterial attachment and the differential production of type 1 fimbriae in E. coli K1 (39, 44), the etiological agent of meningitis in neonates, or in the adhesion process involving other relevant human bacterial pathogens, such as enterohemorrhagic E. coli (47), nontypeable Haemophilus influenzae (40), as well as the animal pathogen Mannheimia haemolytica (20). Accordingly, in silico analysis of the predicted structure of A. baumannii 19606 OmpA shows structural and amino acid sequence features that are central to the adhesion role of this protein (42), predictions which are being tested by amino acid deletions or changes using site-directed approaches.
Our data also present strong experimental evidence showing that infection of A549 human alveolar epithelial cells leads to apoptotic death, an observation that is in agreement with the results obtained after infection or incubation of Hep-2 human laryngeal epithelial cells with A. baumannii 19606 cells or purified OmpA (11). Our observation that A. baumannii 19606 cells also attach to and kill C. albicans tup1 strain filaments but not SC5314 yeast cells also resembles data from a recent report by Peleg et al. (33). However, the killing of tup1 strain filaments by apoptosis is a novel observation. Furthermore, our data show that the death of both types of eukaryotic cells depends largely on the production of OmpA. Also novel is our observation that the A. baumannii 19606 cytotoxic effect is independent of bacterial attachment to epithelial cells and fungal filaments via CsuA/BABCDE pili. The A. baumannii 19606 144 derivative, which harbors an insertion in csuE that abolishes pilus assembly mediated by this secretion system (45), attaches to and kills fungal filaments and epithelial cells as well as 19606 parental cells. Taken together, all these observations indicate that factors other than these pili, which remain to be identified and characterized, play a role in the interaction of A. baumannii with eukaryotic cells.
Finally, the observation that cell-free supernatants of A. baumannii 19606 cells cultured in the absence of eukaryotic cells have cytotoxic activity indicates that this pathogen secretes cytotoxic factors independently of the presence of fungal filaments or epithelial cells. Although this response is independent of the production of CsuA/BABCDE pili, it is apparent that OmpA plays a role in the production of secreted unknown cytotoxic compounds that proved to be heat and protease sensitive. Furthermore, the fact that the response obtained with culture supernatants is lower than that detected with bacterial cells indicates that contact between bacteria and eukaryotic cells could be an important factor. The possibility of the production of cell contact-dependent factors is supported by the observation that the A. baumannii 17978 genome (41) has eight genes located in a pathogenicity island, which could code for putative functions related to the Legionella-Coxiella type IV secretion apparatus (41). These genes together with additional chromosomal conjugation functions may play a role in the virulence of A. baumannii, as was previously described for other human bacterial pathogens (8). Interestingly, preliminary experiments showed that strain 17978 has a response similar to that displayed by 19606 when tested under the same experimental conditions.
In conclusion, this report extends previous observations regarding the nature and effects of the interactions of A. baumannii 19606 with abiotic and biotic surfaces. It is apparent that the ability of A. baumannii to form biofilms is multifactorial and diverse as well as adaptable to the nature of surfaces with which cells are interacting. Similarly relevant is the production of bacterial factors that also determine the outcome of these interactions when particular types of biotic surfaces are involved. The elucidation of these complex interactions is important not only for a better understanding of the pathobiology of A. baumannii but also for the identification of novel targets for future antimicrobial strategies as the age of antibiotics begins to wane.
Acknowledgments
This work was supported by funds from Public Health Service grant AI070174, NSF grant 0420479, and Miami University research funds.
We are grateful to R. Kolter (Harvard Medical School) and D. Frank (Medical College of Wisconsin) for providing the C. albicans and P. aeruginosa PAO1 strains used in this work. We also thank E. Lafontaine (College of Veterinary Medicine, University of Georgia) for providing the A549 cell line.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Actis, L. A., S. A. Potter, and J. H. Crosa. 1985. Iron-regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J. Bacteriol. 161736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barcak, J. G., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204321-432. [DOI] [PubMed] [Google Scholar]
- 3.Barrios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93188-200. [DOI] [PubMed] [Google Scholar]
- 4.Bergogne-Berezin, E. 2001. The increasing role of Acinetobacter species as nosocomial pathogens. Curr. Infect. Dis. Rep. 3440-444. [PubMed] [Google Scholar]
- 5.Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36228-240. [Google Scholar]
- 6.Bratu, S., D. Landman, D. A. Martin, C. Georgescu, and J. Quale. 2008. Correlation of antimicrobial resistance with β-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York City. Antimicrob. Agents Chemother. 522999-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277105-109. [DOI] [PubMed] [Google Scholar]
- 8.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC. 2004. Acinetobacter baumannii infections among patients at military medical facilities treating U.S. service members, 2002-2004. MMWR Morb. Mortal. Wkly. Rep. 531063-1066. [PubMed] [Google Scholar]
- 10.Choi, C. H., S. H. Hyun, J. Y. Lee, J. S. Lee, Y. S. Lee, S. A. Kim, J. P. Chae, S. M. Yoo, and J. C. Lee. 2008. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell. Microbiol. 10309-319. [DOI] [PubMed] [Google Scholar]
- 11.Choi, C. H., E. Y. Lee, Y. C. Lee, T. I. Park, H. J. Kim, S. H. Hyun, S. A. Kim, S. K. Lee, and J. C. Lee. 2005. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 71127-1138. [DOI] [PubMed] [Google Scholar]
- 12.Davis, K. A., K. A. Moran, C. K. McAllister, and P. J. Gray. 2005. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 111218-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5939-951. [DOI] [PubMed] [Google Scholar]
- 14.Dorsey, C. W., A. P. Tomaras, and L. A. Actis. 2002. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 686353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta, N., A. Haque, G. Mukhopadhyay, R. P. Narayan, and R. Prasad. 2005. Interactions between bacteria and Candida in the burn wound. Burns 31375-378. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 2962229-2232. [DOI] [PubMed] [Google Scholar]
- 17.Hunger, M., R. Schmucker, V. Kishan, and W. Hillen. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 8745-51. [DOI] [PubMed] [Google Scholar]
- 18.Jawad, A., H. Seifert, A. M. Snelling, J. Heritage, and P. M. Hawkey. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 361938-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S. A., S. M. Yoo, S. H. Hyun, C. H. Choi, S. Y. Yang, H. J. Kim, B. C. Jang, S. I. Suh, and J. C. Lee. 2008. Global gene expression patterns and induction of innate immune response in human laryngeal epithelial cells in response to Acinetobacter baumannii outer membrane protein A. FEMS Immunol. Med. Microbiol. 5445-52. [DOI] [PubMed] [Google Scholar]
- 20.Kisiela, D. I., and C. J. Czuprynski. 2009. Identification of Mannheimia haemolytica adhesins involved in binding to bovine bronchial epithelial cells. Infect. Immun. 77446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, H. W., Y. M. Koh, J. Kim, J. C. Lee, Y. C. Lee, S. Y. Seol, and D. T. Cho. 2008. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 1449-54. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. C., H. Koerten, P. van den Broek, H. Beekhuizen, R. Wolterbeek, M. van den Barselaar, T. van der Reijden, J. van der Meer, J. van de Gevel, and L. Dijkshoorn. 2006. Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res. Microbiol. 157360-366. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. C., J. Y. Oh, K. S. Kim, Y. W. Jeong, J. C. Park, and J. W. Cho. 2001. Apoptotic cell death induced by Acinetobacter baumannii in epithelial cells through caspase-3 activation. APMIS 109679-684. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. S., J. W. Kim, C. H. Choi, W. K. Lee, H. Y. Chung, and J. C. Lee. 2008. Anti-tumor activity of Acinetobacter baumannii outer membrane protein A on dendritic cell-based immunotherapy against murine melanoma. J. Microbiol. 46221-227. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. S., J. C. Lee, C. M. Lee, I. D. Jung, Y. I. Jeong, E. Y. Seong, H. Y. Chung, and Y. M. Park. 2007. Outer membrane protein A of Acinetobacter baumannii induces differentiation of CD4+ T cells toward a Th1 polarizing phenotype through the activation of dendritic cells. Biochem. Pharmacol. 7486-97. [DOI] [PubMed] [Google Scholar]
- 26.Leong, S. A., G. S. Ditta, and D. R. Helinski. 1982. Heme biosynthesis in Rhizobium: identification of a cloned gene coding for delta-aminolevulininc acid synthetase from Rhizobium meliloti. J. Biol. Chem. 2578724-8730. [PubMed] [Google Scholar]
- 27.Loehfelm, T. W., N. R. Luke, and A. A. Campagnari. 2008. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 1901036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lower, B. H., R. Yongsunthon, F. P. Vellano III, and S. K. Lower. 2005. Simultaneous force and fluorescence measurements of a protein that forms a bond between a living bacterium and a solid surface. J. Bacteriol. 1872127-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meade, H. M., S. R. Long, G. B. Ruvkum, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olmsted, J. B. 1981. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J. Biol. Chem. 25611955-11957. [PubMed] [Google Scholar]
- 31.Orme, R., C. W. Douglas, S. Rimmer, and M. Webb. 2006. Proteomic analysis of Escherichia coli biofilms reveals the overexpression of the outer membrane protein OmpA. Proteomics 64269-4277. [DOI] [PubMed] [Google Scholar]
- 32.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peleg, A. Y., E. Tampakakis, B. B. Fuchs, G. M. Eliopoulos, R. C. Moellering, Jr., and E. Mylonakis. 2008. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 10514585-14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 513471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, A. J., I. Sudbery, and M. Ramsdale. 2003. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA 10014327-14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Baño, J., S. Marti, S. Soto, F. Fernandez-Cuenca, J. M. Cisneros, J. Pachón, A. Pascual, L. Martinez-Martinez, C. McQueary, L. A. Actis, and J. Vila. 2008. Biofilm formation in Acinetobacter baumannii: associated features and clinical implications. Clin. Microbiol. Infect. 14276-278. [DOI] [PubMed] [Google Scholar]
- 37.Salmanian, A. H., F. Siavoshi, F. Akbari, A. Afshari, and R. Malekzadeh. 2008. Yeast of the oral cavity is the reservoir of Helicobacter pylori. J. Oral Pathol. Med. 37324-328. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Shin, S., G. Lu, M. Cai, and K. S. Kim. 2005. Escherichia coli outer membrane protein A adheres to human brain microvascular endothelial cells. Biochem. Biophys. Res. Commun. 3301199-1204. [DOI] [PubMed] [Google Scholar]
- 40.Sirakova, T., P. E. Kolattukudy, D. Murwin, J. Billy, E. Leake, D. Lim, T. DeMaria, and L. Bakaletz. 1994. Role of fimbriae expressed by nontypeable Haemophilus influenzae in pathogenesis of and protection against otitis media and relatedness of the fimbrin subunit to outer membrane protein A. Infect. Immun. 622002-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, M. G., T. A. Gianoulis, S. Pukatzki, J. J. Mekalanos, L. N. Ornston, M. Gerstein, and M. Snyder. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21601-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, S. G., V. Mahon, M. A. Lambert, and R. P. Fagan. 2007. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 2731-11. [DOI] [PubMed] [Google Scholar]
- 43.Takano, T., S. Tsutsumi, W. Tomisato, T. Hoshino, T. Tsuchiya, and T. Mizushima. 2002. Geranylgeranylacetone protects guinea pig gastric mucosal cells from gastric stressor-induced apoptosis. Dig. Dis. Sci. 471546-1553. [DOI] [PubMed] [Google Scholar]
- 44.Teng, C.-H., Y. Xie, S. Shin, F. Di Cello, M. Paul-Satyaseela, M. Cai, and K. S. Kim. 2006. Effects of ompA deletion on expression of type 1 fimbriae in Escherichia coli K1 strain RS218 and on the association of E. coli with human brain microvascular endothelial cells. Infect. Immun. 745609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 1493473-3484. [DOI] [PubMed] [Google Scholar]
- 46.Tomaras, A. P., M. J. Flagler, C. W. Dorsey, J. A. Gaddy, and L. A. Actis. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 1543398-3409. [DOI] [PubMed] [Google Scholar]
- 47.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 714985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Towner, K. J. 1995. Biology of Acinetobacter spp., p. 13-36. In E. Bergogne-Berezin, M. L. Joly-Guillou, and K. J. Towner (ed.), Acinetobacter: microbiology, epidemiology, infections, management. CRC Press, Inc., Boca Raton, FL.
- 49.Towner, K. J., E. Bergogne-Berezin, and C. A. Fewson. 1991. Acinetobacter: portrait of a genus, p. 1-24. In K. J. Towner, E. Bergogne-Berezin, and C. A. Fewson (ed.), The biology of Acinetobacter. Plenum Press, New York, NY.
- 50.Vashist, J., and M. R. Rajeswari. 2006. Structural investigations on novel porin, OmpAb from Acinetobacter baumannii. J. Biomol. Struct. Dyn. 24243-253. [DOI] [PubMed] [Google Scholar]
- 51.Vidal, R., M. Dominguez, H. Urrutia, H. Bello, A. Garcia, G. Gonzalez, and R. Zemelman. 1997. Effect of imipenem and sulbactam on sessile cells of Acinetobacter baumannii growing in biofilm. Microbios 9179-87. [PubMed] [Google Scholar]
- 52.Vidal, R., M. Dominguez, H. Urrutia, H. Bello, G. Gonzalez, A. Garcia, and R. Zemelman. 1996. Biofilm formation by Acinetobacter baumannii. Microbios 8649-58. [PubMed] [Google Scholar]
- 53.Villegas, M. V., and A. I. Hartstein. 2003. Acinetobacter outbreaks, 1977-2000. Infect. Control Hosp. Epidemiol. 24284-295. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Y. 2002. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292396-401. [DOI] [PubMed] [Google Scholar]
- 55.Wargo, M. J., and D. A. Hogan. 2006. Fungal-bacterial interactions: a mixed bag of mingling microbes. Curr. Opin. Microbiol. 9359-364. [DOI] [PubMed] [Google Scholar]
- 56.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33103-109. [DOI] [PubMed] [Google Scholar]
- 57.Zapor, M. J., and K. A. Moran. 2005. Infectious diseases during wartime. Curr. Opin. Infect. Dis. 18395-399. [DOI] [PubMed] [Google Scholar]